Abstract
The lactococcal group II intron Ll.ltrB interrupts the ltrB relaxase gene within a region that encodes a conserved functional domain. Nucleotides essential for the homing of Ll.ltrB into an intronless version of ltrB are found exclusively at positions required to encode amino acids broadly conserved in a family of relaxase proteins of gram-positive bacteria. Two of these relaxase genes, pcfG from the enterococcal plasmid pCF10 and the ORF4 gene in the streptococcal conjugative transposon Tn5252, were shown to support Ll.ltrB insertion into the conserved motif at precisely the site predicted by sequence homology with ltrB. Insertion occurred through a mechanism indistinguishable from retrohoming. Splicing and retention of conjugative function was demonstrated for pCF10 derivatives containing intron insertions. Ll.ltrB targeting of a conserved motif of a conjugative element suggests a mechanism for group II intron dispersal among bacteria. Additional support for this mechanism comes from sequence analysis of the insertion sites of the E.c.I4 family of bacterial group II introns.
Group II introns are self-splicing and mobile RNAs (14) encoded within the genomes of bacteria (27) and in the organelle DNAs of plants, fungi, and protists (3). These RNAs have a conserved secondary structure with six sets of base-pairing regions, domains I through VI, arranged radially around a central circle (29). Much of domain IV is looped out of the secondary structure and determines the synthesis of an intron-encoded protein (IEP) with maturase, endonuclease, and reverse transcriptase (RT) activities (28, 33). Maturase assists in splicing, probably by enhancing the formation or stabilization of a catalytic ribozyme structure which mediates splicing (28) via a mechanism very similar to that of eukaryotic spliceosomal introns (38).
Group II introns home at high frequency into intronless alleles of the same exon gene and transpose at low frequency into ectopic sites with sequence similarity to the natural site (26). Since group II intron mobility occurs through an RNA intermediate, the terms retrohoming and retrotransposition are used to designate these events (9, 21). During retrohoming, the IEP, which is associated with the spliced-out intron RNA, recognizes nucleotides in the distal 5′ portion of the DNA target and EBS/δ ribonucleotides in the intron base pair with IBS/δ′ nucleotides in the DNA target site. This process is followed by reverse splicing of the intron RNA into target DNA (20, 28, 34, 40). The IEP then interacts with the 3′ portion of the DNA target, and the IEP endonuclease nicks the antisense strand of recipient DNA a few base pairs downstream of the insertion site (20, 28). Using the 3′ end of the nicked target DNA as a primer, IEP RT reverse transcribes the intron RNA via a mechanism reminiscent of non-long terminal repeat retrotransposon target DNA-primed reverse transcription (14, 28). In retrohoming by yeast mitochondrial group II introns aI1 and aI2, completion of the insertion event generally occurs by double-strand break repair recombination between cDNA and the target site (15, 16). In contrast, retrohoming by the Lactococcus lactis group II intron Ll.ltrB is independent of homologous recombination (9).
Ll.ltrB was the first group II intron in bacteria that was shown to splice (31) and home (30) in vivo. Ll.ltrB interrupts a putative relaxase gene, ltrB, in the conjugative plasmid pRS01 (31). Relaxase protein nicks conjugative plasmid DNA in a site-specific, strand-specific manner at the origin of transfer (oriT). Only after nicking can a plasmid be transferred to a recipient (7, 24). Genetic data indicate that Ll.ltrB splicing is required for LtrB relaxase expression and for pRS01 conjugation (31).
LtrB and a number of other relaxase proteins encoded by conjugative elements of gram-positive bacteria share three conserved motifs with relaxases in gram-negative bacteria (36, 41). Motif I contains the enzymatically active site. Motif II may play a role in DNA recognition. Motif III contains a histidine that is thought to activate a motif I tyrosine residue by proton abstraction. Motif III has the most amino acid identity among diverse relaxases (36, 41).
Ll.ltrB was independently identified in pRS01 from L. lactis subsp. lactis ML3 and the nearly identical sex factor of the closely related strain L. lactis subsp. lactis 712 (31, 39). It is likely that the two identical introns represent a single insertion event in a common ancestor strain (25). Another intron that is 99.8% identical to Ll.ltrB is present in pAH82 of L. lactis subsp. lactis biovar diacetylactis DPC220 (35). The interrupted gene, mobD, is only about 50% identical to ltrB, although the predicted MobD peptide sequence does include the relaxase structural motifs discussed previously. Although Ll.ltrB family members have so far been identified only in L. lactis, both the IEP and the noncoding RNA are more closely related to yeast mitochondrial introns than they are to other bacterial introns (31, 43, 50).
Biochemical (34, 40) and genetic (19) studies have identified nucleotides in the Ll.ltrB target site that are critical for Ll.ltrB insertion. Substitutions of several nucleotides in the IBS/δ′ region (positions −12 to −8 and −6 to + 3 with respect to intron insertion), a region which base pairs with intron RNA, severely reduced reverse splicing in vitro. Nucleotides outside the IBS/δ′ region that interact with LtrA were also important, including T-23, G-21, and A-20 for reverse splicing and T+5 for antisense strand cleavage.
A more recent study by Zhong et al. suggests that additional positions (positions −23, −19, −17, −15, and −14) may also be important for Ll.ltrB targeting of insertion sites (48). However, since that study utilized an Ll.ltrB-derived gene-targeting vector with randomized EBS and δ sequences to target the Escherichia coli genome, it may not be completely relevant to determining the target site specificity of the wild-type Ll.ltrB in L. lactis.
To test whether Ll.ltrB might target other relaxase genes, we introduced a marked Ll.ltrB construct into cells containing potential Ll.ltrB targets and looked for the transfer of the marker to the relaxase gene targets. Our results indicate that (i) Ll.ltrB does target relaxase genes of other conjugative elements, (ii) the insertion occurs within DNA encoding conserved motif III, and (iii) Ll.ltrB mobility can be observed in Enterococcus faecalis as well as in L. lactis. The results of our study suggest a model for group II intron dispersal in bacteria.
MATERIALS AND METHODS
Computer analyses.
Protein and DNA searches and sequence alignments utilized online software at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). Sequences of relaxase proteins and encoding sequences were accessed there and at the Washington University Pfam database of protein families (http://pfam.wustl.edu/). Multiple-sequence alignments of DNA and protein were produced by using a Seqweb version of GCG (Wisconsin Package) and online alignment tools available at the Baylor College of Medicine website (http://searchlauncher.bcm.tmc.edu/multi-align) or the Canadian Bioinformatics Resource website (http://www.cbr.nrc.ca).
