Abstract
After unsuccessful attempts to recover a viable RecA-deficient mutant of the Lyme borreliosis agent Borrelia burgdorferi, we characterized the functional activities of RecA of B. burgdorferi, as well as RecA of the relapsing fever spirochete Borrelia hermsii and the free-living spirochete Leptospira biflexa, in a recA mutant of Escherichia coli. As a control, E. coli RecA was expressed from the same plasmid vector. DNA damage repair activity was assessed after exposure of the transgenic cells to UV light or the radiomimetic chemicals methyl methanesulfonate and mitomycin C. Recombination activity in the cells was assessed by using an assay for homologous recombination between repeats in the chromosome and by measuring the ability of the cells to foster lytic growth by red gam mutant bacteriophage λ. Overall, we found that transgenic cells with recA genes of B. burgdorferi, B. hermsii, and L. biflexa had approximately equivalent activities in promoting homologous recombination in the lacZ duplication assay, but cells with B. burgdorferi recA and, most notably, B. hermsii recA were significantly less capable than cells with L. biflexa recA or E. coli recA in responding to DNA damage or in facilitating plaque formation in the phage assay. The comparatively poor function of Borrelia recA in the latter set of assays may be the consequence of impaired coordination in the loading of the transgenic RecA by RecBCD and/or RecFOR in E. coli.
Borrelia burgdorferi, a cause of Lyme borreliosis, and Borrelia hermsii, an agent of relapsing fever, are spirochetes, a group of bacteria that are phylogenetically distinct from other major bacterial groups, such as the firmicutes, actinomycetes, proteobacteria, and cyanobacteria (10). The spirochete clade itself is deep, and its genera are highly diverse (34). Besides parasitic genera like Borrelia and Treponema, the spirochete group includes a large number of free-living species, such as Leptospira biflexa. Members of the genus Borrelia are not free living but instead cycle between two sets of hosts: arthropod vectors and mammalian or avian reservoirs (3). Compared to what is known about some other types of bacteria, such as Escherichia coli and Bacillus subtilis, little is known about the genetics and physiology of spirochetes.
The B. burgdorferi genome, which comprises the 1-Mb chromosome and several linear and circular plasmids, has been sequenced (6, 15). The limited biosynthetic capabilities demonstrated by the genome sequence are consistent with the obligately parasitic existence of B. burgdorferi in nature and the high nutritional requirements for cultivation in the laboratory (2). B. hermsii has a similarly limited genome size and restricted ecologic niche (3, 14). A cross-hybridization array study of the genomic DNA of B. hermsii indicated that this species has much the same genetic complement as B. burgdorferi (55).
The genus Leptospira includes both free-living and parasitic species, and in one study a parasitic species, like Leptospira interrogans, was more susceptible to DNA-damaging agents than a free-living species, like L. biflexa (42). As a constant resident of the interiors of either ticks or vertebrates, neither B. burgdorferi nor B. hermsii is likely to have much exposure to DNA-damaging UV radiation compared to the exposure of the facultatively free-living species E. coli and L. biflexa. B. burgdorferi thrives best under microaerophilic conditions (53), an environment in which oxidative damage to the DNA in the organism should be limited (33). As a consequence of this comparatively sheltered lifestyle, the DNA repair functions of the RecA protein may be less critical for the genus Borrelia than for bacteria that exist in the environment. On the other hand, another RecA function, homologous recombination, may be very important for the persistence of B. burgdorferi and B. hermsii in their mammalian hosts. Both species exhibit antigenic variation due to frequent gene conversion events involving the multiple alleles for surface lipoproteins (reviewed in reference 1). Similar antigenic variation through gene conversion in Neisseria gonorrhoeae, the cause of human gonorrhea, has been shown to be largely mediated by RecA (22).
The RecA protein of B. burgdorferi was identified previously and was shown to have many of the conserved motifs of other bacterial RecA proteins (11). To assess the role of RecA in the frequency of recombination at the VlsE locus in B. burgdorferi (54), we attempted to produce a knockout of the recA gene of this species by insertional inactivation (4). While experiments with positive controls demonstrated that transformation and insertional inactivation of another gene could be achieved in B. burgdorferi, no viable clones with insertions into the recA gene were obtained after multiple attempts (Putteet-Driver, unpublished data). This suggested to us that inactivation of recA of B. burgdorferi was either lethal or highly debilitating to the cell. Although recA mutants with insertional inactivation have been obtained for a number of bacterial species, such as Brucella abortus (48), N. gonorrhoeae (21), and Xanthomonas oryzae (28), these organisms have usually been proteobacteria. In these cases both DNA damage repair and homologous recombination were usually impaired in the recA mutant cells (27). The much less common examples of organisms with RecA knockouts that either are apparently lethal or result in extremely slow growth have been members of other bacterial groups, such as the actinomycete Streptomyces lividans (29) or the spirochete L. biflexa (49).
Lacking a means of producing a recA mutant line of B. burgdorferi or B. hermsii, we instead characterized the function of spirochete RecA in E. coli. A previous study by Stamm et al. demonstrated that complementation of E. coli recA mutants with the recA gene of L. biflexa was feasible (43), even with the considerable genetic difference between a proteobacterium like E. coli and spirochetes (10). In the present study, recA mutant E. coli cells were transformed with plasmids expressing recombinant recA genes of the spirochetes B. burgdorferi, B. hermsii, and L. biflexa, and the abilities of the recombinant proteins to complement the recombination and repair functions of the host E. coli cells were assessed. While the three spirochete recA genes complemented defects in E. coli in a homologous recombination assay with nearly equal activities, B. burgdorferi RecA and B. hermsii RecA were less able to complement DNA repair defects in E. coli than either recombinant E. coli RecA or L. biflexa RecA was.
MATERIALS AND METHODS
Strains and growth conditions.
B. burgdorferi strain B31 (= ATCC 35210), B. hermsii strain HS1 (36), Borrelia coriaceae (23), and Borrelia turicatae strain Oz1 (5) were grown in BSK II broth medium (2). For the genomic library of B. hermsii the spirochetes were first grown in mice, and then the population was expanded once by 1:100 dilution in broth medium as described previously (2). The spirochetes were harvested by centrifugation at 9,000 × g for 20 min at 23°C, and the cells were stored frozen in phosphate-buffered saline (pH 7.4) with 10% dimethyl sulfoxide at −80°C. The E. coli strains used in this investigation were RecA+ strain JM105 with lacIq on F′ (50) and the following RecA− strains: JM109 with recA1 and lacIq on F′ (Stratagene, La Jolla, Calif.), Top10F′ with recA1 and lacIq on F′ (Invitrogen Life Technologies, Carlsbad, Calif.), and JC14604 [araC14 Δ(lacY-lacZ)286 tsx-33 supE44 galK2(Oc) λ− lacZ95 hisG4(Oc) Δ(recA-srl)306 srlR301::Tn10 rpsL31(Strr) xylA5 mtl-1 argE3(Oc) thi-1 hsdR4] (20). The E. coli cells were grown either in Luria-Bertani (LB) broth or on LB agar plates, which contained 50 μg of carbenicillin per ml as needed. The doubling times at 37°C in LB medium were determined by measuring the absorbance of the cultures at 600 nm. E. coli JC14604 cells were grown on M9 salts (47 mM Na2HPO4, 22 mM KH2PO4, 8.5 mM NaCl, 18.7 mM NH4Cl; pH 7.0) agar plates supplemented with arginine (20 μg/ml), histidine (20 μg/ml), thiamine hydrochloride (50 ng/ml), and either 0.4% lactose or 0.4% glucose as the carbohydrate. The red gam bacteriophage lambda was the lambda DASH II vector (Stratagene), which contained, as an irrelevant insert, 12 kb of the 18S and 28S rRNA operon of the ixodid tick Amblyomma americanum (18).
