Abstract
Ticks are one of the most important blood-sucking vectors for infectious microorganisms in humans and animals. When feeding they inject saliva, containing microbes, into the host to facilitate the uptake of blood. An understanding of the microbial populations within their salivary glands would provide a valuable insight when evaluating the vectorial capacity of ticks. Three tick species (Ixodes ovatus, I. persulcatus and Haemaphysalis flava) were collected in Shizuoka Prefecture of Japan between 2008 and 2011. Each tick was dissected and the salivary glands removed. Bacterial communities in each salivary gland were characterized by 16S amplicon pyrosequencing using a 454 GS-Junior Next Generation Sequencer. The Ribosomal Database Project (RDP) Classifier was used to classify sequence reads at the genus level. The composition of the microbial populations of each tick species were assessed by principal component analysis (PCA) using the Metagenomics RAST (MG-RAST) metagenomic analysis tool. Rickettsia-specific PCR was used for the characterization of rickettsial species. Almost full length of 16S rDNA was amplified in order to characterize unclassified bacterial sequences obtained in I. persulcatus female samples. The numbers of bacterial genera identified for the tick species were 71 (I. ovatus), 127 (I. persulcatus) and 59 (H. flava). Eighteen bacterial genera were commonly detected in all tick species. The predominant bacterial genus observed in all tick species was Coxiella. Spiroplasma was detected in Ixodes, and not in H. flava. PCA revealed that microbial populations in tick salivary glands were different between tick species, indicating that host specificities may play an important role in determining the microbial complement. Four female I. persulcatus samples contained a high abundance of several sequences belonging to Alphaproteobacteria symbionts. This study revealed the microbial populations within the salivary glands of three species of ticks, and the results will contribute to the knowledge and prediction of emerging tick-borne diseases.
Introduction
Ticks (Acari: Ixodida) are globally one of the most important arthropod vectors of infectious diseases in animals and humans. They carry and transmit a range of bacterial, protozoan and viral pathogens, which are often zoonotic [1], [2], [3]. Opportunities for ticks to come into contact with humans and animals are increasing as their habitats change and their distribution widens. The incidence of tick-borne cases, including those emerging diseases, is on the rise [3], [4]. Recent studies have identified microorganisms, such as Leptospira and Chlamydia, which had previously not been associated with ticks [5], [6].
Ixodid ticks have four stages to their life cycle (egg, larva, nymph, and adult), and each post-embryotic phase requires blood to grow and molt. During feeding, ticks inject saliva into host animals to facilitate the uptake of blood. Tick saliva contains bioactive components that inhibit blood coagulation and the host immune system, and it also assists individuals to make a strong attachment to the skin of the host [7], [8]. Tick-borne pathogens, such as Rickettsia, Ehrlichia and Anaplasma, concentrate within the salivary glands, and are transferred into the host animal during feeding on blood [9], [10], [11], [12]. Electron microscopy has revealed the presence of non-pathogenic symbionts, such as Coxiella-like bacteria, in the salivary glands of ticks [13]. Symbionts have also been identified by molecular methods; however, the functions of these bacteria within tick salivary glands remain unclear [14], [15], [16], [17], [18]. Thus, the investigation of the microbial populations in the tick salivary glands may lead to a better understanding of the microbes that could be transmitted to the mammalian hosts together with tick saliva or that could play roles in tick biology such as by facilitating blood feeding.
High throughput sequencing has provided an insight to the diversity of microbes associated with ticks [5], [19], [20], [21]. The analysis of 16S ribosomal DNA (16S rDNA) amplicons by pyrosequencing is a high throughput technique that can be used to detect non-culturable microbes, and reveal entire populations of tick-borne microbes. The analysis of microbial populations found within tick salivary glands can provide important information to help predict emerging pathogens, assess potential risks, and understand the interactions between tick symbionts and pathogens.
In this study, tick samples were collected in the Shizuoka Prefecture, Japan, where Japanese spotted fever is endemic [22]. The aim of this work was to elucidate the microbial populations (including pathogens) associated with salivary glands of ticks using 16S rDNA amplicon pyrosequencing technology.
Materials and Methods
Sample collection and DNA preparation
Adult host-questing ticks of the species Ixodes ovatus, I. persulcatus, and Haemaphysalis flava were collected by flagging in mountainous areas of Shizuoka Prefecture from 2008 to 2010. No specific permissions were required for these locations and activities. Our field activities did not involve endangered or protected species. Table S1 indicates information about the sampling sites. The sample numbers of I. ovatus, I. persulcatus, and H. flava used for this study were 24 (14 female, 10 male), 12 (6 female, 6 male), and 5 (female only), respectively. Each tick was split into two parts (anterior and posterior) at the area between coxa 1 and coxa 2 using a microtome. The anterior part was then removed from the posterior part using sterile forceps. Since the salivary glands were attached with the anterior part, they could be removed from the tick carcass without damaging the midgut. The salivary glands were then collected into a sterile 1.5 ml tube using sterile forceps, followed by washing with sterile PBS in order to minimize bacterial contamination. All dissection steps were performed under a stereomicroscope with great care to avoid the contamination of the midgut fluid. Genomic DNA was individually extracted using QIAamp DNA Mini kit (QIAGEN, Tokyo, Japan) according to the manufacturer’s instructions, and stored at −20°C. Samples of I. ovatus female and male, I. persulcatus female and male, and H. flava female are indicated by IOf, IOm, IPf, IPm, and HFf, respectively, throughout (Table 1).
Table 1. Sequence results and number of detected genera.