Bacterial strains and growth conditions.
E. coli DH5α (Gibco-BRL) was used in plasmid cloning and as an indicator strain for assessing the frequency of Ll.ltrB insertion into artificial target plasmids. All intron insertion experiments (regardless of the target) were conducted with Enterococcus faecalis strains. Enterococcus faecalis UV202 is resistant to rifampin and fusidic acid and is recombination deficient (47). Three plasmid-free Enterococcus faecalis strains derived from OG1 (13, 18) were also used in this study. OG1RF and OG1SSp are resistant to rifampin and fusidic acid (13) and streptomycin and spectinomycin (12), respectively. OG1ES, which is resistant to erythromycin and streptomycin, was developed in this study. It was constructed by plating out OG1 on Todd-Hewitt broth (THB) agar plates containing 50 times the MIC of erythromycin. (We determined that the erythromycin MIC was ∼0.16 μg/ml.) An erythromycin-resistant strain (for which the MIC is ≥160 μg/ml) was then used for the selection of an erythromycin- and streptomycin-resistant mutant, termed OG1ES. For strain OG1ES, the erythromycin MIC is >100 μg/ml and the streptomycin MIC is >1,000 μg/ml. Because of the high degree of natural resistance of other OG1 strains to streptomycin (the streptomycin MIC for OG1RF is between 100 and 1,000 μg/ml), erythromycin selection alone was typically used in experiments involving OG1ES.
Strain OG1RF::Tn5252 was created by filter mating of the broad-host-range conjugative transposon Tn5252 from Streptococcus pneumoniae SP1000 (1). SP1000 and OG1RF were grown to the stationary phase and then diluted 1:3 and 1:5, respectively, and grown for about 2 h. One hundred microliters of OG1RF and 1,000 μl SP1000 were mixed and passed via a syringe through a 0.2-μm-pore-size membrane filter (Gelman Sciences, Ann Arbor, Mich.) with a diameter of 13 mm. The filter was placed on an agar plate containing brain heart infusion broth supplemented with 0.2% bovine serum albumin (BHI-BSA), overlaid with soft agar containing BHI-BSA, and incubated overnight. The filter was rinsed in 1 ml of potassium phosphate-buffered saline (1.68 mM KH2PO4, 8.0 mM K2HPO4, 150 mM NaCl [pH 7.4]), and the rinse solution was plated out on THB agar supplemented with rifampin, fusidic acid, and chloramphenicol to select for OG1RF::Tn5252.
Enterococcus faecalis was grown in THB, E. coli was grown in Luria broth, and S. pneumoniae was grown in BHI-BSA. For Enterococcus faecalis, antibiotics were used at the following concentrations: chloramphenicol, 25 μg/ml; erythromycin, 10 μg/ml for plasmid and 100 μg/ml for chromosomal marker; fusidic acid, 25 μg/ml; kanamycin, potency of 750 to 1,000 μg/ml; rifampin, 200 μg/ml; spectinomycin, potency of 600 to 1,000 μg/ml for plasmid and 150 to 250 μg/ml for chromosomal marker; streptomycin, potency of 767 μg/ml; tetracycline, 10 μg/ml. For E. coli, antibiotic concentrations were as follows: chloramphenicol, 30 μg/ml; erythromycin, 300 μg/ml; kanamycin, potency of 38 to 50 μg/ml; spectinomycin, potency of 30 to 50 μg/ml. For S. pneumoniae, chloramphenicol was used at 5 μg/ml. All strains were grown at 37°C. E. coli was generally grown with aeration, whereas Enterococcus faecalis and S. pneumoniae were not.
Extrachromosomal genetic elements.
Two conjugative plasmids, pCF10 (11) and its derivative, pCF11 (12), were used in this study. They encode a putative relaxase, PcfG, as well as tetracycline resistance. pCF11 contains a point mutation that results in a constitutively elevated transfer phenotype (12). The conjugative transposon Tn5252 encodes chloramphenicol resistance (1) and a biochemically characterized relaxase protein (encoded by ORF4) (41).
Three Ll.ltrB-Kan donor plasmids were used in this study. pLEIItd+KR" was jointly constructed by us and the Belfort laboratory. The pLE vector encodes chloramphenicol resistance, while 3′,5"-aminoglycoside phosphotransferase type III kanamycin resistance (44) is encoded by a gene that was cloned in Ll.ltrB just downstream of ltrA. A self-splicing group I intron, tdΔ1-3, was cloned between ltrA and the kanamycin resistance gene. The presence of the kanamycin resistance gene and tdΔ1-3 did not affect the splicing efficiency of Ll.ltrB in a previous construct (9). The modified Ll.ltrB, flanked by ltrB exon fragments, was cloned under the control of the nisin-inducible promoter PnisA. While the two-component system for nisin induction is absent in Enterococcus faecalis, the PnisA promoter is somewhat leaky in Enterococcus faecalis (6). Plasmid pLEIItd+KR"ΔORF is identical to pLEIItd+KR" except that we replaced ltrA with an internally deleted version from pLI2(7)ΔORF, in which over 88% of ltrA is deleted. In a previous construct, this deletion abolished splicing and severely reduced homing (9). A third Ll.ltrB donor plasmid, shuttle plasmid pCOMT-Kan, is a modified version of pCOM9 (49). pCOMT-Kan contains the pRS01 oriT (32) and a spectinomycin resistance gene within its vector sequence. The kanamycin resistance gene that encodes 3′,5"-aminoglycoside phosphotransferase type III (44) is cloned into the Ll.ltrB insert downstream of ltrA.
Nisin-inducible Ll.ltrB target plasmids were constructed by cloning a 271-bp PCR product amplified from a region spanning the intron target site in pcfG (positions −179 to +92 with respect to the Ll.ltrB insertion) under the control of the PnisA promoter of pMSP3535 (6). Primers used for PCR amplification incorporated SpeI and XbaI sites, simplifying insertion into the pMSP3535 multicloning site. See Table 1 in the supplemental material (http://www.micab.umn.edu/faculty/Dunny.html) for the oligonucleotide primer sequences. Plasmids were electroporated into DH5α. Orientation and sequences were verified by PCR and automated dideoxynucleotide sequencing (Advanced Genetic Analysis Center, University of Minnesota) before electroporation into Enterococcus faecalis. pMSP3535-FT contained the target in the forward orientation with respect to transcription, while the target in pMSP3535-RT was in the reverse orientation.
Conjugation.