DNA isolation.
For PCR and Southern blot analysis total DNA was isolated from B. burgdorferi, B. hermsii, B. turicatae, and E. coli by using a Qiaquick genomic DNA kit obtained from Qiagen (Valencia, Calif.). Plasmids were isolated from E. coli by using a HighPure plasmid isolation kit (Roche Applied Science, Indianapolis, Ind.) for screening of library clones and a Qiaquick Midi Prep kit (Qiagen) for DNA sequencing. Genomic DNA from Borrelia miyamotoi strain HT31 (17) was provided by M. Fukunaga (Fukuyama University, Fukuyama, Japan). Genomic DNA from L. biflexa serovar patoc was provided by R. Zuerner (United States Department Agriculture, Ames, Iowa). A genomic library of B. hermsii HS1 was constructed by SeqWright (Houston, Tex.) from randomly sheared DNA that was blunt ended and ligated into the vector pUC18. For the library total DNA was extracted from B. hermsii by using sodium dodecyl sulfate, proteinase K, and phenol as described previously (26). The ligation reaction products were transformed into E. coli XL1-Blue (SeqWright). The insert sizes ranged from 5 to 10 kb, and the final library contained 2,306 clones that were individually stored at −20°C in 96-well microtiter plates in 50% LB broth-50% glycerol.
PCR.
All PCR for amplification were performed with the GeneAmp PCR System 2400 (Perkin-Elmer, Norwalk, Conn.). The reaction mixtures (50 μl) contained either 1.25 U of Pwo polymerase or 2.5 U of Taq polymerase (Roche Applied Science), each deoxynucleoside triphosphate (Roche Applied Science) at a concentration of 0.4 mM, 2.0 mM MgSO4, each primer at a concentration of 0.2 μM, and up to 200 ng of template DNA. The PCR conditions were as follows: an initial step of 95°C for 5 min; 30 cycles of 95°C for 15 s, 5°C below the calculated melting temperature for 30 s, and 72°C for 1 min/kb; and a final step of 72°C for 8 min. PCR products were cleaned by using a Qiaquick PCR purification kit (Qiagen), the sizes were confirmed by agarose electrophoresis, and the concentrations were determined by spectrophotometery.
Southern blot analysis.
DNA was digested with a restriction enzyme and then was separated on a 0.8% agarose gel and transferred to an Immobilon-Ny+ membrane (Millipore, Bedford, Mass.) by capillary transfer in 0.4 M NaOH. The DNA was cross-linked to the membrane by UV irradiation. The blots were incubated first in a hybridization buffer containing 4% (wt/vol) AlkPhos blocking reagent (Amersham Biosciences, Piscataway, N.J.) and 0.5 M NaCl for ∼3 h without the probes. The probes labeled with alkaline phosphatase (AlkPhos direct labeling; Amersham Biosciences) were then added, and hybridization was carried at 50°C. The labeled probes were detected by using the chemiluminescent detection reagent CDP-Star (Amersham Biosciences) and X-ray film.
Colony blot hybridization.
Library clones were plated in a grid on plates, and the resultant colonies were transferred to Immobilon-Ny+ membranes (Millipore) as described previously (39). The membranes were prehybridized in 0.12 M Na2PO4 (pH 7.2)-0.25 M NaCl-7% sodium dodecyl sulfate in a shaking water bath at 60°C for 2 to 3 h. They were then probed with a PCR product of the B. burgdorferi recA gene, which spanned nucleotide positions 50 to 922 of the coding sequence (accession number AE001124) and was radiolabeled with a MegaPrime random prime labeling kit (Amersham Biosciences) and [α-32P]ATP in the same buffer at 60°C. The membranes were washed as described previously (37) and then exposed to X-ray film.
Northern blot analysis.
Total RNA was extracted with the TRIzol reagent (Invitrogen Life Technologies) from E. coli Top10F′ cells treated with isopropyl-β-d-thiogalactopyranoside (IPTG) as described above. One microgram of total RNA was separated on a 1% agarose denaturing gel, the bands were transferred to an Immobilon-Ny+ membrane (Millipore), and the blot was prehybridized in hybridization buffer as described previously (16). The probes for the blots were PCR products of positions 234 to 850 of B. burgdorferi recA (accession number AE001124), positions 227 to 840 of B. hermsii recA (AF395125), and the bla gene of the pBluescript II vector from position 2217 to position 2807. PCR products were radiolabeled by using a MegaPrime random prime labeling kit (Amersham Biosciences) and [α-32P]ATP. The membranes were hybridized in hybridization buffer at 42°C and then washed as described previously (16). The blots were exposed to a phosphor screen (Molecular Dynamics) and scanned with a Personal FX phosphorimager (Bio-Rad Laboratories, Hercules, Calif.).
Cloning of recA fragments of other Borrelia species.
The partial recA sequences of other Borrelia species were obtained by using primers whose sequences were conserved in the recA genes of B. burgdorferi and B. hermsii. The forward and reverse primers corresponded to positions 234 to 254 and 850 to 829, respectively, of the B. burgdorferi recA gene and were used under the conditions described above. The PCR products were ligated into a pCR 2.1 TA cloning vector (Invitrogen Life Technologies) and were transformed into competent E. coli Top10F′ cells.
DNA sequencing and analysis.
Inserts of the plasmids were sequenced on both strands with a 377 automated sequencer (Applied Biosystems, Foster City, Calif.) by using M13 primers and custom primers based on the sequence.
Expression of recA genes in E. coli.
An NdeI restriction site was inserted into the pBluescript II KS plasmid (Stratagene, La Jolla, Calif.) by using the following primers: 5′GTAATCATGGTCATATGTGTTTCCTGTGTG3′ and 5′GCCAAGCGCGCAATTAACCCTCACT3′. The recA coding regions of B. burgdorferi, B. hermsii, E. coli, and L. biflexa (accession number AF410431 [49]) were amplified from genomic DNA by using the following forward and reverse primers with NdeI and BamHI restriction sites (underlined), respectively, at their ends: for B. burgdorferi, 5′GGAATTCCATATGTCAAAGTTAAAGGAAAAAAGA3′ and 5′CGGGATCCTACTAACATATTCAAAAAAAAGCTC3′; for B. hermsii, 5′GGAATTCCATATGTCAAAATTAAAAGATAACCCAG3′ and 5′CGGGATCCTACAAACAATCAAATAATCATCTAA3′; for E. coli, 5′GGAATTCCATATGGCTATCGACGAAAACAAACAGA3′ and 5′CGGGATCCGTTGATTCTGTCATGGCATATCCTT3′; and for L. biflexa, 5′GGAATTCCATATGAAAAAAGAGAAAGCTGACAAG3′ and 5′CGGGATCCCGGTCAATGGTTCCTAAAAAGA3′. After digestion with NdeI and BamHI, the PCR products were ligated to an NdeI-BamHI-digested cloning vector, and the ligation mixture was transformed into E. coli Top10F′ cells.
DNA damage and repair assays.
E. coli Top10F′ isolates with the different plasmids were grown in LB broth containing carbenicillin at 37°C with shaking to an optical density at 600 nm (OD600) of 0.5 to 0.6, at which point IPTG was added to a final concentration of 10 μM. The cultures were grown for an additional 1 h and then serially 10-fold diluted in LB broth. For the UV radiation sensitivity assays, 10-μl portions of the suspensions were spotted in triplicate on LB agar plates with carbenicillin. The plates were then exposed to up to 80 mJ of UV irradiation (254 nm) in a Stratalinker UV light box (Stratagene) and then incubated overnight at 37°C in the dark. The percentages of survival were then determined by using nonirradiated plates as the reference (44). To determine susceptibility to chemical DNA-damaging agents, serial dilutions of the cell suspensions were spotted in triplicate on LB agar plates containing various concentrations of methyl methanesulfonate (MMS) or mitomycin C (Sigma-Aldrich, St. Louis, Mo.) and incubated for 48 h at 37°C in the dark, after which colonies were counted and compared to the colonies on plates without either agent.
