Abstract
Functions of the Borrelia burgdorferi RecA protein were investigated in Escherichia coli recA null mutants. Complementation with B. burgdorferi recA increased survival of E. coli recA mutants by 3 orders of magnitude at a UV dose of 2,000 μJ/cm2. The viability at this UV dose was about 10% that provided by the homologous recA gene. Expression of B. burgdorferi recA resulted in survival of E. coli at levels of mitomycin C that were lethal to noncomplemented hosts. B. burgdorferi RecA was as effective as E. coli RecA in mediating homologous recombination in E. coli. Furthermore, E. coli λ phage lysogens complemented with B. burgdorferi recA produced phage even in the absence of UV irradiation. The level of phage induction was 55-fold higher than the level in cells complemented with the homologous recA gene, suggesting that B. burgdorferi RecA may possess an enhanced coprotease activity. This study indicates that B. burgdorferi RecA mediates the same functions in E. coli as the homologous E. coli protein mediates. However, the rapid loss of viability and the absence of induction in recA expression after UV irradiation in B. burgdorferi suggest that recA is not involved in the repair of UV-induced damage in B. burgdorferi. The primary role of RecA in B. burgdorferi is likely to be a role in some aspect of recombination.
The spirochete Borrelia burgdorferi, the causative agent of Lyme disease, has a unique genomic organization consisting of a 910,725-bp linear chromosome and at least 21 linear and circular plasmids (6, 13). Less than one-half of the 1,689 annotated open reading frames have an identifiable database match (6, 13). In addition, very few genes have been characterized biochemically. Functional studies of B. burgdorferi genes in the postgenomic era have been hampered by this spirochete's slow growth in culture and by the limited availability of genetic tools. A potentially useful strategy for circumventing these difficulties is complementation of mutants in well-characterized bacteria, such as Escherichia coli. This strategy has been employed to elucidate the functions of a number of B. burgdorferi gene products (1, 5, 7, 8, 15, 20, 26, 32, 44, 49, 50).
Genetic recombination may play a role in B. burgdorferi persistence by generation of antigenic variation (2, 31, 37, 45, 47, 52, 56, 57). RecA is the key protein linking genetic recombination to DNA replication and repair in bacteria (23, 24). In E. coli, RecA promotes pairing and strand exchange between a single-stranded DNA molecule and a homologous double-stranded DNA molecule through a complex multistep pathway that requires a DNA-dependent ATPase function (29). In addition to this recombinase activity, an ATP-dependent coprotease activity has also been ascribed to RecA. In enterobacteria and Bacillus sp., the latter activity is part of a specialized and regulated form of DNA repair designated SOS repair (3). In E. coli, the SOS response involves the transcriptional repressor LexA, whose autocatalytic cleavage is enhanced by activated RecA following DNA damage (19). This results in the coordinate expression of approximately 30 different genes, including recA and lexA.
The B. burgdorferi recA gene was originally described from isolate Sh-2-82 by Dew-Jager et al. (10). An orthologue of this gene from type strain B31MI has been annotated in the B. burgdorferi genome as BB0131 (13). The gene encodes a 365-amino-acid protein that exhibits 56% identity and 77% similarity with E. coli RecA. As a first step in elucidating the mechanism of recombination in B. burgdorferi, several activities of the B. burgdorferi RecA protein were studied in E. coli recA null mutants.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The following E. coli K-12 strains were employed in this study: C600 (leuB6 thi-1 thr-1), DH5α (recA1 gyrA thi-1 relA1), HB101 (recA13 leuB6 thi-1 proA), and 25404 (wild type). A pUC18-based clone containing B. burgdorferi recA was obtained from the American Type Culture Collection (ATCC 631007). This plasmid contains a 1,410-bp DNA insert encoding the entire B. burgdorferi recA gene (1,095 bp), including 291 bp of 5′ flanking sequences and 24 bp of 3′ flanking sequences. This plasmid is designated pUC18-recABb. An insertionally inactivated derivative of pUC18-recABb was constructed by cleaving the plasmid at a unique EcoRV site in recA and inserting a 1,190-bp kanamycin cassette derived from transposon Tn1545. This antibiotic resistance cassette was PCR amplified from a kanamycin-resistant derivative of plasmid pGK12 (38) by using 5′GGGAATTCTGTTATGAGCCATATTCAACG3′ and 5′GATTAGAAAAACTCATCGAGC3′ as the forward and reverse primers, respectively. A DNA fragment containing the E. coli recA gene along with 223 bp of 5′ flanking sequence and 92 bp of 3′ flanking sequence was generated by PCR by using wild-type E. coli C600 (recA+) DNA as the template and 5′GAGAAGCCTGTCGGCACCGTCTGG3′ and 5′CTTGCTCACTGTGATCGCGCACCG3′ as the forward and reverse primers, respectively. This fragment was cloned into the unique SmaI site of pUC18 (54), and the resulting plasmid was designated pUC18-recAEc.