Cultures of donor and plasmid-free recipient Enterococcus faecalis strains were grown overnight in THB plus antibiotics. Donor and recipient bacteria were washed in THB to remove antibiotics and then combined in THB with a 1:10 donor-to-recipient ratio and incubated for 30 min to 2 h at 37°C, followed by quantitative plating on THB agar with relevant selective antibiotics. Transconjugant-to-donor ratios were calculated by dividing the number of CFU per milliliter resistant to both a recipient chromosomal marker and a conjugative plasmid-mediated marker by the number of CFU resistant to donor antibiotic markers.
Plasmid electroporation.
Electroporation of plasmids into E. coli and Enterococcus faecalis was done by using the Gene Pulser (1.7 kV, 200 Ω, 25 μF, 1-mm-diameter cuvettes; Bio-Rad, Philadelphia, Pa.). Electrocompetent E. coli cells were prepared according to the manufacturer's instructions, while electrocompetent Enterococcus faecalis cells were prepared according to the protocol of Bae et al. (2). Following electroporation, E. coli cells were added to 1 ml of SOC (2% Bacto Tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, 20 mM glucose) and incubated with aeration at 37°C for 2 h before plating on Luria broth plates plus antibiotics. Following electroporation of Enterococcus faecalis, the slurry was added to 200 μl of THB supplemented with 0.5 M sucrose and incubated for about 2 h before plating on THB plates with antibiotics.
DNA and RNA analyses of Ll.ltrB-Kan insertions in pCF11 and Tn5252.
Plasmid isolation was done with miniprep kits (QIAGEN, Valencia, Calif.). Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.) or Promega (Madison, Wis.) and used according to the manufacturer's instructions. PCR was done by using the GeneAmp PCR system 2400 (Perkin-Elmer, Wellesley, Mass.) or a Mastercycler (Eppendorf, Hamburg, Germany) with Bio-X-Act polymerase (Bioline, Randolph, Mass.). DNA sequences were determined by automated dideoxynucleotide sequencing (Advanced Genetic Analysis Center, University of Minnesota). Detailed descriptions of PCR protocols used to analyze Ll.ltrB-Kan insertions in pCF11 and Tn5252 are presented in the supplemental Materials and Methods and Table 2 in the supplemental material (http://www.micab.umn.edu/faculty/Dunny.html). Oligonucleotide primer sequences are given in Table 3 in the supplemental material. RT PCR analysis of pcfG splicing was performed by using SuperScript II reverse transcriptase and Bio-X-Act polymerase (Bioline) in a two-step RT PCR as described in the supplemental Materials and Methods.
Examination of Ll.ltrB-Kan insertion into an inducible target.
OG1RF was electroporated with pMSP3535-FT, pMSP3535-RT, or pMSP3535, followed by electroporation with pCOMT-Kan. Colonies resistant to erythromycin (pMSP3535 marker) and spectinomycin (pCOMT-Kan marker) were grown overnight with continued antibiotic selection in the presence or absence of nisin induction (10 ng/ml). Plasmid was isolated and electroporated into DH5α. Electrotransformant colonies resistant to erythromycin were patched to kanamycin and spectinomycin. Ll.ltrB-Kan insertion into the target plasmids was calculated as the fraction of erythromycin-resistant and spectinomycin-sensitive DH5α colonies that were resistant to kanamycin. Experiments were done in triplicate, and about 50 erythromycin-resistant and spectinomycin-sensitive colonies from each DH5α electrotransformation were examined. DH5α plasmid preparations from each OG1RF experiment that resulted in genetically determined Ll.ltrB-Kan insertions were further analyzed by restriction enzyme digestion. DNA sequences of intron-exon junctions from single examples of each Ll.ltrB-Kan insertion type were determined.
Real-time RT PCR verification of nisin induction.
Nisin induction of RNA transcription from pMSP3535-FT and pMSP3535-RT was confirmed by one-step real-time RT PCR by using the iCycler (Bio-Rad) according to the manufacturer's instructions. OG1RF/pMSP3535-FT,pCOMT-Kan and OG1RF/pMSP3535-RT,pCOMT-Kan were grown up overnight with erythromycin and spectinomycin selection in the presence and absence of nisin. RNA was isolated with the RNeasy minikit (QIAGEN). Primers for RT PCR were designed to anneal between the nisin promoter and the intron insertion site such that Ll.ltrB insertion is not predicted to affect RT PCR efficiency. As a control for RNA loading levels, erm cDNA was also amplified by real-time RT PCR. The erm gene codes for erythromycin resistance.
Sequencing of pcfG.
The pcfG gene was amplified by PCR with primers that correspond to the sequence of pTEF2, a pCF10-like plasmid in Enterococcus faecalis V583 sequenced by the Institute for Genomic Research (37). The sequences of both strands were determined by automated dideoxynucleotide sequencing (Advanced Genetic Analysis Center and MicroChemical Facility, University of Minnesota).
Nucleotide sequence accession numbers.
The GenBank sequence accession number for the pcfG gene is AY288104. GenBank sequence accession numbers for the plasmids used in this study are as follows: pLEIItd+KR", AY303236;pLEIItd+KR"ΔORF, AY303237; pCOMT-Kan, AY303238; pMSP3535,AY303239; pMSP3535-FT, AY303240; pMSP3535-RT, AY303241.
RESULTS
Comparative analysis of Ll.ltrB insertion sites.
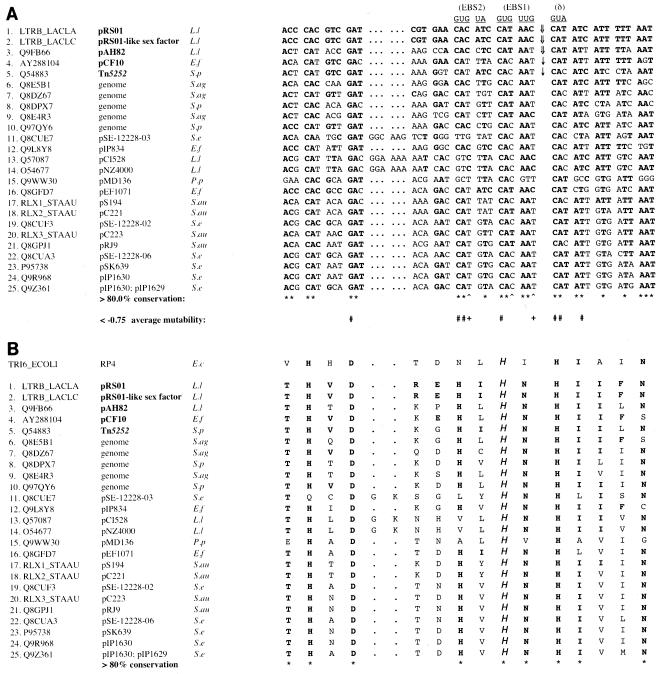
Nearly identical group II introns are present in divergent relaxase genes of the lactococcal conjugative elements pRS01 (31) and pAH82 (35). We noted that the intron insertions in these elements are at the same nucleotide within a sequence that encodes a conserved motif (Fig. 1B). The motif includes a conserved histidine residue shown to be catalytically active in the homologous TraI relaxase encoded by the RP4 plasmid found in gram-negative bacteria (36).