λ Bacteriophage plaque assay.
The titers of bacteriophage were determined with RecA+ JM105 cells as described previously (38). Cultures of JM109 cells containing one of the plasmids were cultivated in LB broth supplemented with 0.2% maltose and 10 mM MgSO4 at 37°C to an OD600 of 0.5 to 0.6, IPTG was added, and cultivation was continued for another 1 h. The cells were harvested by centrifugation at 4,000 × g for 20 min and then resuspended in 10 mM MgSO4 to a final OD600 of ∼0.5. A known number of phage particles was added to each suspension and allowed to adsorb for 15 min at 37°C with gentle shaking. The cells were mixed with molten NZY agar containing 10 μM IPTG to form a top agar on the plates. After overnight incubation at 37°C, the phage plaques were counted after 16 h.
Recombination assay.
E. coli strain JC14604 cells with the different recA plasmids were grown in 1 liter of LB broth at 37°C to an OD600 of 0.5 to 0.6. IPTG was added to some of the cultures, and cultivation was continued for another 1 h. Then the cells were harvested by centrifugation and resuspended in 10 ml of M9 salts and serially diluted. The number of CFU at each dilution was determined by plating 1 ml in triplicate on nonselective M9 minimal medium plates with glucose or on selective M9 plates with lactose. The plates were incubated at 37°C. Colony counting for M9 minimal medium plates with glucose was performed after 48 h of incubation, and colonies on M9 minimal medium plates with lactose were counted after 7 days of incubation.
DNA array data.
The normalized hybridization data were data from a study of B. burgdorferi and B. hermsii that was previously described in part by Zhong and Barbour (55). The genomic DNA arrays of B. burgdorferi on membranes were described by Ojaimi et al. (31). Total RNA from B. burgdorferi strain B31 cells cultivated at 23 or 35°C was provided by Patricia Rosa (Rocky Mountain Laboratories, Hamilton, Mont.); two membranes were used for each of the temperature conditions, as described previously. Total RNA from B. hermsii strain HS1 cells was obtained for four replicates of cells growing in BSK II medium and three replicates of cells growing in the blood of mice. The temperature, pH, and cell density of the in vitro cultures were adjusted so that the conditions were equivalent to the in vivo conditions. cDNA was produced and radiolabeled, and the probes were hybridized with membranes as described previously (55). The DNA ArrayVision 6.0 software package (Research Imaging, Inc., St. Catherines, Ontario, Canada) was used to record the pixel density of each spot on the blots. The background value for control spots without DNA was subtracted from the values for duplicate spots containing DNA representing each open reading frame (ORF). The mean of the values was then normalized by dividing each value by the total number of pixels for all spots on a given membrane. Mean expression levels were compared by using a two-tailed t test.
Nucleotide sequence accession numbers.
The following GenBank accession numbers have been assigned for the complete and partial recA sequences described here: B. hermsii recA with flanking regions, AF395125; B. coriaceae, AY340864; B. miyamotoi, AY340866; and B. turicatae, AY340865.
RESULTS
Cloning of recA genes of spirochetes in E. coli.
In a preliminary study a recombinant plasmid containing the B. burgdorferi recA gene with its native promoter did not complement an E. coli recA mutant in the UV survival assay (data not shown). Accordingly, we used an expression plasmid with the LacZ promoter for inducible expression of the recombinant recA genes in an E. coli recA mutant strain.
The different plasmids with recombinant RecA were constructed by using PCR-amplified coding regions from genomic DNA of B. burgdorferi, E. coli, and L. biflexa. NdeI sites were added before the start codon of each gene, and BamHI sites were added at the 3′ ends of the PCR products 30 to 90 nucleotides after the stop codons. The products were cloned into the modified vector and then transformed into E. coli Top10F′ cells. The inserts were confirmed to be recA genes of the appropriate species by sequencing. The plasmids were designated pBb:recA, pEc:recA, and pLb:recA for the recA genes of B. burgdorferi, E. coli, and L. biflexa, respectively.
Cloning and sequencing of recA of B. hermsii and other relapsing fever species.
The recA gene of B. hermsii had not been identified and cloned previously, and, accordingly, these steps were carried out in the present study. Genomic DNA of B. hermsii was digested with SpeI, and a Southern blot of the digested DNA that was probed with the PCR product of B. burgdorferi recA revealed a single hybridizing band at ∼5 kb. Under the same hybridization conditions with the B. burgdorferi recA probe, we screened a threefold redundant genomic library of B. hermsii in the pUC18 vector. Southern blot analysis of three hybridizing transformants revealed that the clones contained overlapping fragments. Sequencing of these inserts and subsequent database searches identified the recA gene of B. hermsii within the inserts.
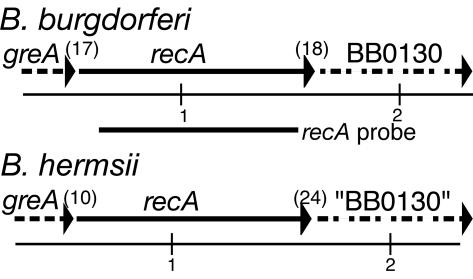
Figure 1 shows the recA gene and its flanking regions in B. hermsii. Upstream of recA on the same strand we identified a partial ORF corresponding to the 3′ end of the transcription elongation factor gene greA. This partial sequence was 76% identical to the corresponding greA gene upstream of recA in B. burgdorferi. Downstream of the recA stop codon is an ORF that over its length is 55% identical to the partial sequence of BB0130, an ORF of B. burgdorferi, and 24% identical to TP0691, an orthologous ORF of Treponema pallidum. All three spirochete ORFs are similar to the ORFs encoding the COG1354 family of proteins with unknown functions in bacteria and archaea (http://www.ncbi.nlm.nih.gov/Structure/cdd/). In both genomes there were only a few nucleotides between the coding sequences for the recA genes and the flanking ORFs.
FIG. 1.
Physical map of B. burgdorferi and B. hermsii recA genes and flanking regions. The numbers in parentheses are the numbers of nucleotides between ORFs, the numbers below the map are the lengths (in kilobases), and the arrows indicate the direction of transcription. The solid lines represent the complete gene sequence, while the dashed lines represent partial sequences. The approximate extent of the B. burgdorferi recA probe used for library screening is shown. The partial sequence of the BB0130 ORF of B. hermsii is homologous to ORF BB0130 of B. burgdorferi.
As was done with the other recA genes, the recA coding region of B. hermsii was amplified by PCR, the product was cloned into the modified pBluescript vector to produce the pBh:recA plasmid, and the sequence of the insert was verified.
On the basis of the conserved sequence in the recA genes of B. burgdorferi and B. hermsii, we designed degenerate primers to amplify the partial recA sequences of other relapsing fever and related species. With these primers we amplified ∼0.6-kb fragments from B. coriaceae, B. miyamotoi, and B. turicatae. These fragments were then cloned into the pCR 2.1 TA cloning vector, and sequencing revealed a 572-bp insert for each of the three species. The fragments corresponded to amino acid residues 86 to 275 of B. hermsii RecA and residues 88 to 278 of B. burgdorferi RecA.
Comparison of deduced RecA proteins of Borrelia spp.