The gene orientation in the inserts relative to the pUC18 backbone and the sequences of the inserted fragments were determined by DNA sequencing (Davis Sequencing, LLC, Davis, Calif.). In all constructs, the lacZ promoter was located upstream of and in the same orientation as the inserted recA gene.
UV sensitivity measurements.
E. coli DH5α clones complemented with various plasmid constructs were tested for UV sensitivity by using the method of Miller (28). Cultures of these clones were grown overnight at 37°C in Luria-Bertani broth (LB) supplemented with 100 μg ampicillin per ml (LB-Amp). A 1:50 dilution of each culture was regrown in LB-Amp to an A600 of 0.8. Samples (0.1 ml) of the E. coli cells were spread on LB-Amp agar plates at a 10−6 dilution and were subsequently irradiated with increasing doses of UV light (254 nm) with a Spectrolinker XL-1000 UV cross-linker (Spectronics, Westbury, N.Y.). The UV-treated bacteria were protected from room light, and colonies were counted after overnight incubation at 37°C. Identical plates which were not irradiated were used to measure the viable counts of the starting cultures.
Measurement of MMC sensitivity.
Five-milliliter portions of exponentially growing E. coli cells were incubated for 12 h at 37°C in the presence of increasing amounts of mitomycin C (MMC) (Sigma) in LB-Amp. Prior to plating, the cultures were serially diluted in LB-Amp to obtain concentrations 104, 105 and 106 cells/ml, and 0.1-ml portions of each dilution were plated on LB-Amp plates in duplicate and incubated overnight at 37°C. MMC sensitivity was measured by counting the surviving colonies. Untreated cells grown in an identical manner served as controls.
UV induction of lambda lysogens of E. coli.
Wild-type bacteriophage lambda lysogens were produced by cross-streaking E. coli DH5α preparations containing the various recA plasmid constructs with a high-titer phage lysate on LB agar (4). Colonies at the crossing points were purified and cross-streaked against both a wild-type phage lysate and a phage cI repressor-insensitive mutant phage (λvir) lysate. Colonies that were resistant to wild-type phage superinfection but sensitive to λvir lysis were employed in subsequent experiments.
Lysogenic E. coli strains were grown overnight at 37°C in LB-Amp and were serially diluted and plated on LB agar. The plates were exposed to increasing doses of UV light, and this was followed by overlaying with a liquid E. coli C600 culture. Phage plaques were counted after overnight incubation at 37°C. Untreated lysogens served as controls.
P1 transductions.
Phage P1 transductions were performed by using the protocol of Miller (28). Both phage and E. coli 25404 were purchased from the American Type Culture Collection. Stocks of P1 were generated by plate lysis on LB agar. Approximately 106 phage were absorbed to the exponentially growing donor E. coli 25404 (Leu+) in LB supplemented with 5 × 10−3 M CaCl2. The infected cells were plated on three LB agar plates in soft R-top agar. The lysates were scraped off the plates, pooled, and treated with chloroform. Cell debris was removed by centrifugation, and the lysate was stored in the presence of chloroform. The phage yield was measured by using liquid agar overlays and E. coli 25404 as the lawn. A lysate titer of 9.4 × 108 PFU/ml was obtained under these conditions.