FIG. 1.
The Ll.ltrB target site encodes a highly conserved relaxase domain. Sequence identifiers (except the pcfG GenBank identifier) correspond to those given in the Pfam database (http://pfam.wustl.edu/). Sequences of Q9FB66 and LTRB_LACLC are given according to the updated exon-intron junctions of Dai and Zimmerly (10). Calculations of overall nucleotide and amino acid conservation are based exclusively on sequences listed on the Pfam database. E.f, Enterococcus faecalis; L.l, L. lactis; P.p, Pediococcus pentosaceus; S.ag, Streptococcus agalactiae; S.au, Staphylococcus aureus; S.e, Staphylococcus epidermidis; S.p, Streptococcus pneumoniae. (A) Nucleotide sequences homologous to the ltrB insertion site (positions −30 to +15 with respect to the Ll.ltrB insertion). Nucleotides of Ll.ltrB intron RNA that base pair with ltrB during homing are shown above the alignment. Identities with ltrB are indicated with boldface type. Wide arrows (⇓) indicate previously identified Ll.ltrB family intron insertion sites in nature, while narrow arrows (↓) indicate Ll.ltrB insertion sites determined experimentally in this work. Asterisks below the alignment indicate >80% conservation of ltrB-type nucleotides, while carets indicate combined conservation of >80% of C and T at positions that are thought to base pair with specific G residues in Ll.ltrB RNA. Pound signs (#) indicate average mutability values of less than −0.75 for non-ltrB-type nucleotides, as described in the text. Plus signs represent average mutability values of less than −0.75 for A and G nucleotides at positions where wild-type ltrB residues (C or T) base pair with specific G residues in intron RNA. (B) Predicted relaxase polypeptide sequences encoded by the nucleotide sequences shown in panel A. Putative functional histidine residues are indicated by “H.” Asterisks indicate >80% conservation of LtrB-type amino acids. Homologous sequence from the TraI relaxase protein, including its functional histidine, is shown above the alignments.
In a previous study, Guo et al. tested the homing of Ll.ltrB into a plasmid pool containing ltrB target sites with multiple random mutations (19). Each position in the target site (positions −30 to +15 with respect to the Ll.ltrB insertion) was randomly mutated, with an average overall mutation frequency of ∼30% at each position. Using the raw supplemental data available at http://www.sciencemag.org/feature/data/1050641.shl, we recalculated the mutability values with the formulas of Guo et al. and determined an average mutability value for each position. Briefly, mutability values were determined by comparing the ratios (R) of mutant to wild-type nucleotide residues in active DNA target sites with those in the initial recipient pool by using the expression [(Rmut/wt)active/(Rmut/wt)pool] − 1. A mutability value of −1 corresponds to a nucleotide that is completely absent at a particular residue in active DNA target sites yet present at some frequency at that position in the recipient pool. In contrast, an increased frequency of mutant nucleotides in the active DNA target site compared to that in the recipient pool corresponds to a positive mutability value. Positions −21, −12, −11, −6, 1, 2, and 5 had average mutability values of less than −0.75, implying a >75% reduction in homing among target plasmids containing at least one mutation at these seven positions. Since the target plasmids in the recipient pool contained multiple mutations throughout the target site, one cannot determine on the basis of this experiment how single mutations at these sites would affect homing. However, single mutations at positions −21, −12, and +5 reduced homing frequency by 95 to 97% (34). To our knowledge, no single-mutation data are available for the other four positions.
Each of these seven positions in pAH82 is identical to its homologue in pRS01 (Fig. 1A), compared to an overall identity of 71% throughout the 45-bp target sequence. These sites are also conserved in >80% of firmicute bacterial relaxase genes that have this domain and that are listed in the Pfam database (http://pfam.wustl.edu/) (Fig. 1A). Interestingly, these seven nucleotides map in every case to highly conserved first or second positions of codons that encode widely conserved amino acids, including the putative functional histidine residue in motif III (Fig. 1B).
Three nucleotides that map to less-conserved third codon positions (positions −10, −4, and −1) base pair with G nucleotides in the intron EBS sequences during Ll.ltrB homing. Each position contains either a C or a T nucleotide in pRS01 and pAH82 (Fig. 1A). Positions −10, −4, and −1 had average mutability values of −0.635, −0.563, and −0.650, respectively. When we excluded C→T and T→C mutations from our calculations, average mutability values decreased to −0.785, −0.620, and −0.884 for positions −10, −4, and −1, respectively. Intriguingly, all but one of the 24 Pfam database firmicute relaxase genes contain exclusively C or T nucleotides at positions −10, −4, and −1 (Fig. 1A).
Sequencing in our laboratory of the enterococcal conjugative plasmid pCF10 (H. Hirt and E. M. Bryan, unpublished data) revealed a putative relaxase gene, pcfG, which showed a degree of similarity to ltrB of pRS01. The pcfG nucleotide sequence flanking a predicted Ll.ltrB insertion site is identical to the ltrB sequence at the seven critical positions discussed above and also contains either T or C nucleotides at positions −10, −4, and −1 (Fig. 1A).
Ll.ltrB insertion into the pCF10 pcfG gene.
Preliminary experiments (data not shown) indicated that an Ll.ltrB derivative could move from a donor plasmid (pCOMT-Kan) to pcfG in pCF10 when both plasmids were present in the same Enterococcus faecalis host strain. To examine Ll.ltrB insertion into pcfG in detail, we used a conjugation assay in which the intron, expressed from the nonconjugative plasmid pLEIItd+KR", was allowed to transpose into pCF11 (a derivative of pCF10 with an identical pcfG gene whose higher constitutive conjugative transfer frequency simplified genetic analysis). The intron donor and recipient plasmids were maintained in the recombination-deficient host strain UV202. This strain was then mated with a second enterococcal strain, OG1SSp (Fig. 2). A kanamycin resistance gene cloned into Ll.ltrB made genetic selection for the intron possible. Intron insertion prevalence was calculated by dividing the frequency of transfer of kanamycin resistance by the frequency of transfer of pCF11-encoded tetracycline resistance. We observed an insertion prevalence of 1.8 × 10−2 when pLEIItd+KR" served as the intron donor plasmid, whereas insertion was undetectable when we used a donor plasmid carrying the splicing- and homing-deficient Ll.ltrBΔORF-Kan intron. Assuming that approximately 30 generations elapsed between electroporation of pLEIItd+KR" into UV202/pCF11 and conjugation into OG1SSp, the insertion frequency per pCF11 target was approximately 6 × 10−4/generation.