| Sample ID | Tick species | Sex | No. of sequence reads | No. of genera |
| IOf1 | I. ovatus | female | 5,664 | 10 |
| IOf2 | I. ovatus | female | 4,484 | 10 |
| IOf3 | I. ovatus | female | 3,498 | 6 |
| IOf4 | I. ovatus | female | 4,712 | 7 |
| IOf5 | I. ovatus | female | 5,591 | 3 |
| IOf6 | I. ovatus | female | 4,200 | 5 |
| IOf7 | I. ovatus | female | 5,030 | 6 |
| IOf8 | I. ovatus | female | 5,634 | 23 |
| IOf9 | I. ovatus | female | 7,643 | 8 |
| IOf10 | I. ovatus | female | 5,636 | 7 |
| IOf11 | I. ovatus | female | 4,049 | 2 |
| IOf12 | I. ovatus | female | 7,275 | 14 |
| IOf13 | I. ovatus | female | 3,351 | 3 |
| IOf14 | I. ovatus | female | 7,048 | 7 |
| IOm1 | I. ovatus | male | 4,986 | 13 |
| IOm2 | I. ovatus | male | 3,790 | 12 |
| IOm3 | I. ovatus | male | 7,916 | 22 |
| IOm4 | I. ovatus | male | 3,844 | 5 |
| IOm5 | I. ovatus | male | 6,340 | 18 |
| IOm6 | I. ovatus | male | 7,130 | 22 |
| IOm7 | I. ovatus | male | 6,176 | 22 |
| IOm8 | I. ovatus | male | 9,788 | 28 |
| IOm9 | I. ovatus | male | 9,628 | 12 |
| IOm10 | I. ovatus | male | 7,170 | 17 |
| IPf1 | I. persulcatus | female | 8,964 | 38 |
| IPf2 | I. persulcatus | female | 3,599 | 16 |
| IPf3 | I. persulcatus | female | 7,085 | 42 |
| IPf4 | I. persulcatus | female | 8,242 | 40 |
| IPf5 | I. persulcatus | female | 7,943 | 25 |
| IPf6 | I. persulcatus | female | 10,506 | 18 |
| IPm1 | I. persulcatus | male | 7,414 | 55 |
| IPm2 | I. persulcatus | male | 8,173 | 19 |
| IPm3 | I. persulcatus | male | 10,803 | 26 |
| IPm4 | I. persulcatus | male | 10,144 | 34 |
| IPm5 | I. persulcatus | male | 16,117 | 29 |
| IPm6 | I. persulcatus | male | 9,221 | 40 |
| HFf1 | H. flava | female | 6,438 | 3 |
| HFf2 | H. flava | female | 8,339 | 44 |
| HFf3 | H. flava | female | 6,204 | 5 |
| HFf4 | H. flava | female | 10,017 | 18 |
| HFf5 | H. flava | female | 8,294 | 24 |
PCR amplification of V1 to V3 regions for 16S rDNA amplicon libraries
The V1, V2, and V3 hyper-variable regions of bacterial 16S rDNA were amplified by PCR using the universal primers 27F (5′-X-AGAGTTTGATCMTGGCTCAG-3′) and 518R (5′-ATTACCGCGGCTGCTGG-3′), corresponding to positions 27 to 518 of the Escherichia coli 16S rDNA [23], [24]. The 27F primer contained ten bases of a multiplex identifier (MID) tag sequence denoted as ‘X’. Primers 27F and 518R were modified with 5′-adapter A and 5′-adapter B sequences, respectively, for pyrosequencing (Roche, Basel, Switzerland). PCR was performed in a total volume of 50 µl, containing PCR Buffer, 0.2 µl of Platinum Taq DNA polymerase (Life technologies, Tokyo, Japan), 0.2 mM of each primer, 1 µl of 10 mM dNTP mixture, 1.5 µl of 50 mM MgCl2, and 1 µl of template DNA. The PCR reaction was preceded with a 2 min denaturation step at 94°C, followed by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min. The quality of the PCR products (about 500 bp in length) were assessed by agarose (1%) gel electrophoresis, followed by purification using the Wizard SV Gel and PCR Clean-Up System (Promega, Tokyo, Japan). Concentration and quality of the amplicons were assessed with an Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, USA) using a DNA 1000 lab chip (Agilent Technologies).
Pyrosequencing and data analysis
Amplicon libraries with different MID tags were mixed and subjected to pyrosequencing using a 454 Genome Sequencer Junior (GS-Junior; Roche) following the manufacturer’s protocol. The pyrosequencing data were deposited in DDBJ with accession no. DRA001731. The resulting data files (standard flowgram format, .sff files) were converted to FASTA files and sorted according to sample-specific MID tags using CLC Genomics Work Bench (CLC Bio, Tokyo, Japan). MID tag barcodes and primers were trimmed, then low-quality and short sequence reads (<150 bp) were removed. DECIPHER’s Find Chimeras web tool (http://decipher.cee.wisc.edu/FindChimeras.html) was used to remove chimeric sequences. The remaining reads were phylogenetically classified with the assistance of the Ribosomal Database Project (RDP) 16S Classifier version 10 (http://rdp.cme.msu.edu/index.jsp), which can accurately and rapidly provide assignments for domains to the genus level. A comparative analysis of each sample was performed using the MG-RAST metagenomics analysis server employing the RDP dataset (http://metagenomics.anl.gov/). Alpha diversity of each sample was also calculated using the MG-RAST server. Data sets were represented as the mean ± standard deviations (S.D.) after the Smirnov-Grubbs outlier test (α = 0.05).
Conventional PCR methods
Rickettsia-specific PCR amplification of the citrate synthase gene (gltA) using the primers RpCS877p (5′-GGGGGCCTGCTCACGGCGG-3′) and RpCS1273r (5′-CATAACCAGTGTAAAGCTG-3′) [25] was performed on 22 samples that were highlighted by RPD analysis as containing the genus Rickettsia. PCR was performed in a total volume of 25 µl containing PCR Buffer for KOD-Plus-Neo, 0.5 µl of KOD-Plus-Neo DNA polymerase (Toyobo, Tokyo, Japan), 0.3 mM of each primer, 2.5 µl of 2 mM dNTP mixture, 1.5 µl of 25 mM MgSO4, and 1 µl of template DNA. The reaction started with a denaturation step at 94°C for 2 min, followed by 40 cycles of 94°C for 15 sec, 54°C for 30 sec, and 68°C for 30 sec, and a final extension step at 68°C for 2 min. PCR products were purified using ExoSap-IT (Affymetrix, Tokyo, Japan) according to the manufacturer’s instructions. Cycle sequencing was performed using BigDye v3.1 terminator chemistry (Applied Biosystems, Tokyo, Japan) and the same primers. Sequencing products were assessed using a 3130xl Genetic Analyzer (Life Technologies, Tokyo, Japan).
RDP analysis did not classify all sequences to the genus level. To characterize these sequences, the near full-length 16S rDNA gene was obtained from four I. persulcatus female samples by PCR using the universal primers fD1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and Rp2 (5′-ACGGCTACCTTGTTACGACTT-3′) [26]. PCR was carried out in a final volume of 50 µl PCR Buffer for KOD-Plus-Neo, 1 µl of KOD-Plus-Neo DNA polymerase, 0.3 mM of each primer, 5 µl of 2 mM dNTP mixture, 3 µl of 25 mM MgSO4, and 2 µl of template DNA. PCR conditions started with a denaturation step at 94°C for 2 min, followed by 40 cycles of 94°C for 15 sec, 55°C for 30 sec, and 68°C for 45 sec, and a final extension step at 68°C for 2 min. Quality of the PCR products (approx. 1400 bp) was assessed by agarose (1%) gel electrophoresis, followed by purification of the products using the Wizard SV Gel and PCR Clean-Up System (Promega). PCR products were A-tailed and then cloned with TA cloning plasmids pGEM-T Easy (Promega). Ten clones per sample were randomly selected and sequenced.