Portions (0.1 ml) of overnight cultures of E. coli HB101 (Leu−) harboring the various pUC18-based plasmid constructs were transduced with 0.1 ml of lysate, and Leu+ colonies were selected on glucose minimal medium plates containing 100 μg of ampicillin per ml. Lysate controls for bacterial contamination and host cell controls for Leu+ revertants were also included. Colonies which appeared after 48 h of incubation were serially patched three times on glucose minimal medium plates containing 100 μg of ampicillin per ml. All experiments were performed in triplicate.
Determination of B. burgdorferi UV sensitivity.
B. burgdorferi B31MI, an infectious isolate at passage 13, was grown in BSK-H medium at 33°C to a density of 1 × 108 organisms/ml. Cells were harvested by centrifugation and resuspended in 0.1 ml of phosphate-buffered saline (PBS) containing 1 × 107 cells. Spirochetes were rapidly transferred into and spread in culture dishes (32 by 10 mm) and then were exposed to UV irradiation at doses of 800 to 9,000 μJ/cm2. After UV exposure the cells were kept in the dark. Then the cells were recovered by aspiration and serially diluted in BSK-H medium so that each plate contained approximately 100 organisms per 0.1 ml. Each 0.1-ml dilution was added to 2 ml of BSK-H medium containing 0.8% molten agarose and was overlaid onto plates (60 by 15 mm) containing solid BSK-H medium. The agar plates were incubated at 33°C for 9 days in a 5% CO2 incubator, and colonies were counted. B. burgdorferi not exposed to UV irradiation served as a control.
Determination of recA expression by quantitative reverse transcription (RT)-PCR.
A 40-ml culture of B. burgdorferi B31MI was grown to a density of 1 × 108 cells/ml in BSK-H medium at 33°C. Cells were harvested by centrifugation at 12,000 × g for 10 min and were suspended in 1.2 ml of sterile PBS. Then 0.3-ml aliquots were placed into culture dishes (35 by 10 mm) and exposed to increasing doses of UV irradiation. Following irradiation, the cells were kept in the dark, 10 ml of BSK-H medium was added, and the cells were incubated at 33°C for 2 h.
Forty milliliters of LB was inoculated with an E. coli C600 overnight culture by using a 1:50 dilution, and the culture was grown at 37°C to an A600 of 1.0. Cells were harvested by centrifugation at 12,000 × g for 10 min and were suspended in 4 ml of sterile PBS. One-milliliter aliquots were placed into culture dishes (100 by 15 mm) and exposed to increasing doses of UV. Following irradiation, the cells were protected from room light and following addition of 10 ml of LB were incubated at 37°C for 2 h.
B. burgdorferi or E. coli cells were recovered by centrifugation at 12,000 × g for 10 min, and the pellets were washed twice with sterile PBS. RNA extraction was performed individually for each sample by using a PURESCRIPT kit (Gentra Systems, Minneapolis, Minn.) according to the manufacturer's protocol for gram-negative bacteria. Total RNA was treated with DNase (AMBION DNase kit), and RNA concentrations were determined spectrophotometrically at 260 nm. cDNA was generated by RT. Each reaction mixture contained first strand buffer, each deoxynucleoside triphosphate at a concentration of 0.4 mM, 10 U of RNase inhibitor, 500 ng of each random hexamer primer, 500 ng of total RNA, and 100 U of Superscript II. Following 1 h of incubation at 42°C, 5 ng of cDNA was retrieved, and RT-PCR was performed by using SYBR Green and a Lightcycler (Roche Diagnostics). The conditions used for RT-PCR amplification and the recA-specific primer sequences for B. burgdorferi have been described previously (27). The E. coli recA RT-PCR amplification conditions were essentially the same as those reported for B. burgdorferi, with the following exceptions. The PCR primer sequences employed for E. coli recA RT-PCR amplification were 5′ATGTGGAAACCATCTCTACCGG 3′ and 5′GTTGGACTGCTTCAGGTTACC 3′ for the forward and reverse primers, respectively, and an annealing temperature of 48°C was used. Each RT-PCR was performed in triplicate, and control mixtures containing no reverse transcriptase or no cDNA were included.
RESULTS
Complementation of E. coli RecA null mutants with B. burgdorferi RecA.