FIG. 2.
Overview of homing assay. The homing of Ll.ltrB-Kan into pCF11 (illustrated by a curved arrow) was made possible by the transformation of Enterococcus faecalis UV202 cells containing pCF11 with the intron donor plasmid pLEIItd+KR". pCF11 plasmids with integrated Ll.ltrB-Kan were separated from pLEIItd+KR" by conjugative mating to a plasmid-free strain, OG1SSp, for subsequent analysis. Experiments using the pLEIItd+KR"ΔORF control plasmid, which contains the splicing-deficient Ll.ltrBΔORF-Kan construct, utilized identical procedures.
Structural and functional analysis of inserted Ll.ltrB.
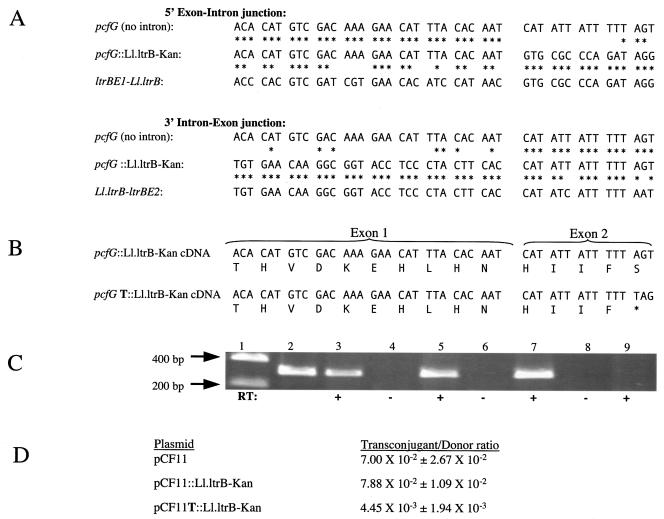
We PCR amplified and sequenced the 5′ and 3′ intron-exon junctions of 11 Ll.ltrB-Kan insertions into pCF11 from independent experiments. As shown in Fig. 3A, Ll.ltrB-Kan insertion into pcfG occurred at precisely the site predicted by homology with the pRS01 insertion site. The resulting plasmid was termed pCF11::Ll.ltrB-Kan. There was no conversion of DNA sequence from the recipient pcfG sequence to the donor exon sequence which flanks Ll.ltrB in pLEIItd+KR".
FIG. 3.
Structural and functional analysis of Ll.ltrB homing into pCF11. (A) 5′ and 3′ exon-intron junctions in pCF11::Ll.ltrB-Kan. Nucleotide identities with intronless pcfG and the relaxase-intron junctions of pRS01 are indicated by asterisks. The 5′ junction is the junction between DNA encoding exon 1 and Ll.ltrB. The 3′ junction is the junction between DNA encoding Ll.ltrB and exon 2. (B) Exon-exon junctions of cDNA. In 1 of 11 homed products analyzed (pCF11T::Ll.ltrB-Kan), a thymidine insertion resulted in the incorporation of a stop codon (*). (C) Gel electrophoresis of products of RT PCR of spliced exon in the presence (+) or absence (−) of RT. Lane 1, DNA ladder; lane 2, PCR control of intronless pCF11; lanes 3 and 4, pCF11::Ll.ltrB-Kan; lanes 5 and 6, pCF11T::Ll.ltrB-Kan; lanes 7 and 8, intronless pCF11; lane 9, plasmid-free OG1SSp. (D) Conjugation frequencies of pCF11, pCF11::Ll.ltrB-Kan, and pCF11T::Ll.ltrB-Kan. Mean transconjugant-to-donor ratios ± standard deviations for three independent matings are given.
In 1 of the 11 Ll.ltrB insertions in pCF11 that we examined, an extra thymidine was inserted into the pentathymidine tract found 3′ from the insertion site in pcfG. This plasmid was termed pCF11T::Ll.ltrB-Kan. We also observed this thymidine insertion in a preliminary experiment involving Ll.ltrB insertion into pCF10. The insertion is predicted to result in a stop codon and truncation of the PcfG protein, even if the intron was spliced from the mRNA (Fig. 3B).
To analyze the precision of intron splicing, we performed RT PCR of spliced pcfG RNA from enterococcal transconjugants containing either pCF11::Ll.ltrB-Kan or pCF11T::Ll.ltrB-Kan. As demonstrated in Fig. 3C, Ll.ltrB remains splicing proficient in the context of pcfG whether or not pcfG contains a thymidine insertion in the 3′ exon. Sequences of the cDNA around the exon-exon junction were identical to the cognate DNA sequences, including the presence or absence of the thymidine insertion. While pCF11::Ll.ltrB-Kan plasmids displayed unimpaired conjugation frequency relative to pCF11, pCF11T::Ll.ltrB-Kan conjugation frequency was decreased more than 10-fold (Fig. 3D).
In order to examine whether Ll.ltrB could insert into a relaxase gene of another type of conjugative element, we examined insertion into the conjugative transposon Tn5252. Tn5252 contains a relaxase-encoding sequence, open reading frame 4 (ORF4) (41), that includes a region highly homologous to the Ll.ltrB insertion target (Fig. 1A). Tn5252 was originally isolated from S. pneumoniae, but it can transfer by conjugative transposition to the chromosomes of various gram-positive bacterial species (46). We introduced pLEIItd+KR" into an Enterococcus faecalis strain into which we had previously introduced Tn5252 by conjugation from S. pneumoniae and propagated the strain for ∼40 generations. We subsequently observed PCR products consistent with insertion of the intron into the expected location within ORF4. The sequences of Ll.ltrB-ORF4 junctions from one experiment were determined and revealed an absence of sequence coconversion or other changes to the flanking exons. The extremely low transfer frequency (∼10−9/donor) of Tn5252 from the Enterococcus faecalis strain used for these studies prevented us from carrying out quantitative conjugation-based assays of intron insertion frequency. However, there is no evidence to suggest that the mechanism of insertion is any different from that observed when pCF11 was used as the target.