Sanger sequencing data analysis
Sanger sequencing data were analyzed using GENETYX version 9.1 (GENETYX Corporation, Tokyo, Japan). The GenBank accession numbers for the gltA sequences are AB911107 to AB911109, and the 16S rDNA sequences AB906824 to AB906829. Sequences were compared with those in public databases using nucleotide BLAST at NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Phylogenetic analysis was conducted using MEGA version 6.05 [27]. The universal 16S rDNA sequences were aligned with those of closely related bacteria in GenBank using ClustalW and a maximum likelihood phylogram was constructed.
Results
Classification and quantification of bacterial taxa
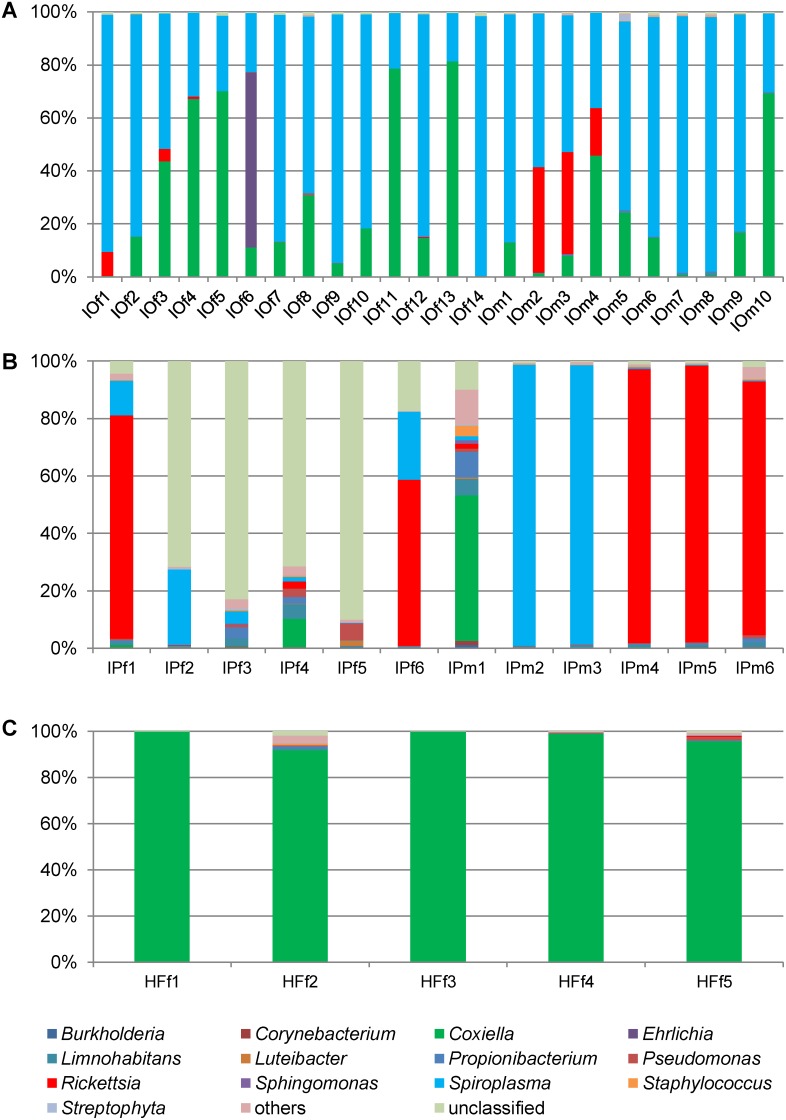
Between 3,351 and 9,788 sequences were obtained for individual I. ovatus, of which almost 98% were assigned to the genus level (Table 1) (Figure 1A). A total of 71 bacterial genera were detected in I. ovatus, with 59 found in males and 37 in females. The two dominant bacterial genera were Spiroplasma and Coxiella, and these accounted for more than 90% of the bacterial community in ticks, except for a single I. ovatus female and 3 I. ovatus males (Figure 1A). Rickettsia (genus contains known tick-borne pathogens R. japonica and R. helvetica) was detected in ten samples, and Ehrlichia (genus contains known tick-borne pathogens E. chaffeensis and E. muris) was detected in two samples.
Figure 1. Relative abundances of different bacterial genera in the salivary glands of (A) I. ovatus, (B) I. persulcatus and (C) H. flava.
All genera with less than 1.0% contribution were pooled into one group and labelled “others”.
Between 3,599 and 16,117 sequences were recorded for individual I. persulcatus, with almost 82% assigned to the genus level (Table 1) (Figure 1B), except for those of four I. persulcatus females. Over 80% of the reads in these samples were unclassified at the genus level. At the phylum level, these reads were classified as Proteobacteria or Alphaproteobacteria by the RDP classifier (data not shown). There were 127 different bacterial genera detected in I. persulcatus, of which 92 were detected in males, and 81 in females. Rickettsia was detected in nine I. persulcatus (4 female, 5 male) individuals and Ehrlichia was detected in a single I. persulcatus male (IPm5).
Between 6,204 and 10,017 sequences were obtained for individual H. flava, of which almost 97% were identified to the genus level (Table 1) (Figure 1C). A total of 59 different bacterial genera were detected, and Coxiella accounted for more than 90% of the microbial population in all samples. Spiroplasma was not detected in any individuals of H. flava, despite appearing in all Ixodes samples. Rickettsia spp. were detected in three H. flava females, and no sample contained Ehrlichia spp. The details of microbial population analysis for each sample are presented in Table S2.