The B. hermsii recA gene was 1,071 bp long, compared to the 1,095-bp recA gene of B. burgdorferi, and the nucleotide sequence was 78% identical to the B. burgdorferi recA sequence. The pairwise difference value for the nucleotide sequences of these two species of Borrelia is equal to the values obtained for different genera belonging to the gamma division of the class Proteobacteria. For example, the recA gene of E. coli K-12 (accession number NC_000913) is 79 and 77% identical to the orthologs in Yersinia pestis (NC_004835) and Shewanella oneidensis (NC_004347), respectively. The predicted 356-amino-acid protein was 78% identical to B. burgdorferi RecA, 54% identical to T. pallidum RecA, 51% identical to L. biflexa RecA, and 53% identical to E. coli RecA. Alignment of the partial RecA sequences of the five Borrelia spp. showed that the levels of pairwise amino acid identity between B. burgdorferi and the relapsing fever group of species were 89 to 91% and that the levels of pairwise identity between members of the relapsing fever group were 92 to 95%. The codon usages for the B. burgdorferi and B. hermsii recA genes were very similar, and like other Borrelia genes, these genes showed a bias toward A- and T-rich codons.
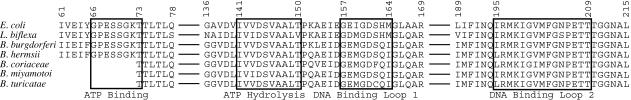
Figure 2 shows an alignment of conserved functional sites, as summarized by Lusetti and Cox (25), of the RecA sequences of E. coli (accession number NP_417179), L. biflexa, B. burgdorferi, B. hermsii, B. coriaceae, B. miyamotoi, and B. turicatae. The ATP-binding sites (residues 66 to 73 of E. coli RecA) of all the RecA proteins were identical, and the ATP hydrolysis sites (residues 141 to 150) were the same except for an I142V substitution in B. miyamotoi and L. biflexa RecA and a V150T substitution in L. biflexa RecA. The sequences of the primary DNA binding sites (residues 157 to 164) were more divergent; compared to the E. coli and L. biflexa RecA proteins, there were three substitutions, M159I, Q163H, and I164 M, in the Borrelia spp. RecA proteins. The secondary DNA binding sites (residues 195 to 209) of Borrelia spp. RecA proteins have polymorphisms at some positions, but these polymorphisms are conservative substitutions, such as an arginine at position 198 in B. burgdorferi in place of the lysine in other Borrelia spp. A less conservative substitution was the substitution of glutamine for lysine at position 152. This position is not in a defined functional site, but this junction between the ATP hydrolysis site and the primary DNA binding site is highly conserved among RecA proteins (19).
FIG. 2.
Alignment of partial amino acid sequences of the RecA proteins of E. coli, L. biflexa, B. burgdorferi, B. hermsii, B. coriaceae, B. miyamotoi, and B. turicatae. The boxes indicate the identified functional sites of the E. coli RecA protein (25). The numbering of residues is the numbering for the full-length E. coli RecA protein (accession number NC_000913).
UV survival assays.
The ability of the Borrelia spp. recA genes to complement a defective recombinational DNA repair pathway was investigated by using E. coli Top10F′ cells containing plasmids pBluescript, pBb:recA, pBh:recA, pEc:recA, and pLb:recA. The cells were grown in the presence of IPTG before exposure to various doses of short-wavelength UV light after plating on solid agar without IPTG. For all the cells containing recombinant RecA, the survival of cells without IPTG induction was 5- to 10-fold less than the survival of cells treated with IPTG before UV exposure.
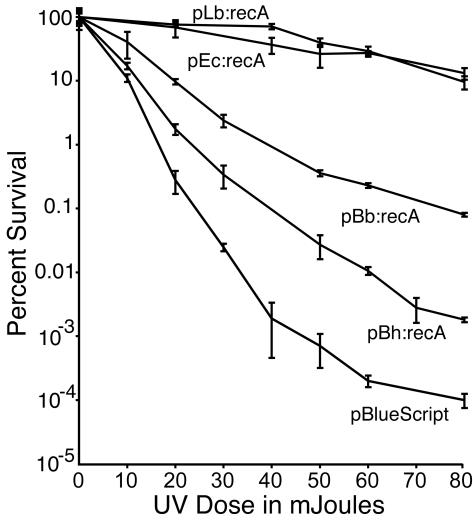
Figure 3 shows the mean levels of survival of the different cultures after different exposures. The survival of the recA mutant E. coli complemented by the L. biflexa recA gene was essentially the same as the survival of cells with E. coli recA provided on a plasmid. Although both B. burgdorferi recA and B. hermsii recA resulted in increased survival of the recA mutant host, the benefit was 103- to 104-fold less than the benefit provided by L. biflexa recA. The survival of E. coli cells with B. hermsii recA was 100-fold less than the survival of cells with B. burgdorferi recA and only 10- to 20-fold greater than the survival of the recA mutant host alone. The doses estimated to kill 99% of the cells were 17, 24, and 40 mJ for E. coli harboring plasmids pBluescript, pBh:recA, and pBb:recA, respectively.
FIG. 3.
Survival after exposure to UV light of recA mutant E. coli strain Top10F′ containing cloned recA genes. The doses of UV light (254 nm) delivered to the bacteria on plates are indicated. The number of colonies on the untreated control plate was defined as 100% survival, and the mean percentages of survival on treated plates were determined by comparison of colony counts with the control colony counts. The error bars indicate the 95% confidence interval at each dose based on three to six replicates. Data for cells with plasmids containing E. coli recA (pEc:recA), L. biflexa recA (pLb:recA), B. burgdorferi recA (pBb:recA), and B. hermsii recA (pBh:recA), as well as an empty vector control (pBluescript), are shown.
MMS and mitomycin C susceptibility assays.
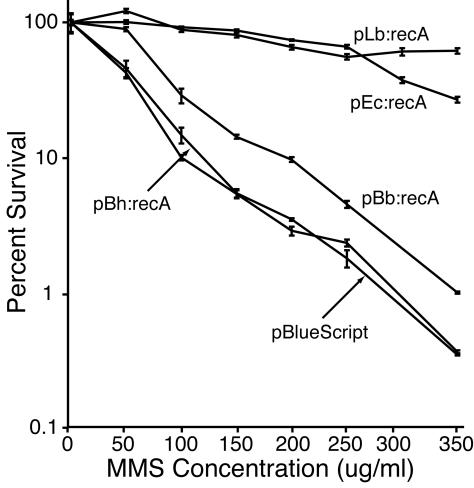
recA mutant cells are sensitive to the radiomimetic effects of MMS and mitomycin C (13). To determine if Borrelia spp. RecA proteins were capable of participation in the repair of DNA damage due to these chemicals, complementation studies with E. coli Top10F′ cells were preformed. IPTG-induced cells harboring one of the recA expression plasmids were grown in the presence of increasing doses of MMS or mitomycin C in the absence of IPTG. Figure 4 shows the survival and growth of colonies observed with different concentrations of MMS. In this experiment, the cells expressing L. biflexa RecA complemented the recA-deficient host as well as the E. coli recA-bearing plasmid. Even at the highest concentration (350 μg/ml), the plating efficiency for pLb:recA-bearing cells was 90% of that for cells plated on agar without MMS. The population of cells with the B. hermsii recA gene was indistinguishable from the null plasmid control population in terms of susceptibility to MMS. The estimated concentrations of MMS necessary to reduce the numbers of colonies by 99% were 291, 286, and 350 μg/ml for pBluescript, pBh:recA, and pBb:recA, respectively.
FIG. 4.
Survival of E. coli recA mutant strain Top10F′ containing cloned recA genes grown in the presence of different concentrations of MMS. The number of colonies on the plates without MMS was defined as 100% survival, and the mean percentages of survival and 95% confidence intervals were determined by using three to six replicates. Data for cells with plasmids containing E. coli recA (pEc:recA), L. biflexa recA (pLb:recA), B. burgdorferi recA (pBb:recA), and B. hermsii recA (pBh:recA), as well as an empty vector control (pBluescript), are shown.