Mechanistic analysis of Ll.ltrB insertion into pcfG.
The presence of a self-splicing group I intron, tdΔ1-3, cloned into Ll.ltrB-Kan of pLEIItd+KR" (Fig. 4A) allowed us to distinguish between RNA- and DNA-mediated Ll.ltrB insertion events. The insertion of Ll.ltrB DNA directly into the target site should result in the retention of tdΔ1-3, whereas if an RNA intermediate is involved, tdΔ1-3 should be lost in at least some cases (9). PCR of eight independent Ll.ltrB insertion events into pCF11 demonstrated the loss of tdΔ1-3 in every case (Fig. 4B). Therefore, Ll.ltrB insertion into pCF11 occurred through an RNA intermediate.
FIG. 4.
Analysis of tdΔ1-3 splicing from Ll.ltrB during events of Ll.ltrB-Kan insertion into pCF11. (A) Ll.ltrB-Kan-tdΔ1-3 construct. PCR primers used to amplify tdΔ1-3 anneal to flanking ltrA and kan sequences. (B) PCR products of the tdΔ1-3 region. Lane 1, DNA ladder; lanes 2 to 9, PCR products (arrows) from strains representing eight independent Ll.ltrB insertion events; lane 10, pLEIItd+KR" control. Plasmid figures (not to scale) shown on the right illustrate predicted sizes in the presence and absence of tdΔ1-3.
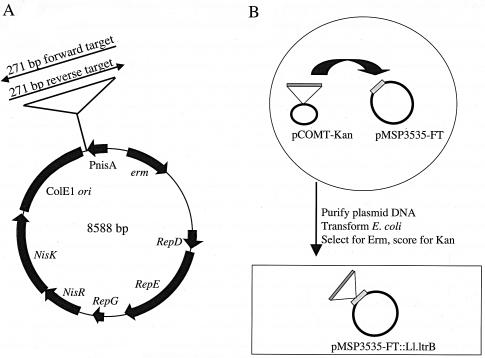
Proposed mechanisms for retrotransposition once focused upon insertion into single-stranded RNA followed by homologous recombination of cDNA with the target DNA (8). More recent evidence suggests that retrotransposition occurs by reverse splicing directly into DNA, with a propensity for targeting the template for the lagging strand of DNA replication (21). To determine whether Ll.ltrB insertion into the pcfG target site is dependent upon either RNA transcription or target site orientation with respect to DNA replication, we cloned a 271-bp target site from pcfG in both orientations under the control of the nisin promoter of pMSP3535 (Fig. 5A). Since pMSP3535 contains the well-characterized pAMβ1 unidirectional theta replication origin (4, 5), we were able to determine target site orientations with respect to the direction of DNA replication. The pMSP3535 vector served as a negative control.
FIG. 5.
Genetic analysis of the frequency of Ll.ltrB-Kan insertion into the pCF10 Ll.ltrB target. (A) Map of pMSP3535 target showing forward and reverse orientations of the insertion site from pcfG cloned downstream from the nisin-inducible promoter. (B) Schema for genetic analysis of the frequency of Ll.ltrB insertion into the target plasmids. The intron donor plasmid, pCOMT-Kan, was electroporated into Enterococcus faecalis cells containing one of the target plasmids. E. coli cells, illustrated by the rectangular box, were transformed with purified plasmid DNA from Enterococcus faecalis cells and used to assess the frequency of the homing of Ll.ltrB-Kan into the target plasmids. See Materials and Methods for more details.
Each construct was introduced into Enterococcus faecalis, followed by the introduction of the Ll.ltrB-Kan donor plasmid, pCOMT-Kan. Intron insertions were analyzed following the introduction of the plasmids containing intron insertions into E. coli, as described in Fig. 5B and Materials and Methods. We carried out three independent experiments per target construct. If RNA is the target of Ll.ltrB insertion, nisin induction should greatly increase the frequency of Ll.ltrB insertion into the forward target while not increasing the frequency of insertion into the reverse target. However, if Ll.ltrB inserts into DNA and there is a bias for the template for the lagging strand, the reverse target, which is encoded on this strand, should be favored, and nisin induction should have little or no effect. Our genetic analysis indicated that Ll.ltrB-Kan inserted into 100% of target plasmids containing the 271-bp pcfG fragment, irrespective of target orientation and nisin induction. No insertion into a vector plasmid lacking the target sequence was detected. Restriction enzyme analysis and sequencing confirmed Ll.ltrB-Kan insertion into precisely the same site as that observed in pCF11. Real-time RT PCR analysis indicated that nisin increased transcription of the Ll.ltrB target between 13- and 37-fold in the two constructs (data not shown).
DISCUSSION
Group II introns in bacteria are often found to be associated with mobile genetic elements, which may facilitate lateral transfer to other bacteria (22). In a recent study, 23 of 31 full-length bacterial group II introns identified in computer searches were found in mobile DNAs, especially plasmids (10). Interestingly, the three introns identified in the Ll.ltrB family (>99.8% identical) are found only in a conserved domain of relaxase genes. In this work, we showed experimentally that Ll.ltrB is able to target similar relaxase genes of other mobile genetic elements and that insertion can occur in Enterococcus faecalis at a relatively high efficiency. Since multiple conjugative elements may be present in the same strain, the targeting of a conserved conjugative transfer gene is predicted to facilitate lateral transfer between conjugative elements. Because Ll.ltrB splices out of RNA, its presence in an essential conjugation gene should not impair the mobility of the targeted conjugative element. In fact, the use of this target site might provide a selection for precise insertion events that do not mutate the region flanking the insertion site.
Not all relaxase genes have this domain. For example, two relaxases, TraX and Orf8, encoded by Enterococcus faecalis pheromone plasmids pAD1 and pAM373, respectively, appear to completely lack the three-histidine signature of motif III (17). A third example is the recently characterized MbeA relaxase encoded on plasmid ColE1, which contains a histidine, a glutamate, and an asparagine in place of the typical three-histidine motif (45). Nevertheless, relaxase genes encoding the three-histidine motif III remain the rule. Our observations suggest that the Ll.ltrB family is adapted to target relaxase genes of conjugative elements found in a broad spectrum of gram-positive bacteria.