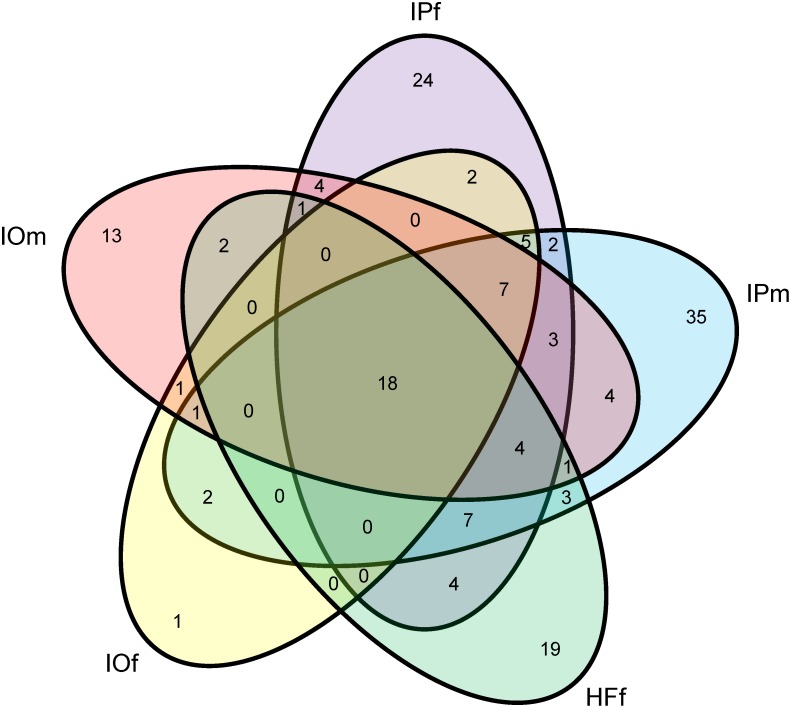
A summarized diagram of the number of bacterial genera detected in each tick group is presented in Figure 2. Out of 163 different genera identified, 18 were detected in all tick groups. These were Acinetobacterium, Arcicella, Burkholdelia, Corynebacterium, Coxiella, Cryobacter, Curvibacterium, Flavobacterium, Limnohabitas, Methylobacterium, Novosphingomonas, Polynucleobacter, Propionilbacterium, Pseudomonas, Rickettsia, Sphingomonas, Staphylococcus, and Sterptophyta. Some bacterial genera were uniquely associated with tick species or sex, i.e., IOf (1 genus), IOm (13 genera), IPf (24), IPm (35) and HFf (19).
Figure 2. Venn diagram of all 163 identified genera distributed across the tick species and sex.
Comparison of microbiomes in salivary glands between tick species
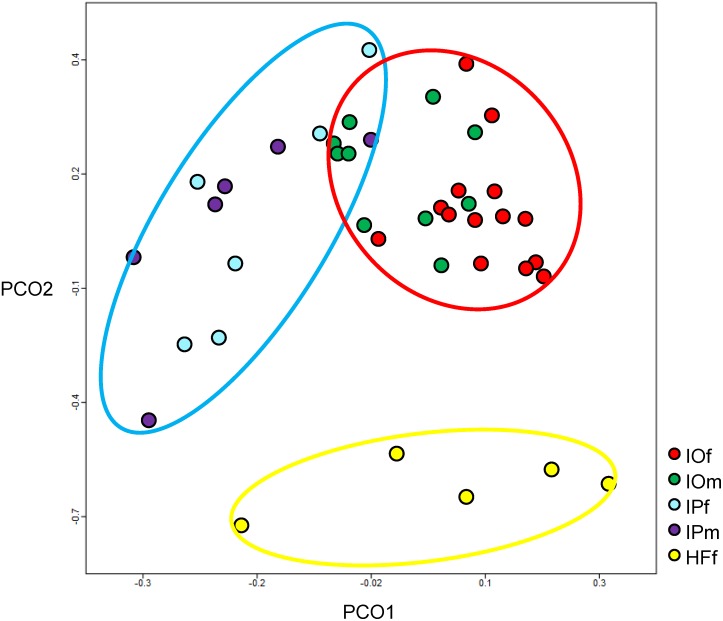
PCA was performed using the MG-RAST server with normalized values and Bray-Curtis distance (Figure 3) for each tick sample. The microbial community composition of each sample clustered approximately according to tick species. The microbial populations of Ixodes and Haemaphysalis were completely separated by PCO2. The microbial community composition of Haemaphysalis ticks was broadly distributed along PCO1; however, in I. ovatus and I. persulcatus microbial populations were more distinct, but with some overlap within this component.
Figure 3. Principal component analysis of the bacterial composition in each tick sample.
The plots were generated using the MG-RAST server. Each tick sample is shown in a different color depending on the species and sex of the tick; IOf, IOm, IPf, IPm, and HFf are respectively, shown in red, green, blue, purple, and yellow. The plots derived from the same tick species are highlighted in circles; I. ovatus (IO), I. persulcatus (IP), and H. flava (HF) are, respectively, highlighted in red, blue, and yellow circles.
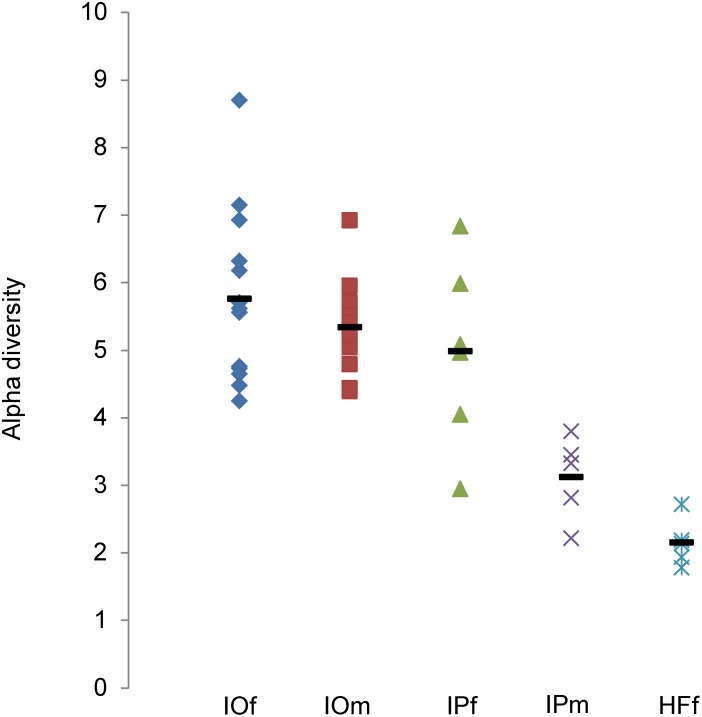
Alpha diversity for each sample was calculated using the MG-RAST server (Figure 4). Smirnov-Grubbs’s outlier test (α = 0.05) was used before the calculation of means and S.D. IPm1 was identified as an outlier and removed in the calculation for the mean value of IPm alpha diversity. Mean values were 5.75±1.19 (IOf), 5.33±0.72 (IOm), 4.97±1.25 (IPf), 3.11±0.55 (IPm) and 2.14±0.32 (HFf).
Figure 4. Alpha diversity calculated for each tick sample.