With mitomycin C at concentrations less than 1.0 μg/ml there was no difference in survival and growth of the cells with the different plasmids. At a mitomycin C concentration of 1.0 μg/ml, the number of colonies of the recA mutant host strain with pBluescript was 0.05% of the number of colonies on plates without the agent, and the percentage of surviving colonies with the pLb:recA plasmid (0.31%) was more than 10-fold lower than the percentage of surviving colonies for the cells with E. coli recA (4.9%). The percentage of colonies with B. burgdorferi recA (0.29%) was approximately the same as the percentage of surviving colonies for the cells with pLb:recA, but no discernible increase in survival was provided by the B. hermsii recA plasmid.
To assess whether the observed differences between transgenic cells with B. burgdorferi recA and transgenic cells with B. hermsii recA in the DNA damage assays was attributable to pleiotropic effects of the plasmids, we measured the generation times of the different cell lines in the broth medium. The doubling times for cultures of Top10F′ cells harboring pBluescript, pBb:recA, pBh:recA, pLb:recA, and pEc:recA were 66, 63, 66, 49, and 51 min, respectively. The difference between the doubling times for cells with B. burdorferi recA and cells with B. hermsii recA was not statistically significant.
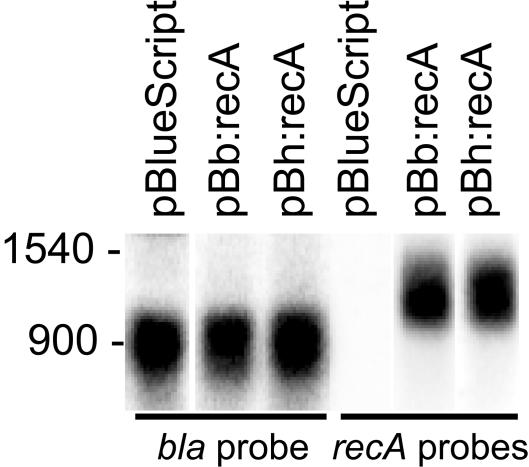
To determine if the lower level of complementation of the E. coli recA mutant by recombinant B. hermsii RecA was due to decreased expression of the gene in E. coli, a Northern blot analysis was performed with total RNA from E. coli Top10F′ cells containing plasmid pBb:recA, pBh:recA, or pBluescript. The blots were probed with either a PCR product of the plasmid's beta-lactamase gene, bla, or a 1:1 mixture of equivalently labeled PCR products of the recA genes of B. burgdorferi and B. hermsii. The results obtained with IPTG-induced cells are shown in Fig. 5. When we normalized for bla expression, there was not a detectable difference between transcription of the B. burgdorferi recA gene and transcription of the B. hermsii recA gene in E. coli. Similar results were obtained with uninduced E. coli cells (data not shown).
FIG. 5.
Northern blot analysis of transgenic E. coli Top10F′ cells containing plasmids pBluescript, pBb:recA, and pBh:recA. Total RNA was separated on a 1% agarose gel with formaldehyde. The blots were probed with radiolabeled PCR products of the beta-lactamase (bla) gene of the plasmid (left) or a mixture of the recA genes of B. burgdorferi and B. hermsii (right). The positions of single-strand size markers (in nucleotides) are indicated on the left.
red gam λ phage plaque production.
The results of the chemical susceptibility assays and the UV survival assay indicated that Borrelia RecA is capable of participating in the DNA repair pathway in E. coli but it participates at much lower levels than the RecA protein of L. biflexa. We next compared the activities of the different recombinant RecA proteins in recombination assays. We used red gam mutant λ phage, which in the absence of a suppressing recB or recC mutation lack the ability to form plaques on recA mutant E. coli strains (12). For the experiment, red gam mutant phage were adsorbed to E. coli JM109 cells harboring the different recA plasmids, and the efficiency of plaque production was observed.
Table 1 shows the efficiency of plaque production in cells complemented with each of the recombinant recA plasmids. With IPTG-induced cells containing the pEc:recA plasmid, an estimated 65% of the phage added were capable of forming plaques, while 25% of the phage incubated with cells containing plasmid pLb:recA formed plaques. No plaques were seen on plates of cells containing either pBb:recA, pBh:recA, or pBluescript, even when the number of phage adsorbed was increased 10-fold. This was an indication that multimers of the mutant phage genomes were not produced from monomer circles in the cells with Borrelia RecA.
TABLE 1.
Efficiencies of plating of red gam λ bacteriophage on RecA+ (JM105) and RecA− (JM109) E. coli cells containing expression plasmids bearing E. coli, L. biflexa, B. burgdorferi, and B. hermsii recA genes
| Strain/plasmid | recA gene | No. of PFU/platea | Mean no. of plaques (95% CI)b | % Compared to JM105 |
|---|---|---|---|---|
| JM105/none | E. coli | 500 | 514 (454-572) | 100 |
| JM109/pBluescript | None | 15,000 | 0 | <0.006 |
| JM109/pEc:recA | E. coli | 1,500 | 979 (866-1092) | 65 |
| JM109/pLb:recA | L. biflexa | 1,500 | 374 (331-415) | 25 |
| JM109/pBb:recA | B. burgdorferi | 15,000 | 0 | <0.006 |
| JM109/pBh:recA | B. hermsii | 15,000 | 0 | <0.006 |
Estimated number of PFU per plate.
95% CI, 95% confidence interval.
Homologous recombination assay.
The second recombination assay called for intramolecular homologous recombination between approximately 2.5-kb inverted repeats to produce a functional LacZ ORF in E. coli strain JC14604 (20). This strain contains a duplication of the lacZ region, and each copy possesses a different lacZ missense mutation. In the experiment the colonies growing on minimal agar plates with lactose as the carbon source were counted in triplicate after 7 days of incubation, and the frequency of recombinants was estimated by comparing the number of colonies on lactose plates with the number of colonies on glucose plates. Table 2 shows the results. Recombinants were undetectable on plates with lawns of the recA mutant cells with the vector plasmid alone. For cells bearing the pEc:recA plasmid the recombination frequency was approximately 25-fold higher than that for the cells bearing either pBb:recA or pBh:recA. In contrast to the DNA damage assays and phage recombination assay, this recombination assay demonstrated that the complementation provided by Borrelia spp. RecA was nearly as great as that provided by L. biflexa RecA.
TABLE 2.
Frequencies of lactose-fermenting (Lac+) recombinants of E. coli strain JC14604 cells containing expression plasmids bearing E. coli, L. biflexa, B. burgdorferi, and B. hermsii recA genes
| Plasmid | recA gene | Frequency of Lac+ colonies (10−8) (95% CI)a |
|---|---|---|
| pBluescript | None | <0.006 |
| pEc:recA | E. coli | 48 (42-55) |
| pLb:recA | L. biflexa | 5.9 (5.6-6.2) |
| pBb:recA | B. burgdorferi | 2.3 (2.1-2.5) |
| pBh:recA | B. hermsii | 2.6 (2.4-2.8) |
95% CI, 95% confidence interval.
Expression of recA in Borrelia species.