The frequency of Ll.ltrB insertion into the pcfG target site may be somewhat lower than that of Ll.ltrB insertion into the ltrB target site. In work done with L. lactis, Mills et al. reported that approximately 60% of a high-copy-number recipient plasmid, pMN1343 (based on the shuttle vector pDL278), contained Ll.ltrB homed from the pRS01 derivative pM2036 (30). Based upon an estimate of 30 generations between the introduction of pMN1343 into L. lactis/pM2036 and pMN1343 plasmid isolation, we calculated an insertion frequency of 3 × 10−2/generation, two orders of magnitude greater than that observed with the pCF11 target in the present work. However, when the pcfG target was cloned into pMSP3535, Ll.ltrB insertion frequency approximated that observed with the ltrB target. Given the different bacterial hosts, Ll.ltrB donor plasmids, and target plasmids used in the two studies, a precise comparison of homing efficiencies into the two targets cannot be made at this time.
In 1 out of 11 sequences we examined, an extra thymidine was inserted into pCF11 just downstream of the Ll.ltrB insertion site in pcfG. While this could be an artifact of PCR, the identical sequences obtained from RT PCR, combined with the reduction in the conjugation frequency of pCF11T::Ll.ltrB-Kan compared to that of pCF11::Ll.ltrB-Kan, suggest that the thymidine insertion is real. During Ll.ltrB retrohoming, endonuclease nicks the antisense strand at position +9 with respect to Ll.ltrB insertion. These nine nucleotides are regenerated by the RT transcribing from the opposite DNA strand. This region contains a run of five thymidines (positions +8 to +12), which might allow RT frameshifting due to DNA slippage by the Streisinger frameshift mechanism (42).
The mechanism of Ll.ltrB insertion into an intronless site has been the subject of considerable research. While retrotransposition was once believed to occur through intron reverse splicing into RNA rather than DNA (8), subsequent work showed that both retrotransposition and retrohoming target DNA, at least in the majority of insertions (21). Retrotransposition does show some specificity for the template for the lagging strand of DNA synthesis, suggesting that a single-stranded DNA target may be favored (21). In the present work, Ll.ltrB targeting of the pcfG target site occurred with 100% efficiency regardless of the orientation of the target with respect to plasmid replication and in the absence or presence of nisin-induced target RNA transcription. Thus, while we could not determine whether or not target site transcription or target orientation increases intron insertion frequency, neither target site transcription nor a particular target site orientation with respect to replication is essential for Ll.ltrB insertion.
In their comprehensive review, Dai and Zimmerly (10) identified two bacterial group II intron families whose members are inserted within ORFs: the Ll.ltrB family and the E.c.I4 family. Members of the latter family include E.c.I4, S.f.I1, and Y.p.F1 from E. coli, Shigella flexneri, and Yersinia pestis, respectively. Since the target sites are only 57% identical (17 of 30 bp upstream of the insertion sites), compared to 93% homology among the introns, lateral gene transfer events, rather than vertical descent from a common ancestor, appear to be responsible for the presence of E.c.I4 family members in these sites (10).
We deduced the amino acid sequences encoded by the DNA flanking E.c.I4 intron family insertions and found that in each case in which intron-exon junctions could be determined, the intron insertion site was in a domain encoding a conserved transposase functional domain of insertion sequence elements (Fig. 6). Interestingly, this domain, termed the D,D(35)E region, is conserved in transposases of insertion sequence elements of eubacteria, as well as in integrases of retrotransposons and even eukaryotic retroviruses, including human immunodeficiency virus type 1 (HIV-1) (23). While the HIV1 integrase gene may not be an efficient target for E.c.I4 family introns, its similar sequence does illustrate the widespread conservation of another group II intron target site in nature (Fig. 6C).
FIG. 6.
Target sites of E.c.I4 family group II introns in insertion sequence (IS) elements of gram-negative bacteria identified by Dai and Zimmerly (10). Nucleotide and amino acid identities are indicated by asterisks. (A) DNA sequences spanning E.c.I4, S.f.I1, and Y.p.F1. [⇓], intron insertion site. Because of a deletion, the downstream intron-exon junction of Y.p.F1 cannot be determined. (B) Amino acid residues encoded by intron-spanning sequences. Aspartate (D, in boldface type) corresponds to the first conserved aspartate of the D,D(35)E region. The residue is conserved in transposases of bacterial IS elements, retrotransposons, and retroviruses. (C) Homologous subregion of HIV-1 integrase (accession number P04585), with amino acids conserved among retroviral and IS element transposases underlined.
In conclusion, we have demonstrated experimentally that when given the opportunity, Ll.ltrB is able to efficiently insert into a conserved relaxase gene motif on mobile genetic elements. These experimental data shed light on the observation that Ll.ltrB and E.c.I4 family group II introns are found exclusively within conserved DNA. We suggest that the targeting of conserved sites on mobile genetic elements may be a recurring theme in group II intron dispersal in bacteria.
Acknowledgments
This work was supported by grants GM49530 and GM58279 from the NIH to G.M.D. J.H.S. was supported by NIH training grants (T32AI07421, T32GM08347, and MSTP grant T32GM08244). E.M.B. was supported by an NIH training grant (T32AI07421).
We gratefully acknowledge Marlene Belfort and Ben Cousineau for their assistance in the construction of plasmids pLEIItd+KR" and pLEIItd+KR"ΔORF, Helmut Hirt for help in sequencing pcfG, Moses Vijayakumar for S. pneumoniae strain SP1000, Larry McKay for lactococcal strains containing Ll.ltrB, and Tim Leonard for his help in preparing the manuscript.