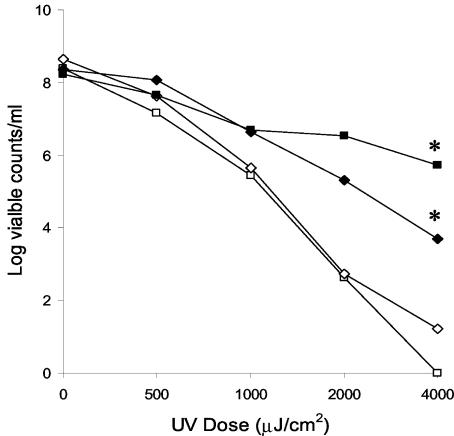
The activity of the B. burgdorferi RecA protein was assessed by measuring survival of an E. coli recA null mutant complemented with plasmid-borne B. burgdorferi recA after UV irradiation. The data in Fig. 1 show that B. burgdorferi recA increased the survival of an E. coli recA mutant by 3 orders of magnitude at a UV dose of 2,000 μJ/cm2. This complementation was about 10% that provided by the homologous E. coli recA gene expressed from the same vector construct at comparable UV doses. Complementation with a plasmid containing a variant of B. burgdorferi recA inactivated by insertion of a Kmr cassette did not result in increased survival. This clearly indicates that the spirochetal recA is functional in E. coli.
FIG. 1.
Survival of DH5α complemented with various pUC18-recA constructs after UV irradiation. ▪, pUC18-recAEc; ♦, pUC18-recABb; □, pUC18; ⋄, pUC18-recABb: Kmr. The enhanced survival of pUC18-recAEc- or pUC18-recABb-complemented E. coli at a UV dose of 4,000 μJ/cm2 relative to the survival of pUC18-complemented E. coli was statistically significant at a P value of 0.002 (indicated by asterisks).
Complementation of MMC-treated E. coli by B. burgdorferi recA.
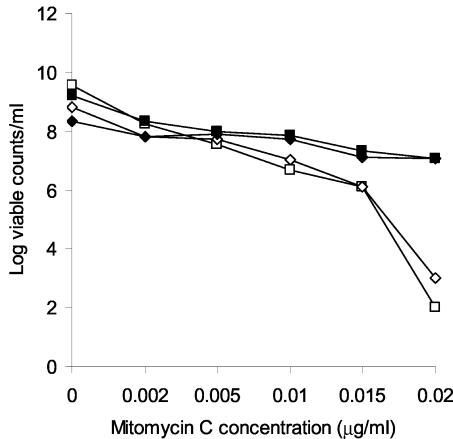
UV irradiation results in mostly intrastrand thymine dimers in DNA, and repair of this damage reflects nucleotide excision (39). In order to assess the ability of B. burgdorferi recA to promote repair of interstrand cross-links, similar survival studies were performed with cells exposed to increasing concentrations of MMC. Complementation of E. coli DH5α with either wild-type E. coli recA or B. burgdorferi recA resulted in a 6-log increase in survival at the highest concentration of MMC (0.02 μg/ml) (Fig. 2). In this case, the insertionally inactivated B. burgdorferi recA gene did not provide any protection. Together, the findings obtained with UV irradiation and MMC exposure demonstrated that B. burgdorferi recA can mediate repair of both intra- and interstrand DNA damage.
FIG. 2.
Survival of DH5α containing various pUC18-recA plasmids after 12 h of exposure to different doses of MMC. ▪, pUC18-recAEc; ♦, pUC18-recABb; □, pUC18; ⋄, pUC18-recABb:Kmr. Viable cells were counted by plating dilutions of MMC-treated and control cells on LB-Amp plates.
Prophage induction by B. burgdorferi RecA.
RecA activates the SOS response by cleavage of the LexA transcriptional repressor. Another substrate for the RecA coprotease activity is the λ phage cI repressor protein. In contrast to the rapid LexA cleavage by RecA, autocleavage of cI and other lambdoid phage repressors is slow and occurs at the high Mg2+ concentrations (>10 mM) often found in moribund cells (19, 36).