Expression of recA in B. burgdorferi and B. hermsii was examined next. The recA genes of B. hermsii and B. burgdorferi are preceded by σ70-type promoters. The putative −10 element was TATAAA in B. burgdorferi and B. hermsii. The sequences of the corresponding −35 elements were CCCCGA and TCCTGA in B. burgdorferi and B. hermsii, respectively. We first attempted to document the expression of recA in B. burgdorferi by Northern blot analysis using the flaB gene as a positive control, but the level of transcription was below the level of detection of the assay (Putteet-Driver, unpublished data). We then analyzed the raw data from DNA array experiments to document recA expression in B. burgdorferi and B. hermsii and to assess whether the level of expression was affected by changes in environmental conditions (55). In these experiments a membrane-based DNA array of the ORFs of the genome of B. burgdorferi was probed with cDNA produced from RNA of B. burgdorferi growing at 23 or 35°C or RNA of B. hermsii growing in culture medium or the blood of infected mice. The raw data were adjusted for differences between ORFs in terms of the amount of hybridization by genomic DNA of either B. burgdorferi or B. hermsii and for differences in nucleotide identity.
Table 3 shows the results of this analysis. The ospC gene of B. burgdorferi was used a positive control for a temperature-regulated gene (40), and the expression of the flaB gene of the two species was known not to vary at different temperatures or when in vitro cultivation conditions and in vivo cultivation conditions were compared (55). The recA genes of both B. burgdorferi and B. hermsii were expressed under all growth conditions examined but at a lower level than the flaB genes of the species. Unlike the ospC gene, which showed eightfold-higher transcription at the higher temperature, the expression of recA in B. burgdorferi was not different at the two temperatures; these results confirm those of Ojaimi et al. (32). There was no detectable difference between the level of expression of recA in B. hermsii under in vitro conditions and the level of expression of recA in B. hermsii under in vivo conditions.
TABLE 3.
B. burgdorferi genome array analysis of recA expression in B. burgdorferi cells under two temperature conditions and in B. hermsii cells growing in culture medium or in the blood of infected mice
| Organism | Gene | Growth conditions | Relative expression (95% CI)a | P valueb |
|---|---|---|---|---|
| B. burgdorferi | flaB | 23°C | 37 (29-45) | 0.2 |
| flaB | 35°C | 49 (31-67) | ||
| recA | 23°C | 5.2 (2.3-8.0) | 0.2 | |
| recA | 35°C | 6.8 (6.1-7.4) | ||
| ospC | 23°C | 6.5 (5.1-7.8) | <10−7 | |
| ospC | 35°C | 53 (49-58) | ||
| B. hermsii | flaB | Medium | 330 (13.8-31.0) | 0.1 |
| flaB | Mouse | 211 (9.4-19.3) | ||
| recA | Medium | 3.9 (0.18-0.40) | 0.2 | |
| recA | Mouse | 2.5 (0.16-0.21) |
95% CI, 95% confidence interval.
Determined by two-tailed t test.
DISCUSSION
RecA was first identified in 1965 by Clark and Margulies through characterization of E. coli cells deficient in recombination (8). Soon after this, these and other investigators demonstrated that recA mutant cells were highly sensitive to UV irradiation, thus revealing that there is an association between the recombination and DNA repair processes (7). Subsequent studies over the last three decades, almost entirely with E. coli, have resulted in further characterization of RecA and its three major activities (reviewed in references 9 and 25). First, the RecA protein directly participates in recombination catalyzing DNA strand exchange reactions. Recombination occurs in conjunction with either of the two other recombination pathways: RecBCD, which mainly participates in repair of double strand breaks; and RecFOR, which mainly participates in repair of nicks or other damage present at stalled replication forks (25). Second, RecA promotes the autocatalytic cleavage of repressor proteins, such as LexA of the SOS response in E. coli and cI of bacteriophage λ. Third, RecA in conjunction with polymerase V allows mutagenic translesion synthesis of DNA after DNA damage.
With the exception of the genome of the endosymbiont Buchnera aphidicola (47), a recA ortholog has been identified in all bacterial genomes sequenced to date (http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=Genome), and the presence of a recA ortholog has been demonstrated in several other bacterial species belonging to diverse taxa. RecA function in spirochetes was first identified in the free-living spirochete L. biflexa by Stamm et al. through functional complementation of a recA mutant of E. coli (43). In that study, immunoprecipitation of cellular extracts of B. burgdorferi and T. pallidum with rabbit antiserum to E. coli RecA failed to detect RecA protein in these spirochetes. A recA gene of B. burgdorferi was later found by Dew-Jager et al., who showed that the deduced sequence of B. burgdorferi's RecA protein had several substitutions, which, when individually present in E. coli RecA, altered or eliminated the function in that organism (11).
The sequence of the genome of B. burgdorferi confirmed the presence of a single ortholog of recA on the chromosome, as well as the presence of genes of the RecBCD pathway for double-strand break repair (15). These findings revealed the possible enzymatic basis for such recombination-mediated phenomena in B. burgdorferi as intramolecular gene conversions between multiple alleles of the vlsE sequences (54). On the other hand, the genome sequence did not reveal orthologs for the following genes: lexA, the gene for the SOS response repressor; umuC and umuD, the genes encoding the subunits of DNA polymerase V; and the recF, recO, or recR genes of the RecFOR pathway for gap repair (15). The DNA repair and recombination capabilities of B. hermsii are expected to be similar to those of B. burgdorferi since the genome sizes (14) and genetic complements of these organisms are similar (55). In the present study we found that the same genes that flank recA in B. burgdorferi also flank recA in B. hermsii.
To further define the functional capabilities of the RecA protein of Borrelia spp., we attempted to introduce gene disruptions within recA of B. burgdorferi, but we were unable to recover clones that contained the integrated plasmid (Putteet-Driver, unpublished data). This finding was the consequence of the lethality of such an event, as observed in Streptomyces lividans (51), or was obtained because the growth rate was greatly reduced, as occurred in the more robust organism L. biflexa (49). As an alternative approach, we characterized the DNA repair and recombination capabilities of recombinant RecA of B. burgdorferi and B. hermsii in recA mutant E. coli cells and compared these cells with cells expressing E. coli RecA or L. biflexa RecA from the same plasmid constructs under control of the Lac promoter. In addition, we identified, sequenced, and cloned into the same plasmid vector the recA gene of B. hermsii. The different plasmid constructs were compared with each other, as well as with recA mutant cells in which the E. coli RecA protein was expressed from the same plasmid vector. We demonstrated that recA was detectably transcribed in both B. burgdorferi and B. hermsii, but there was not a discernible difference in transcription at two different temperatures or between growth in broth medium and growth in mouse blood.
The recA genes of both B. hermsii and B. burgdorferi enhanced the ability of the host recA mutant E. coli cells to recover from UV irradiation and to carry out intramolecular recombination between inverted repeats to utilize the sole carbohydrate source in the medium. However, neither of these recA genes could match the transgenic recA of another spirochete, L. biflexa, or the recA gene of E. coli in either of these assays or in the promotion of plaque formation by red gam mutant λ bacteriophage. Overall, the RecA proteins of B. burgdorferi and B. hermsii appeared to be more competent to complement a recA mutant in an assay for intramolecular homologous recombination than in DNA repair assays.
In addition, we found significant differences between cells bearing B. hermsii recA and cells with B. burgdorferi recA in the DNA damage assays despite similar expression levels. While transgenic recA genes of both Borrelia spp. had equivalent activities in the assay of recombination between lacZ duplications, the transgenic cells with the B. hermsii recA gene exhibited only a 10-fold increase in survival after UV exposure compared to the survival of RecA− cells and no detectable increase in survival in the presence of the radiometic chemicals MMS and mitomycin C. The recA gene of B. burgdorferi was less effective than the recA gene of free-living L. biflexa in these assays but was still substantially more effective than the ortholog of B. hermsii.
