Abstract
Screening a Clostridium difficile strain collection for the chimeric element CdISt1, we identified two additional variants, designated CdISt1-0 and CdISt1-III. In in vitro assays, we could prove the self-splicing ribozyme activity of these variants. Structural comparison of all known CdISt1 variants led us to define four types of IStrons that we designated CdISt1-0 through CdISt1-III. Since CdISt1-0 encodes two complete transposase-like proteins (TlpA and TlpB), we suggest that it represents the original genetic element, hypothesized before to have originated by fusion of a group I intron and an insertion sequence element.
Recently, the genetic element CdISt1 was identified in the genome of Clostridium difficile strain C34 (1, 5) and was shown to combine features of group I introns and insertion sequence (IS) elements. To distinguish between individual CdISt1 variants from different strains, the strain C34 element is herein designated CdISt1-C34, since the designation CdISt1 includes all variants combining features of the group I intron and the IS elements of the IS605 type. The 434 bp at the 5′ end of CdISt1-C34 show the typical structures and key features of group I introns, while the 3′ part harbors two open reading frames (ORFs) coding for a truncated (tlpA) and a complete (tlpB) protein. The putative proteins TlpA and TlpB show high homology to transposases, which are characteristic products of composite IS elements of the IS605 type (Fig. 1) (4). In contrast to classical group I introns, CdISt1-C34 is found in protein-encoding genes of a bacterial chromosome (1, 2). Sequence analysis of CdISt1-C34, several of its variants (CdISt1a-C34 through CdISt1j-C34), and their integration sites revealed that these genetic elements show all features characteristic of an IS605-like mobility mechanism (1, 3). Therefore, we assumed that the IS element component mediates the spread of CdISt1 while the intron component is responsible for efficient splicing of CdISt1 from precursor mRNA. Due to their chimeric nature, we called these elements IStrons.
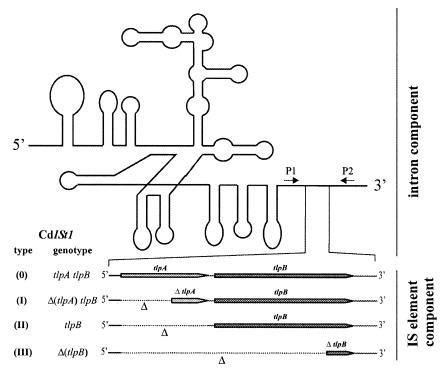
FIG. 1.
Schematic drawing of CdISt1 IStron variants types 0 to III. The four CdISt1 types are depicted; the intron component (upper part) shows little variation, whereas the IS element component (lower part) varies as indicated. All variants have conserved ribozyme activities, and deletions of the putative TlpA and TlpB proteins are found in all but type 0 variants. Arrows labeled P1 (5′-CGACAACCTCAAAAATGATAAA-3′) and P2 (5′-TCTTAATCCTTCTTTTAATATATTT-3′) indicate the primers for PCR amplification of the four IStron types.
Previously, two types of IStrons (CdISt1-I and CdISt1-II) were identified in C. difficile, and these types differ in the IS element component (Fig. 1) (1). We have grouped all characterized CdISt1 IStron variants into four types (CdISt1-0 to CdISt1-III) based on their structural features. Within each type, the variants differ in their genomic integration sites and the strains in which they are located. Compared to CdISt1-I IStrons, CdISt1-II IStrons show a deletion of about 100 bp covering nearly the entire tlpA gene (Fig. 1). To verify the proposal that CdISt1 was generated by fusion of a group I intron and an IS element from the IS605 family, we searched for additional CdISt1 types without deletion of the IS element component. C. difficile isolates were screened for CdISt1-related sequences by using two internal primers (P1, CGACAACCTCAAAAATGATAAA, and P2, TCTTAATCCTTCTTT TAATATATTT) that amplify a major part of the CdISt1 sequence. PCR conditions were as follows: 30 cycles of 10 s at 94°C, 30 s at 42°C, and 4 min at 68°C. Using this PCR assay, we detected CdISt1-related sequences in the genomes of all C. difficile isolates tested but not in those of other clostridial species such as C. acetobutylicum, C. bifermentans, C. botulinum, and C. perfringens.
Six different patterns of PCR amplification products were obtained with C. difficile chromosomal DNA as the template (Fig. 2). PCR amplicons of 1,397 bp identified type I variants, while amplicons of 1,318 bp identified type II variants in the bacterial genome. The amplicon sizes in four of the six patterns differed remarkably from those corresponding to CdISt1 types I and II (Fig. 2). This indicated the existence of yet unknown CdISt1 variants in the genomes of the respective C. difficile strains. One of these PCR products (Fig. 2, lanes 5 and 6) was approximately 250 bp larger than the PCR product representing CdISt1-I, and the other (Fig. 2, lanes 1, 4, and 6) was more than 1.3 kb smaller than the PCR product representing CdISt1-II. Sequencing of the two unusual PCR products verified that they actually represent two novel CdISt1 types. The larger was designated type 0 (CdISt1-0), and the smaller was designated type III (CdISt1-III) (Fig. 1).
FIG. 2.
PCR patterns representing CdISt1 variants from different C. difficile isolates. Primers P1 and P2 were used for PCR analysis of genomic DNA prepared from different C. difficile strains, as indicated in the text. Lanes are as follows: M, 1-kb ladder (MBI Fermentas); C, negative control (PCR run without template DNA); 1 to 6, chromosomal DNA template derived from strains C34 (1), ST1322/96 (2), 37444 (3), 46307 (4), SE918 (5), and CH3859 (6). The double band at 1,397 and 1,318 bp identifies CdISt1 type I and type II variants, respectively. Bands at approximately 1,650 bp in lanes 5 and 6 correspond to PCR products derived from CdISt1 type 0 variants; bands with sizes of 150 bp (lanes 1 and 6) and ∼210 bp (lane 4) correspond to CdISt1 type III PCR products. The size difference results from deletions of different portions of tlpB in various isolates.