Phage λ cI repressor cleavage by B. burgdorferi RecA was measured by phage induction of E. coli lysogens. In this assay, the number of plaques obtained under each condition directly reflected the number of host cells that contained cI repressor cleaved by RecA and that were subsequently converted into lytic centers, thereby producing detectable plaques. Table 1 shows the efficiencies of phage induction in E. coli DH5α complemented with plasmids containing the various recA constructs. In uncomplemented E. coli DH5α, no active phage production was observed since RecA is absolutely required for this process. Complementation with a plasmid containing either E. coli recA or B. burgdorferi recA resulted in production of active phage. Furthermore, exposure to increasing doses of UV irradiation resulted in significantly more plaques, as expected. Interestingly, even prior to UV irradiation, there was a 55-fold enhancement of λ phage induction in B. burgdorferi recA-complemented lysogens compared with that observed in cells complemented with the E. coli recA gene. This suggests that the B. burgdorferi RecA protein is able to induce the autocatalytic activity of the cI repressor even more efficiently than the homologous RecA is.
TABLE 1.
Production of λ infective centers in E. coli DH5α, complemented with plasmids containing recAa
| Plasmid | Viable counts (cells/ml)b | PFU/ml with:
|
||
|---|---|---|---|---|
| No UV irradiation | 4,000 μJ of UV/cm2 | 8,000 μJ of UV/cm2 | ||
| pUC18 | 4.0 × 108 ± 1.4 × 108 | 0 | 0 | 0 |
| pUC18-recABb | 5.8 × 108 ± 3.0 × 108 | 8.8 × 107 ± 4.5 × 107c | 5.7 × 108 ± 1.3 × 108d | 4.0 × 108 ± 1.3 × 108 |
| pUC18-recABb:Kmr | 4.6 × 108 ± 2.1 × 108 | 0 | 0 | 0 |
| pUC18-recAEc | 8.3 × 108 ± 2.3 × 108 | 1.4 × 106 ± 5.3 × 105c | 6.9 × 107 ± 4.4 × 107d | 1.8 × 108 ± 9.2 × 107 |
| pUC18 (C600)e | 1.7 × 109 ± 2.2 × 108 | 4.2 × 107 ± 1.8 × 107 | 5.0 × 108 ± 4.0 × 107 | 6.0 × 108 ± 1.3 × 108 |
The results are averages for three independent experiments.
Average ± standard deviation.
With no irradiation the difference between the value for pUC18-recABb and the value for pUC18-recAEc was statistically significant at a P value of 0.0005.
With 4,000 μJ/cm2 the difference between the value for pUC18-recABb and the value for pUC18-recAEc was statistically significant at a P value of 0.0108.
An E. coli C600 (RecA+) λ lysogen harboring vector pUC18 was employed to estimate the contribution of chromosomal recA to λ phage induction by UV irradiation.
Recombinational activity of the B. burgdorferi RecA protein.
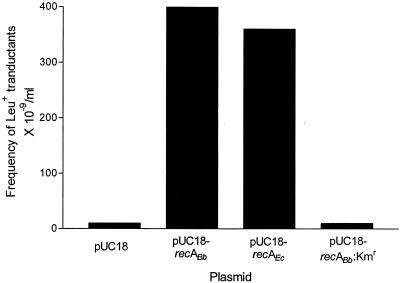
The ability of B. burgdorferi RecA to promote homologous recombination in E. coli was assayed by generalized P1 transduction for leucine prototrophs. In this case, HB101 was used as a surrogate host because it has a Leu− phenotype. HB101 complemented with pUC18 did not yield any Leu+ transductants. In contrast, complementation with plasmids containing either wild-type E. coli recA or B. burgdorferi recA resulted in viable Leu+ recombinants (Fig. 3). In this case, B. burgdorferi recA disrupted with a kanamycin cassette could not provide the necessary RecA activity.
FIG. 3.
Phage P1 transduction frequencies of HB101 complemented with various pUC18 recA constructs. The frequency was measured by determining the ratio of the number of Leu+ transductants to the total number of viable cells recovered at comparable multiplicities of infection.
Effect of UV irradiation on B. burgdorferi viability.