The alpha diversity of each tick sample was calculated using the MG-RAST server. The mean value obtained for each tick group is represented by the horizontal line. Mean alpha diversity values: IOf (5.75), IOm (5.33), IPf (4.97), IPm (3.11), and HFf (2.14).
Sequencing of gltA
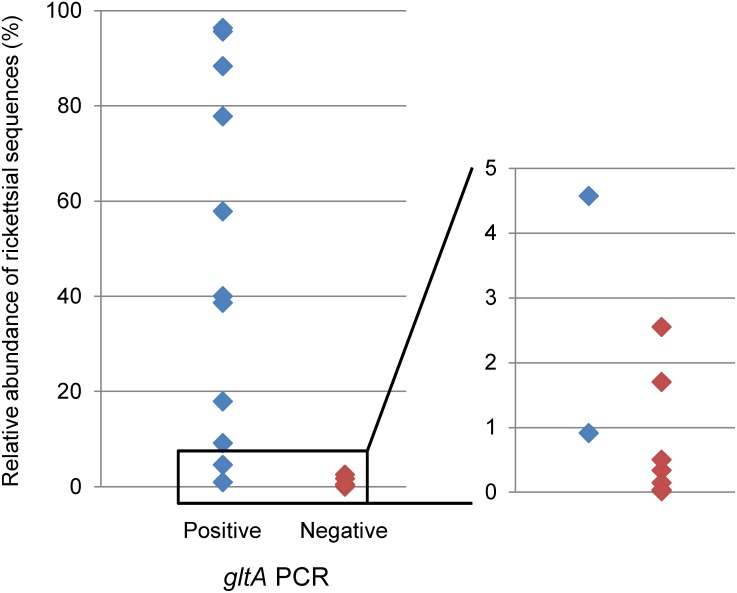
The gltA gene was detected in 11 out of 22 samples previously identified as containing the genus Rickettsia. Samples that were gltA-positive tended to have a greater abundance of rickettsial bacteria than those that were negative (Figure 5). All gltA-positive samples were subjected to sequencing analysis. Each gltA-positive sample contained only one sequence type, indicating that individual ticks harbored bacteria carrying a single gltA allele. From 11 tick samples, three different gltA sequences were identified, and BLAST searches showed the highest identities (99.8% to 100%) with R. asiatica, R. helvetica, and uncultured Rickettsia sp. (Table 2).
Figure 5. Comparison of the relative abundance of rickettsial sequences estimated by 16S amplicon analysis and the results of gltA PCR.
Vertical axis represents the relative abundance of rickettsial sequences calculated from the data obtained from 16S amplicon analysis. Blue dots represent samples in which Rickettsia was detected by both 16S amplicon analysis and gltA PCR. Red dots represent samples in which Rickettsia was detected by 16S amplicon analysis but not by gltA PCR. The plots with relative abundance values between 0% and 5% are shown in the magnified graph provided in the right column.
Table 2. Summary of gltA sequencing.
| SequenceID | Tick sampleID | Identity withreference (no.matched/no.nucleotides) | ReferenceGenBank no. | Rickettsiaspecies | GenBankno. |
| gltA_IOf1 | IOf1 | 99.8% (438/439) | AB297808 | R. asiatica | AB911107 |
| gltA_IOf3 | IOf3 | 99.8% (438/439) | AB297808 | R. asiatica | AB911107 |
| gltA_IOf4 | IOf4 | 99.8% (438/439) | AB297808 | R. asiatica | AB911107 |
| gltA_IOm2 | IOm2 | 99.8% (438/439) | AF394901 | R. asiatica | AB911107 |
| gltA_IOm3 | IOm3 | 99.8% (438/439) | AF394901 | R. asiatica | AB911107 |
| gltA_IOm4 | IOm4 | 99.8% (438/439) | AF394901 | R. asiatica | AB911107 |
| gltA_IPf1 | IPf1 | 99.8% (438/439) | U59723 | R. helvetica | AB911108 |
| gltA_IPf6 | IPf6 | 100% (394/394) | JN849396 | UnculturedRickettsia sp. | AB911109 |
| gltA_IPm4 | IPm4 | 99.8% (438/439) | U59723 | R. helvetica | AB911108 |
| gltA_IPm5 | IPm5 | 99.8% (438/439) | U59723 | R. helvetica | AB911108 |
| gltA_IPm6 | IPm6 | 99.8% (438/439) | U59723 | R. helvetica | AB911108 |
Sequencing of unclassified bacterial 16S rDNA
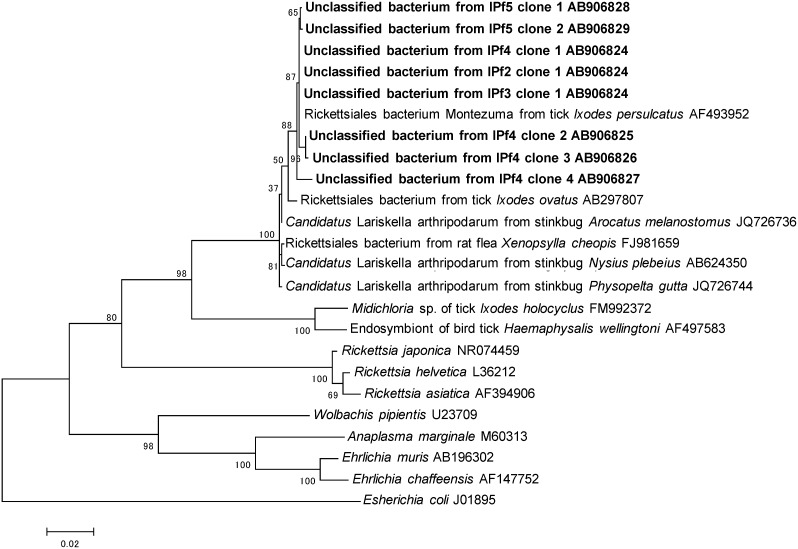
PCR fragments (1400 bp) were generated using universal primers to resolve the identities of sequences detected in four individuals of I. persulcatus. Between six and nine clones per sample were classified into Alphaproteobacteria (data not shown) based on BLASTn similarity searches. All the clones analyzed from two individual ticks were the same sequence type. There were four different sequence types in one individual, and two in another. These showed highest identities (99.5% to 99.7%) with uncultured Rickettsiales previously reported from I. persulcatus (GenBank accession number AF497583).