Our findings for the expression of recombinant RecA of B. burgdorferi in E. coli were generally similar to those of Liveris et al. (24). In both studies Borrelia RecA proteins mediated homologous recombination more efficiently than they mediated recombinational DNA repair in E. coli. The P1 transduction assay of Liveris et al. demonstrated that B. burgdorferi RecA is able to promote intermolecular recombination in E. coli, while our studies of recombination between lacZ duplications demonstrated that intramolecular recombination is also promoted by Borrelia spp. RecA in E. coli. In both studies, the increase in survival compared with the survival of the vector control was 103- to 104-fold at the highest dose of UV irradiation. While we observed that recombinant B. burgdorferi RecA conferred a modest increase in mitomycin C resistance compared with that of the vector control, Liveris et al. observed complete or nearly complete complementation of a recA mutant E. coli strain in their mitomycin C assay. The difference may be attributable to the continuing presence of mitomycin C during cultivation in our assay and the postexposure removal of mitomycin C by Liveris et al. before cultivation. Liveris et al. also observed that complementation with B. burgdorferi recA leads to induction of λ prophage and provided a measure of RecA's coprotease activity; cleavage of the cI repressor protein permitted excision of the phage genome and subsequent entry into the lytic cycle.
The comparatively poorer complementation by Borrelia spp. recA than by L. biflexa recA in DNA damage repair assays could be attributable in part to lower expression of the borrelial RecA proteins in E. coli, but we do not think that this was a major determinant of the results. For one thing, all the recA genes were provided the same promoter at the same nucleotide distance from the start codon in the plasmid constructs, and we did not observe a difference in plasmid copy numbers between the cell lines (unpublished data). The Borrelia spp. recA genes have different codon usages than E. coli recA has, but even when the codon usages were the same, as they were for B. hermsii and B. burgdorferi, we observed a significant difference in activity between the recA genes of different species in some assays. Moreover, L. biflexa is as evolutionarily distant as either Borrelia species from E. coli (10, 34), yet its recA was demonstrably more effective in the E. coli background. Finally, the recA genes of all three spirochete species functioned about the same in the homologous recombination assay. Thus, we concluded that B. burgdorferi RecA and especially B. hermsii RecA have relatively effective abilities to carry out homologous recombination but not DNA damage repair when they are expressed in a distantly related host species.
A discordance between homologous recombination and DNA repair was also observed with the recA gene of another obligate pathogen, N. gonorrhoeae, a member of the beta subdivision of the Proteobacteria (44). Stohl et al. reported that after UV irradiation E. coli cells expressing N. gonorrhoeae RecA had 103- to 104-fold lower levels of survival than cells expressing E. coli RecA had. In contrast, assays of homologous recombination with P1 transduction and Hfr conjugation showed that the activity of transgenic cells with N. gonorrhoeae recA was the same as that of cells with E. coli recA. Stohl et al. concluded that RecA of N. gonorrhoeae was able to interact with the RecBCD protein of E. coli as efficiently as E. coli RecA was. The findings of our lacZ duplication assay and the P1 transduction assay of Liveris et al. for homologous recombination are consistent with this interpretation (24).
However, the failure of the red gam mutant λ to produce plaques on cells expressing recombinant Borrelia spp. RecA and the results of the recombination assays with strain JC14604 suggest that the lack of plaque production by these phage is dependent upon more than the restoration of recombination and the interaction between RecA and RecBCD. Although the mechanism for the lack of plaque formation by red gam mutant λ in RecA-deficient E. coli remains to be established, one hypothesis is that recA-deficient cells cannot provide sufficient recombination for the production of multimers of the phage genome needed for packaging and successful multiplication of the phage (41). The finding that recBC mutations suppress recA mutations in assays of plaque formation by red gam mutant λ indicates that transduction and plaque assays assess different aspects of the interaction between RecA and RecBCD (52).
Stohl et al. attributed the poor complementation of the recA mutant in DNA damage assays to a failure of the RecA protein from N. gonorrhoeae to induce the SOS response as efficiently as E. coli RecA induces this response (44). B. burgdorferi does not appear to have an ortholog of the LexA repressor, and it is possible that Borrelia RecA has coprotease activity for cI, as demonstrated by Liveris et al. (24), but does not have activity for LexA. An alternative or supplementary explanation is that there is alteration of loading of Borrelia or Neisseria RecA by RecFOR or another interaction between RecA and the RecFOR pathway in E. coli. B. burgdorferi does not have detectable orthologs of recF, recO, or recR (15). N. gonorrhoeae appears to have orthologs of recO and recR but not of recF (accession number NC_002946). It is possible that the RecA proteins of Borrelia and Neisseria cannot function in the key interaction between RecA and RecF in the SOS response in E. coli (35); consequently, these proteins would be expected to ineffectively complement a recA mutation in assays that mainly assess DNA damage repair.
We do not know why the RecA protein of B. hermsii is less efficacious than B. burgdorferi RecA in DNA damage repair in the E. coli genetic background. One possible explanation is the K152Q substitution found in the RecA protein of B. hermsii and three other relapsing fever species (Fig. 2). Among all other RecA sequences in the accessible databases, only the sequences of the RecA proteins of T. pallidum (accession number AAC65660) and Chlorobium tepidum (AAM73149) have a substitution at this position. This residue is located between the ATP hydrolysis site and a DNA binding site of E. coli RecA, and this region is involved in monomer-monomer interactions (25, 45, 46). Although an E. coli recA mutant with a K152Q substitution exhibited full activity in both DNA repair and coprotease assays (30), it is possible that this substitution has more adverse effects in conjunction with other substitutions in the protein.
Acknowledgments
We thank Chris Crowder for his contributions in preparing the manuscript, Thuy Tran Hoang for assistance with experiments, Masahito Fukunaga for providing B. miyamotoi DNA, Patricia Rosa for providing total RNA from B. burgdorferi, Dionysios Liveris and Ira Schwartz for sharing results before publication, and Richard Zuerner for providing L. biflexa DNA.
This work was supported by NIH grant AI24424.
REFERENCES
- 1.Barbour, A. G. 2003. Antigenic variation in Borrelia: relapsing fever and Lyme borreliosis, p. 319-352. In Antigenic variation. Elsevier Science Ltd., New York, N.Y.