As described by Braun et al. (1), we then used primers P3 (CGGAGCTTACCTGCTGATTG) and P4 (GCCCCTACCGGACACCTCTT) for inverse PCR and determined the integration sites and the complete sequences of CdISt1l-C34 (CdISt1-III variant) and CdISt1a-SE918 (CdISt1-0 variant) in strain SE918. CdISt1l-C34 has a size of 734 bp and is located in an ORF homologous to one from C. perfringens encoding a protein tyrosine phosphatase. CdISt1l-C34 contains the complete ribozyme component showing 97.5% identity to CdISt1-I and CdISt1-II copies. Downstream of the intron component, we found a deletion in CdISt1l-C34 of 1,247 bp covering tlpA and the major part of tlpB (positions 558 to 1804 of CdISt1-C34). The 3′ ends of CdISt1l-C34 and CdISt1-I IStrons are identical and contain the 3′ splice site. Using reverse transcription-PCR and an in vitro splice assay, we verified that CdISt1l-C34 is precisely and efficiently excised from precursor mRNA in vitro and in vivo (data not shown). This demonstrates that all structures associated with the ribozyme activity of the IStron are preserved in CdISt1l-C34.
CdISt1a-SE918 is integrated into an ORF homologous to one from Thermogota maritima encoding a 16S pseudouridylate synthase. Strikingly, in C. difficile strain C34, the same ORF is interrupted by a CdISt1-II variant and not by a CdISt1-0 variant (1). Sequence analysis revealed that CdISt1a-SE918 shows very high homology to CdISt1-I IStrons, but CdISt1a-SE918 contains an additional DNA stretch of 239 bp located upstream of the truncated tlpA of type I IStrons (Fig. 1). Except for a TAA stop codon at position 40, this additional sequence is in frame with the remnant of tlpA. Analyzing C. difficile strains 55767 and R9304, which also contain large CdISt1-0 variants (CdISt1a-55767 and CdISt1a-R9304), we found a CAA sequence instead of this TAA stop codon. Within the three CdISt1-0 IStron variants, we found a potential Shine-Dalgarno sequence 7 bp upstream of the start codon of the tlpA ORF. Thus, tlpA of CdISt1-0 variants seems to code for a complete transposase-like protein. The calculated TlpA size (135 amino acids) and molecular mass (16 kDa) are typical of IS200 homolog transposases. A database search using the translated amino acid sequence of the putative TlpA encoded by CdISt1-0 variants of strains SE918, 55767, and R9304 verified high similarity to IS200 homologous transposases from C. perfringens (accession number, NP_562804), Deinococcus radiodurans (accession number, BAA32389), Methanosarcina mazei strain Goel (MM1729; accession number, NP_633753), and Helicobacter pylori (TnpA; accession number, AAD11513). The ribozyme activity of CdISt1a-SE918 was again checked in an in vitro splice assay. This assay demonstrated that CdISt1a-SE918 is able to carry out self-splicing (data not shown). Obviously, the huge spacing of more than 1,750 bp between the intron core structures and the 3′ splice site in CdISt1a-SE918 does not interfere with the ribozyme activity of this chimeric element.
To conclude, more than 20 CdISt1 copies from different C. difficile strains have been characterized by now (1; this study), and four types have been found: CdISt1-0 (TlpA/TlpB), CdISt1-I (ΔTlpA/TlpB), CdISt1-II (TlpB), and CdISt1-III (ΔTlpB) (Fig. 1). All CdISt1 types share a complete and functional group I intron component but differ in their IS element components. The CdISt1-0 variants alone seem to contain a nearly complete IS element of the IS605 family coding for two transposase-like proteins. We assume that CdISt1-0 variants may represent the original genetic element that—according to a recent proposal—originated by the fusion of a group I intron and an IS element.
Nucleotide sequence accession numbers.
Sequences of CdISt1l-C34, CdISt1a-SE918, CdISt1a-55767, and CdISt1a-R9304 have been deposited in the EMBL database under accession numbers AJ440753, AJ579718, AJ579716, and AJ579717, respectively.
Acknowledgments
This work was supported by grant Ei 206/10-1 and 10-2 from the Deutsche Forschungsgemeinschaft.
C.V.E.-S. expresses his special gratitude to the Johannes Gutenberg University in Mainz for providing his group with laboratory space in the Verfügungsgebäude für Forschung und Entwicklung.
REFERENCES
- 1.Braun, V., M. Mehlig, M. Moos, M. Rupnik, B. Kalt, D. E. Mahony, and C. von Eichel-Streiber. 2000. A chimeric ribozyme in Clostridium difficile combines features of group I introns and insertion elements. Mol. Microbiol. 36:1447-1459. [DOI] [PubMed] [Google Scholar]
- 2.Edgell, D. R., M. Belfort, and D. A. Shub. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kersulyte, D., N. S. Akopyants, S. W. Clifton, B. A. Roe, and D. E. Berg. 1998. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223:175-186. [DOI] [PubMed] [Google Scholar]
- 4.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehlig, M., M. Moos, V. Braun, B. Kalt, D. E. Mahony, and C. von Eichel-Streiber. 2001. Variant toxin B and a functional toxin A produced by Clostridium difficile C34. FEMS Microbiol. Lett. 198:171-176. [DOI] [PubMed] [Google Scholar]