Molecular phylogenetic analysis revealed that the Alphaproteobacteria from four I. persulcatus females clustered together within a single clade. This clade contains Candidatus Lariskella arthropodarum identified in several stinkbug species (Arocatus melanostomus, Nysius plebeius, and Physopelta gutta) and Rickettsiales derived from flea (Xenopsylla cheopis) and ticks (I. ovatus and I. persulcatus) [28], [29], [30], [31] (Figure 6).
Figure 6. Phylogenetic analysis of the 16S rDNA sequences of unclassified bacteria from IPf2, IPf3, IPf4, and IPf5 using maximum likelihood method.
The tree is rooted with the Escherichia coli. All bootstrap values from 1000 replications are shown on interior branch nodes.
Discussion
The aim of this study was to assess and compare the diversity of bacterial populations within the salivary glands of I. ovatus, I. persulcatus, and H. flava. This is the first reported study of bacterial populations found in the salivary glands of different tick species. This metagenomic approach revealed bacterial populations totaling 163 different genera found in tick salivary glands. These included the genera of tick-borne pathogens such as Ehrlichia and Rickettsia. Further identification using species-specific PCR would be needed to clarify the presence of the tick-borne bacterial pathogens, such as E. muris, E. chaffeensis, R. japonica and R. helvetica, in the ticks used in this study [32], [33], [34], [35]. This combination of detection approaches may be useful for the screening and detection of possible pathogens in arthropod vectors.
Rickettsia was detected in 22 of the 41 (53.6%) samples by 16S rDNA amplicon pyrosequencing; however, only half of the 22 positive samples were positive with gltA PCR. This may be attributed to the relative amounts of rickettsial DNA in the PCR templates, where gltA PCR-positive samples tended to contain a higher proportion of rickettsial DNA than those that were negative (Figure 5). However, there were two gltA-negative samples (IPm1 and IPf4) that had higher proportions of rickettsial DNA than a gltA-positive sample (IOf4). This suggests that the sensitivity of conventional gltA PCR may be affected by the other factors such as the resolving power of agarose gel electrophoresis and the presence of PCR inhibitory components in samples [36], [37]. We suggest that a 16S rDNA amplicon pyrosequencing approach is a more sensitive method to detect specific pathogens.
Analysis of the gltA gene sequences from I. ovatus and I. persulcatus revealed that they belonged to R. asiatica and R. helvetica, respectively (Table 2). This result agrees with previous findings about the potential of the ticks to act as vectors for these rickettsia in Japan [38]. R. helvetica belongs to the spotted fever group of rickettsia and is a causative agent of febrile illness. A human case associated with this pathogens has been reported elsewhere [39], [40]. There was a high abundance (>70%) of this rickettsial species in some I. persulcatus samples (Figure 1B), suggesting that it is well adapted to the salivary glands of ticks, and waiting for transmission to mammalian hosts. In addition to pathogenic strains, the genus Rickettsia also contains symbionts associated with ticks. Rickettsia-like symbionts can influence the tick physiology, population dynamics, and the transmission of other pathogenic Rickettsia spp [41], [42].
Coxiella burnetii and Coxiella-like endosymbionts have been identified in several tick genera, including Dermacentor, Ixodes, Haemaphysalis and Rhipicephalus [43], [44], [45], [46]. Coxiella-like endosymbionts have been located at high densities in the salivary glands of the lone star tick (Amblyomma americanum) using fluorescence in situ hybridization [13]. The findings in this study also highlighted the presence of Coxiella in the salivary glands of three species of tick. The dominant presence of Coxiella in the salivary glands of ticks warrants further investigation to resolve their potential roles in tick biology, particularly blood-sucking behavior, and their interaction with other microbes.
The genus Spiroplasma contains a wide diversity of often unnamed or poorly characterized species, including non-pathogenic, symbiotic, and pathogenic organisms associated with a wide variety of arthropods. Symbiotic Spiroplasma has a close association with, and can affect the behavior of, their host arthropods. For example, Hurst et al. (2000) reported the preferential killing of males by Spiroplasma; when female insects (e.g., the butterfly Danaus chrysippus) are infected, the broods are female-biased because the infected male progeny die during embryogenesis [47]. One Spiroplasma sp. has been reported in ticks [48], [49], and it has also been associated with transmissible spongiform encephalopathy in humans and ruminants, although its role in the pathology of the host has not been clarified [50]. In this study, Spiroplasma was detected in Ixodes ticks, and not in H. flava (Figure 1). Previous research reported the genera Spiroplasma and the closely related Mycoplasma in several tick species in Japan [51]. The pathogenicity of Spiroplasma harbored in ticks in Japan is not known yet.
Results from the PCA of sequences indicated that microbial population structures in the salivary glands of ticks were different, and that samples from the same species of tick clustered together (Figure 3). Ticks can acquire microorganisms through a variety of ways, such as transovarial transmission, and from the environment, host animals during blood feeding, and mating partners. For microorganisms to exist in the salivary glands, they need to migrate from the midgut and enter the glands. The establishment of microorganisms within ticks can depend on the interactions between particular microbes, ticks and other symbioses [41], [52], [53]. The differences in the microbial populations within the salivary glands of tick species in this study were attributed to these complicated factors.
Previous studies revealed that tick microbial populations were different between developmental stages (egg, nymph, and adult) [19], [21]. The bacterial compositions also differed between organs, such as between midgut and ovary [19]. Some bacterial species, for instance Borrelia burgdorferi that is a causative agent of Lyme disease, exist in the midgut of the tick, moving into the salivary glands when stimulated by feeding on blood [54], [55]. For better understanding of microbial interactions with ticks as well as the potential pathogens transmitted by ticks, further study should include the comparison of the microbes between salivary glands and other organs. The analysis of the dynamics of microbial community composition during the process of feeding on blood may also uncover the roles of tick microbes.
The mean alpha diversity value (Figure 4) was greater for the female I. ovatus (5.61) than that of male (5.31). This rank order was also recorded for female (5.02) and male (3.38) I. persulcatus ticks, and may imply that some bacterial species preferentially select the gender of ticks. There may be some strategic biological relevance in the transmission of bacteria to mammalian hosts because female ticks feed for a longer period of time than males. The total number of bacterial genera (Table 1) detected in I. persulcatus (127) was greater than in I. ovatus (71).
Several I. persulcatus females contained unclassified bacteria belonging to the Proteobacteria and Alphaproteobacteria (Figure 1B). Based on the analysis of the nearly complete 16S rDNA sequences, the unclassified bacterial were classified into a single phylogenetic clade, which was recently proposed as a “Candidatus L. arthropodarum” clade [30]. This clade also includes Rickettsiales bacterium previously found in blood and biopsy samples of the patients with an acute fever disease, etiologically linked with tick bites [31]. The relationships between these microorganisms and their arthropod hosts are not clear, and their potential to act as causative agents of emerging tick-borne mammalian diseases warrants further investigation.