- 2.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour, A. G., and S. F. Hayes. 1986. Biology of Borrelia species. Microbiol. Rev. 50:381-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cadavid, D., D. D Thomas, R. Crawley, and A. G. Barbour. 1994. Variability of a bacterial surface protein and disease expression in a possible mouse model of systemic Lyme borreliosis. J. Exp. Med. 179:631-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 7.Clark, A. J., and M. Chamberlin. 1966. Abnormal metabolic response to ultraviolet light of a recombination deficient mutant of Escherichia coli K12. J. Mol. Biol. 19:442-454. [DOI] [PubMed] [Google Scholar]
- 8.Clark, A. J., and A. D. Margulies. 1965. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 53:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, M. M. 2001. Historical overview: searching for replication help in all the rec places. Proc. Natl. Acad. Sci. 98:8173-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubin, V., M. Gouy, and G. Perriere. 2002. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 12:1080-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dew-Jager, K., W. Q. Yu, and W. M. Huang. 1995. The recA gene of Borrelia burgdorferi. Gene 167:137-140. [DOI] [PubMed] [Google Scholar]
- 12.Enquist, L. W., and A. Skalka. 1973. Replication of bacteriophage lambda DNA dependent on the function of host and viral genes. I. Interaction of red, gam and rec. J. Mol. Biol. 75:185-212. [DOI] [PubMed] [Google Scholar]
- 13.Favre, D., S. J. Cryz, and J. F. Viret. 1991. Cloning of the recA gene of Bordetella pertussis and characterization of its product. Biochimie 73:235-244. [DOI] [PubMed] [Google Scholar]
- 14.Ferdows, M. S., P. Serwer, G. A. Griess, S. J. Norris, and A. G. Barbour. 1996. Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J. Bacteriol. 178:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. C. Venter, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 16.Frederick, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Preparation and analysis of RNA, p. 4.9.1-4.9.7. In F. M. Frederick, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley and Sons, New York, N.Y. [Google Scholar]
- 17.Fukunaga, M., Y. Takahashi, Y. Tsuruta, O. Matsushita, D. Ralph, M. McClelland, and M. Nakao. 1995. Genetic and phenotypic analysis of Borrelia miyamotoi sp. nov., isolated from the ixodid tick Ixodes persulcatus, the vector for Lyme disease in Japan. Int. J. Syst. Bacteriol. 45:804-810. [DOI] [PubMed] [Google Scholar]
- 18.Jaworski, D. C., A. Jasinskas, C. N. Metz, R. Bucala, and A. G. Barbour. 2001. Identification and characterization of a homologue of the pro-inflammatory cytokine macrophage migration inhibitory factor in the tick, Amblyomma americanum. Insect Mol. Biol. 10:323-331. [DOI] [PubMed] [Google Scholar]
- 19.Karlin, S., and L. Brocchieri. 1996. Evolutionary conservation of RecA genes in relation to protein structure and function. J. Bacteriol. 178:1881-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keener, S. L., K. P. McNamee, and K. McEntee. 1984. Cloning and characterization of recA genes from Proteus vulgaris, Erwinia carotovora, Shigella flexneri, and Escherichia coli B/r. J. Bacteriol. 160:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koomey, J. M., and S. Falkow. 1987. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J. Bacteriol. 169:790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koomey, M., E. C. Gotschlich, K. Robbins, S. Bergstrom, and J. Swanson. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane, R. S., W. Burgdorfer, S. F. Hayes, and A. G. Barbour. 1985. Isolation of a spirochete from the soft tick, Ornithodoros coriaceus; a possible agent of epizootic bovine abortion. Science 230:85-87. [DOI] [PubMed] [Google Scholar]
- 24.Liveris, D., V. Mulay, and I. Schwartz. 2004. Functional properties of Borrelia burgdorferi recA. J. Bacteriol. 186:2275-2280. [DOI] [PMC free article] [PubMed]
- 25.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71:71-100. [DOI] [PubMed] [Google Scholar]
- 26.Meier, J. T., M. I. Simon, and A. G. Barbour. 1985. Antigenic variation is associated with DNA rearrangements in a relapsing fever Borrelia. Cell 41:403-409. [DOI] [PubMed] [Google Scholar]
- 27.Mintz, K. P., C. Brissette, and P. M. Fives-Taylor. 2002. A recombinase A-deficient strain of Actinobacillus actinomycetemcomitans constructed by insertional mutagenesis using a mobilizable plasmid. FEMS Microbiol. Lett. 206:87-92. [DOI] [PubMed] [Google Scholar]
- 28.Mongkolsuk, S., S. Rabibhadana, R. Sukchavalit, and G. Vaughn. 1998. Construction and physiological analysis of a Xanthomonas oryzae pv. oryzae recA mutant. FEMS Microbiol. Lett. 169:269-275. [DOI] [PubMed] [Google Scholar]
- 29.Muth, G., D. Frese, A. Kleber, and W. Wohlleben. 1997. Mutational analysis of the Streptomyces lividans recA gene suggests that only mutants with residual activity remain viable. Mol. Gen. Genet. 255:420-428. [DOI] [PubMed] [Google Scholar]
- 30.Nastri, H. G., and K. L. Knight. 1994. Identification of residues in the L1 region of the RecA protein which are important to recombination or coprotease activities. J. Biol. Chem. 269:26311-26322. [PubMed] [Google Scholar]
- 31.Ojaimi, C., C. Brooks, D. Akins, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katonah, J. Radolf, M. Caimano, J. Skare, K. Swingle, S. Sims, and I. Schwartz. 2002. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 358:165-177. [DOI] [PubMed] [Google Scholar]
- 32.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, E. M., M. K. Shigenaga, P. Degan, T. S. Korn, J. W. Kitzler, C. M. Wehr, P. Kolachana, and B. N. Ames. 1992. Assay of excised oxidative DNA lesions: isolation of 8-oxoguanine and its nucleoside derivatives from biological fluids with a monoclonal antibody column. Proc. Natl. Acad. Sci. USA 89:3375-3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paster, B. J., and F. E. Dewhirst. 2000. Phylogenetic foundation of spirochetes. J. Mol. Microbiol. Biotechnol. 2:341-344. [PubMed] [Google Scholar]
- 35.Rangarajan, S., R. Woodgate, and M. F. Goodman. 2002. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA and RecFOR proteins. Mol. Microbiol. 43:617-628. [DOI] [PubMed] [Google Scholar]
- 36.Restrepo, B. I., T. Kitten, C. J. Carter, D. Infante, and A. G. Barbour. 1992. Subtelomeric expression regions of Borrelia hermsii linear plasmids are highly polymorphic. Mol. Microbiol. 6:3299-3311. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Analysis and cloning of eukaryotic genomic DNA, p. 9.52-9.55. In N. Ford (ed.), Molecular cloning: a laboratory manual, 2nd ed., vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Bacteriaphage lambda vectors, p. 2.60-2.61. In N. Ford (ed.), Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Plasmid vectors, p. 1.98. In N. Ford (ed.), Molecular cloning: a laboratory manual, 2nd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 40.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, G. R. 1983. General recombination, p. 175-209. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 42.Stamm, L. V., and N. W. Charon. 1988. Sensitivity of pathogenic and free-living Leptospira spp. to UV radiation and mitomycin C. Appl. Environ. Microbiol. 54:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamm, L. V., E. A. Parrish, and F. C. Gherardini. 1991. Cloning of the recA gene from a free-living leptospire and distribution of RecA-like protein among spirochetes. Appl. Environ. Mircobiol. 57:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stohl, E. A., L. Blount, and H. Seifert. 2002. Differential cross-complementation patterns of Escherichia coli and Neisseria gonorrhoeae RecA proteins. Microbiology 148:1821-1831. [DOI] [PubMed] [Google Scholar]
- 45.Story, R. M., and T. A. Steitz. 1992. Structure of the recA protein-ADP complex. Nature 355:374-376. [DOI] [PubMed] [Google Scholar]
- 46.Story, R. M., I. T. Weber, and T. A. Steitz. 1992. The structure of the E. coli recA protein monomer and polymer. Nature 355:318-325. [DOI] [PubMed] [Google Scholar]
- 47.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 48.Tatum, F. M., D. C. Morfitt, and S. M. Halling. 1993. Construction of a Brucella abortus RecA mutant and its survival in mice. Microb. Pathog. 14:177-185. [DOI] [PubMed] [Google Scholar]
- 49.Tchamedeu Kameni, A. P., E. Couture-Tosi, I. Saint-Girons, and M. Picardeau. 2002. Inactivation of the spirochete recA gene results in a mutant with low viability and irregular nucleoid morphology. J. Bacteriol. 184:452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 51.Vierling, S., T. Weber, W. Wohlleben, and G. Muth. 2001. Evidence that an additional mutation is required to tolerate insertional inactivation of the Streptomyces lividans recA gene. J. Bacteriol. 183:4374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinstock, G. M. 1987. General recombination in Escherichia coli, p. 1034-1043. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. L. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 53.Whitehouse, C. A., L. R. Williams, and F. E. Austin. 1997. Identification of superoxide dismutase activity in Borrelia burgdorferi. Infect. Immun. 65:4865-4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 55.Zhong, J., and A. G. Barbour. 2004. Cross-species hybridization of a Borrelia burgdorferi DNA array reveals infection- and culture-associated genes of the unsequenced genome of the relapsing fever agent Borrelia hermsii. Mol. Microbiol. 51:729-748. [DOI] [PubMed] [Google Scholar]