REFERENCES
- 1.Ayoubi, P., A. O. Kiliç, and M. N. Vijayakumar. 1991. Tn5253, the pneumococcal Ω (cat tet) BM6001 element, is a composite structure of two conjugative transposons, Tn5251 and Tn5252. J. Bacteriol. 173:1617-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315:995-1007. [DOI] [PubMed] [Google Scholar]
- 3.Bonen, L., and J. Vogel. 2001. The ins and outs of group II introns. Trends Genet. 17:322-331. [DOI] [PubMed] [Google Scholar]
- 4.Bruand, C., S. D. Ehrlich, and L. Janniere. 1991. Unidirectional theta replication of the structurally stable Enterococcus faecalis plasmid pAMβ1. EMBO J. 10:2171-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruand, C., E. Le Chatelier, S. D. Ehrlich, and L. Janniere. 1993. A fourth class of theta-replicating plasmids: the pAMβ1 family from gram-positive bacteria. Proc. Natl. Acad. Sci. USA 90:11668-11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 7.Byrd, D. R., and S. W. Matson. 1997. Nicking by transesterification: the reaction catalysed by a relaxase. Mol. Microbiol. 25:1011-1022. [DOI] [PubMed] [Google Scholar]
- 8.Cousineau, B., S. Lawrence, D. Smith, and M. Belfort. 2000. Retrotransposition of a bacterial group II intron. Nature 404:1018-1021. [DOI] [PubMed] [Google Scholar]
- 9.Cousineau, B., D. Smith, S. Lawrence-Cavanagh, J. E. Mueller, J. Yang, D. Mills, D. Manias, G. Dunny, A. M. Lambowitz, and M. Belfort. 1998. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94:451-462. [DOI] [PubMed] [Google Scholar]
- 10.Dai, L., and S. Zimmerly. 2002. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 30:1091-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270-278. [DOI] [PubMed] [Google Scholar]
- 12.Dunny, G., M. Yuhasz, and E. Ehrenfeld. 1982. Genetic and physiological analysis of conjugation in Streptococcus faecalis. J. Bacteriol. 151:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eickbush, T. H. 1999. Mobile introns: retrohoming by complete reverse splicing. Curr. Biol. 9:R11-R14. [DOI] [PubMed] [Google Scholar]
- 15.Eskes, R., L. Liu, H. Ma, M. Y. Chao, L. Dickson, A. M. Lambowitz, and P. S. Perlman. 2000. Multiple homing pathways used by yeast mitochondrial group II introns. Mol. Cell. Biol. 20:8432-8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eskes, R., J. Yang, A. M. Lambowitz, and P. S. Perlman. 1997. Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell 88:865-874. [DOI] [PubMed] [Google Scholar]
- 17.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 18.Gold, O. G., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 19.Guo, H., M. Karberg, M. Long, J. P. Jones III, B. Sullenger, and A. M. Lambowitz. 2000. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science 289:452-457. [DOI] [PubMed] [Google Scholar]
- 20.Guo, H., S. Zimmerly, P. S. Perlman, and A. M. Lambowitz. 1997. Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J. 16:6835-6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichiyanagi, K., A. Beauregard, S. Lawrence, D. Smith, B. Cousineau, and M. Belfort. 2002. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 46:1259-1272. [DOI] [PubMed] [Google Scholar]
- 22.Klein, J. R., and G. M. Dunny. 2002. Bacterial group II introns and their association with mobile genetic elements. Front. Biosci. 7:d1843-d1856. [DOI] [PubMed] [Google Scholar]
- 23.Kulkosky, J., K. S. Jones, R. A. Katz, J. P. Mack, and A. M. Skalka. 1992. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 12:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 25.Le Bourgeois, P., M. L. Daveran-Mingot, and P. Ritzenthaler. 2000. Genome plasticity among related Lactococcus strains: identification of genetic events associated with macrorestriction polymorphisms. J. Bacteriol. 182:2481-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann, K., and U. Schmidt. 2003. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit. Rev. Biochem. Mol. Biol. 38:249-303. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Abarca, F., and N. Toro. 2000. Group II introns in the bacterial world. Mol. Microbiol. 38:917-926. [DOI] [PubMed] [Google Scholar]
- 28.Matsuura, M., R. Saldanha, H. Ma, H. Wank, J. Yang, G. Mohr, S. Cavanagh, G. M. Dunny, M. Belfort, and A. M. Lambowitz. 1997. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 11:2910-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel, F., K. Umesono, and H. Ozeki. 1989. Comparative and functional anatomy of group II catalytic introns—a review. Gene 82:5-30. [DOI] [PubMed] [Google Scholar]
- 30.Mills, D. A., D. A. Manias, L. L. McKay, and G. M. Dunny. 1997. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol. 179:6107-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mills, D. A., T. G. Phister, G. M. Dunny, and L. L. McKay. 1998. An origin of transfer (oriT) on the conjugative element pRS01 from Lactococcus lactis subsp. lactis ML3. Appl. Environ. Microbiol. 64:1541-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mohr, G., P. S. Perlman, and A. M. Lambowitz. 1993. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 21:4991-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohr, G., D. Smith, M. Belfort, and A. M. Lambowitz. 2000. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev. 14:559-573. [PMC free article] [PubMed] [Google Scholar]
- 35.O'Sullivan, D., R. P. Ross, D. P. Twomey, G. F. Fitzgerald, C. Hill, and A. Coffey. 2001. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility functions to a food-grade selectable marker. Appl. Environ. Microbiol. 67:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pansegrau, W., W. Schröder, and E. Lanka. 1994. Concerted action of three distinct domains in the DNA cleaving-joining reaction catalyzed by relaxase (TraI) of conjugative plasmid RP4. J. Biol. Chem. 269:2782-2789. [PubMed] [Google Scholar]
- 37.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 38.Sharp, P. A. 1991. “Five easy pieces.” Science 254:663. [DOI] [PubMed] [Google Scholar]
- 39.Shearman, C., J. J. Godon, and M. Gasson. 1996. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol. Microbiol. 21:45-53. [DOI] [PubMed] [Google Scholar]
- 40.Singh, N. N., and A. M. Lambowitz. 2001. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol. 309:361-386. [DOI] [PubMed] [Google Scholar]
- 41.Srinivas, P., A. O. Kiliç, and M. N. Vijayakumar. 1997. Site-specific nicking in vitro at oriT by the DNA relaxase of Tn5252. Plasmid 37:42-50. [DOI] [PubMed] [Google Scholar]
- 42.Streisinger, G., Y. Okada, J. Emrich, J. Newton, A. Tsugita, E. Terzaghi, and M. Inouye. 1966. Frameshift mutations and the genetic code. Cold Spring Harbor Symp. Quant. Biol. 31:77-84. [DOI] [PubMed] [Google Scholar]
- 43.Toor, N., G. Hausner, and S. Zimmerly. 2001. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA 7:1142-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 45.Varsaki, A., M. Lucas, A. S. Afendra, C. Drainas, and F. De La Cruz. 2003. Genetic and biochemical characterization of MbeA, the relaxase involved in plasmid ColE1 conjugative mobilization. Mol. Microbiol. 48:481-493. [DOI] [PubMed] [Google Scholar]
- 46.Vijayakumar, M. N., and S. Ayalew. 1993. Nucleotide sequence analysis of the termini and chromosomal locus involved in site-specific integration of the streptococcal conjugative transposon Tn5252. J. Bacteriol. 175:2713-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong, J., M. Karberg, and A. M. Lambowitz. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 31:1656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, L., D. A. Manias, and G. M. Dunny. 2000. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol. Microbiol. 37:639-651. [DOI] [PubMed] [Google Scholar]
- 50.Zimmerly, S., G. Hausner, and X. Wu. 2001. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 29:1238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]