Supporting Information
Longitude and latitude of sampling sites.
(XLSX)
Details of classification results.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Japan Initiative for the Global Research Network on Infectious Diseases (J-GRID) (http://www.crnid.riken.jp/jgrid/en/) and the Program for Leading Graduate Schools “Fostering Global Leaders in Veterinary Science for Contributing to One Health” (http://www.jsps.go.jp/english/index.html) from the Ministry of Education, Culture, Sports, Science and Technology (NEXT). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Berrada ZL, Telford SR (2009) Burden of tick-borne infections on American companion animals. Top Companion Anim Med 24: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gorenflot A, Moubri K, Precigout E, Carcy B, Schetters TP (1998) Human babesiosis. Ann Trop Med Parasitol 92: 489–501. [DOI] [PubMed] [Google Scholar]
- 3. Yu XJ, Liang MF, Zhang SY, Liu Y, Li JD, et al. (2011) Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364: 1523–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parola P, Davoust B, Raoult D (2005) Tick- and flea-borne rickettsial emerging zoonoses. Vet Res 36: 469–492. [DOI] [PubMed] [Google Scholar]
- 5. Nakao R, Abe T, Nijhof AM, Yamamoto S, Jongejan F, et al. (2013) A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks. ISME J 7: 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wójcik-Fatla A, Zając V, Cisak E, Sroka J, Sawczyn A, et al. (2012) Leptospirosis as a tick-borne disease? Detection of Leptospira spp. in Ixodes ricinus ticks in eastern Poland. Ann Agric Environ Med 19: 656–659. [PubMed] [Google Scholar]
- 7. Ribeiro JM (1995) Blood-feeding arthropods: live syringes or invertebrate pharmacologists? Infect Agents Dis 4: 143–152. [PubMed] [Google Scholar]
- 8. Wikel SK (1999) Tick modulation of host immunity: an important factor in pathogen transmission. Int J Parasitol 29: 851–859. [DOI] [PubMed] [Google Scholar]
- 9. Futse JE, Ueti MW, Knowles DP, Palmer GH (2003) Transmission of Anaplasma marginale by Boophilus microplus: retention of vector competence in the absence of vector-pathogen interaction. J Clin Microbiol 41: 3829–3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Popov VL, Korenberg EI, Nefedova VV, Han VC, Wen JW, et al. (2007) Ultrastructural evidence of the ehrlichial developmental cycle in naturally infected Ixodes persulcatus ticks in the course of coinfection with Rickettsia, Borrelia, and a flavivirus. Vector Borne Zoonotic Dis 7: 699–716. [DOI] [PubMed] [Google Scholar]
- 11. Santos AS, Bacellar F, Santos-Silva M, Formosinho P, Grácio AJ, et al. (2002) Ultrastructural study of the infection process of Rickettsia conorii in the salivary glands of the vector tick Rhipicephalus sanguineus . Vector Borne Zoonotic Dis 2: 165–177. [DOI] [PubMed] [Google Scholar]
- 12.Sonenshine DE (2005) The biology of tick vectors of human disease. In: Goodman JL, Dennis DT, Sonenshine DE, editors. Tick-borne diseases of humans. Washington DC: ASM Press. 12–36.
- 13. Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C (2007) Localization and visualization of a coxiella-type symbiont within the lone star tick, Amblyomma americanum . Appl Environ Microbiol 73: 6584–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahantarig A, Trinachartvanit W, Baimai V, Grubhoffer L (2013) Hard ticks and their bacterial endosymbionts (or would be pathogens). Folia Microbiol (Praha) 58: 419–428. [DOI] [PubMed] [Google Scholar]
- 15. Kurtti TJ, Palmer AT, Oliver JH (2002) Rickettsiella-like bacteria in Ixodes woodi (Acari: Ixodidae). J Med Entomol 39: 534–540. [DOI] [PubMed] [Google Scholar]
- 16. Liu L, Li L, Liu J, Hu Y, Liu Z, et al. (2013) Coinfection of Dermacentor silvarum olenev (acari: ixodidae) by Coxiella-Like, Arsenophonus-like, and Rickettsia-like symbionts. Appl Environ Microbiol 79: 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niebylski ML, Peacock MG, Fischer ER, Porcella SF, Schwan TG (1997) Characterization of an endosymbiont infecting wood ticks, Dermacentor andersoni, as a member of the genus Francisella . Appl Environ Microbiol 63: 3933–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Noda H, Munderloh UG, Kurtti TJ (1997) Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol 63: 3926–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andreotti R, Pérez de León AA, Dowd SE, Guerrero FD, Bendele KG, et al. (2011) Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carpi G, Cagnacci F, Wittekindt NE, Zhao F, Qi J, et al. (2011) Metagenomic profile of the bacterial communities associated with Ixodes ricinus ticks. PLoS One 6: e25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Menchaca AC, Visi DK, Strey OF, Teel PD, Kalinowski K, et al. (2013) Preliminary assessment of microbiome changes following blood-feeding and survivorship in the Amblyomma americanum nymph-to-adult transition using semiconductor sequencing. PLoS One 8: e67129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hashimoto S, Kawado M, Murakami Y, Izumida M, Ohta A, et al. (2007) Epidemics of vector-borne diseases observed in infectious disease surveillance in Japan, 2000–2005. J Epidemiol 17 Suppl: S48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y (2012) Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 24: 521–530, e248. [DOI] [PMC free article] [PubMed]
- 24. Kim YS, Kim MC, Kwon SW, Kim SJ, Park IC, et al. (2011) Analyses of bacterial communities in meju, a Korean traditional fermented soybean bricks, by cultivation-based and pyrosequencing methods. J Microbiol 49: 340–348. [DOI] [PubMed] [Google Scholar]
- 25. Roux V, Rydkina E, Eremeeva M, Raoult D (1997) Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol 47: 252–261. [DOI] [PubMed] [Google Scholar]
- 26. Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Erickson DL, Anderson NE, Cromar LM, Jolley A (2009) Bacterial communities associated with flea vectors of plague. J Med Entomol 46: 1532–1536. [DOI] [PubMed] [Google Scholar]
- 29. Fujita H, Takada N, Kawabata H, Ishiguro F, Yamamoto S, et al. (2007) some suggestive records of rickettsiae isolated from ticks in Korea and central China. Annual Report of Ohara General Hospital 47: 4. [Google Scholar]
- 30. Matsuura Y, Kikuchi Y, Meng XY, Koga R, Fukatsu T (2012) Novel clade of alphaproteobacterial endosymbionts associated with stinkbugs and other arthropods. Appl Environ Microbiol 78: 4149–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mediannikov OIu, Ivanov LI, Nishikawa M, Saito R, Sidel’nikov IN, et al.. (2004) Microorganism “Montezuma” of the order Rickettsiales: the potential causative agent of tick-borne disease in the Far East of Russia. Zh Mikrobiol Epidemiol Immunobiol: 7–13. [PubMed]
- 32. Fournier PE, Fujita H, Takada N, Raoult D (2002) Genetic identification of rickettsiae isolated from ticks in Japan. J Clin Microbiol 40: 2176–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kawahara M, Ito T, Suto C, Shibata S, Rikihisa Y, et al. (1999) Comparison of Ehrlichia muris strains isolated from wild mice and ticks and serologic survey of humans and animals with E. muris as antigen. J Clin Microbiol 37: 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahara F (1997) Japanese spotted fever: report of 31 cases and review of the literature. Emerg Infect Dis 3: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shibata S, Kawahara M, Rikihisa Y, Fujita H, Watanabe Y, et al. (2000) New Ehrlichia species closely related to Ehrlichia chaffeensis isolated from Ixodes ovatus ticks in Japan. J Clin Microbiol 38: 1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakao R, Magona JW, Zhou L, Jongejan F, Sugimoto C (2011) Multi-locus sequence typing of Ehrlichia ruminantium strains from geographically diverse origins and collected in Amblyomma variegatum from Uganda. Parasit Vectors 4: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peter TF, Deem SL, Barbet AF, Norval RA, Simbi BH, et al. (1995) Development and evaluation of PCR assay for detection of low levels of Cowdria ruminantium infection in Amblyomma ticks not detected by DNA probe. J Clin Microbiol 33: 166–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fujita H, Fournier PE, Takada N, Saito T, Raoult D (2006) Rickettsia asiatica sp. nov., isolated in Japan. Int J Syst Evol Microbiol 56: 2365–2368. [DOI] [PubMed] [Google Scholar]
- 39. Noji Y, Takada N, Ishiguro F, Fujino S, Aoyama T, et al. (2005) The first reported case of spotted fever in Fukui Prefecture, the northern part of central Japan. Jpn J Infect Dis 58: 112–114. [PubMed] [Google Scholar]
- 40. Ishiguro F, Takada N, Fujita H, Noji Y, Yano Y, et al. (2008) Survey of the vectorial competence of ticks in an endemic area of spotted fever group rickettsioses in Fukui Prefecture, Japan. Microbiol Immunol 52: 305–309. [DOI] [PubMed] [Google Scholar]
- 41. Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, et al. (2013) Update on Tick-Borne Rickettsioses around the World: a Geographic Approach. Clin Microbiol Rev 26: 657–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Socolovschi C, Mediannikov O, Raoult D, Parola P (2009) The relationship between spotted fever group Rickettsiae and ixodid ticks. Vet Res 40: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bernasconi MV, Casati S, Péter O, Piffaretti JC (2002) Rhipicephalus ticks infected with Rickettsia and Coxiella in Southern Switzerland (Canton Ticino). Infect Genet Evol 2: 111–120. [DOI] [PubMed] [Google Scholar]
- 44. Bonnet S, de la Fuente J, Nicollet P, Liu X, Madani N, et al. (2013) Prevalence of tick-borne pathogens in adult Dermacentor spp. ticks from nine collection sites in France. Vector Borne Zoonotic Dis 13: 226–236. [DOI] [PubMed] [Google Scholar]
- 45. Cooper A, Stephens J, Ketheesan N, Govan B (2013) Detection of Coxiella burnetii DNA in wildlife and ticks in northern Queensland, Australia. Vector Borne Zoonotic Dis 13: 12–16. [DOI] [PubMed] [Google Scholar]
- 46. Lee JH, Park HS, Jang WJ, Koh SE, Park TK, et al. (2004) Identification of the Coxiella sp. detected from Haemaphysalis longicornis ticks in Korea. Microbiol Immunol 48: 125–130. [DOI] [PubMed] [Google Scholar]
- 47. Hurst GD, Jiggins FM (2000) Male-killing bacteria in insects: mechanisms, incidence, and implications. Emerg Infect Dis 6: 329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henning K, Greiner-Fischer S, Hotzel H, Ebsen M, Theegarten D (2006) Isolation of Spiroplasma sp. from an Ixodes tick. Int J Med Microbiol 296 Suppl 40: 157–161. [DOI] [PubMed] [Google Scholar]
- 49. Tully JG, Rose DL, Yunker CE, Cory J, Whitcomb RF, et al. (1981) Helical mycoplasmas (spiroplasmas) from Ixodes ticks. Science 212: 1043–1045. [DOI] [PubMed] [Google Scholar]
- 50. Bastian FO, Sanders DE, Forbes WA, Hagius SD, Walker JV, et al. (2007) Spiroplasma spp. from transmissible spongiform encephalopathy brains or ticks induce spongiform encephalopathy in ruminants. J Med Microbiol 56: 1235–1242. [DOI] [PubMed] [Google Scholar]
- 51. Taroura S, Shimada Y, Sakata Y, Miyama T, Hiraoka H, et al. (2005) Detection of DNA of ‘Candidatus Mycoplasma haemominutum’ and Spiroplasma sp. in unfed ticks collected from vegetation in Japan. J Vet Med Sci 67: 1277–1279. [DOI] [PubMed] [Google Scholar]
- 52.Burgdorfer W, Hayes SF, Mavros AJ (1981) Non-Pathogenic Rickettsiae in Dermacentor andersoni: A Limiting Factor for the Distribution of Rickettsia rickettsii. In: Burgdorfer W, Anacker RL, editors. Rickettsiae and Rickettsial Disease. New York: Academic. 585–594.
- 53. Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF (2002) Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia . J Med Entomol 39: 809–813. [DOI] [PubMed] [Google Scholar]
- 54. Benach JL, Coleman JL, Skinner RA, Bosler EM (1987) Adult Ixodes dammini on rabbits: a hypothesis for the development and transmission of Borrelia burgdorferi . J Infect Dis 155: 1300–1306. [DOI] [PubMed] [Google Scholar]
- 55. Kurtenbach K, De Michelis S, Etti S, Schäfer SM, Sewell HS, et al. (2002) Host association of Borrelia burgdorferi sensu lato–the key role of host complement. Trends Microbiol 10: 74–79. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Longitude and latitude of sampling sites.
(XLSX)
Details of classification results.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.