The studies described above demonstrated that the B. burgdorferi genome encodes a RecA protein that can participate in DNA repair and recombination. However, the B. burgdorferi genome apparently does not contain a LexA gene (13). Furthermore, the B. burgdorferi life cycle involves propagation in either a mammalian host or a tick vector and likely does not provide an opportunity for exposure to UV irradiation. In an initial effort to ascertain the possible role(s) of the RecA protein in B. burgdorferi, sensitivity to UV irradiation was measured.
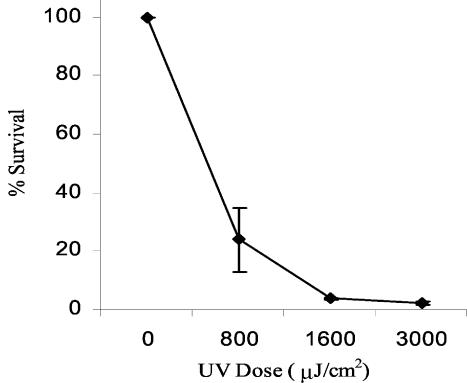
A UV survival curve for B. burgdorferi B31MI is shown in Fig. 4. A UV dose of only 800 μJ/cm2 resulted in a 76% loss of viability. At this UV dose, no reduction in viability was observed for wild-type E. coli; however, the response was similar to that observed with E. coli recA null mutants. Interestingly, a similar UV dose curve has been reported for Leptospira biflexa (48). The rather steep slope of the survival curve indicates that the accumulated damage in B. burgdorferi DNA cannot be effectively repaired.
FIG. 4.
Survival of B. burgdorferi B31MI after UV irradiation. The numbers of viable cells were determined by plating dilutions of UV-treated cells on BSK-H medium plates. The experiment was performed in triplicate.
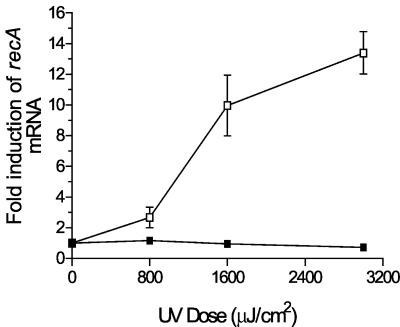
In E. coli, UV irradiation led to a dose-dependent increase in recA transcription (Fig. 5). Irradiation of B. burgdorferi with UV doses as high as 3,000 μJ/cm2 did not lead to any increase in recA expression. These UV exposure studies suggested that recA is not involved in the repair of UV-induced damage in B. burgdorferi.
FIG. 5.
Induction of recA mRNA after UV irradiation as determined by quantitative RT-PCR. □, E. coli C600; ▪, B. burgdorferi B31MI. The level of recA mRNA was measured by real-time RT-PCR as described in Materials and Methods. mRNA levels are expressed as the fold induction relative to the induction of unirradiated cells, which was defined as 1. The values are the averages obtained in three independent experiments.
DISCUSSION
Three different but interrelated functions of the B. burgdorferi RecA protein were measured in a complementation model system by using E. coli recA null mutants as surrogate hosts. The results of the present study suggest that B. burgdorferi RecA functions in the activation of both the E. coli SOS response and λ phage lysogen induction. B. burgdorferi RecA is also able to mediate homologous recombination in E. coli. Similar results for an E. coli complementation model have been reported by Putteet-Driver et al. (33). These findings are not surprising in view of the extensive studies demonstrating complementation of E. coli with heterologous recA (21, 29, 43), which is likely the result of extensive conservation of this protein at the amino acid level. Alignment of the deduced amino acid sequences of the RecA proteins of B. burgdorferi, Treponema pallidum, and L. biflexa and E. coli RecA showed that B. burgdorferi RecA exhibits 56% amino acid identity and 77% similarity with E. coli RecA, 63% identity and 84% similarity with T. pallidum RecA, and 62% identity and 80% similarity with L. biflexa RecA.
The functional domains of the E. coli protein have been assigned by both structural and mutational studies (23). On the basis of sequence alignment and conservation, it is possible deduce the identities of these functional domains in RecA proteins from other organisms. The nucleotide binding and hydrolysis regions defined by E. coli RecA residues G66 to T73 (A site), E96, D100, Y103, and I140 to D144 (B site) are invariant. The two domains designated loop 1 and loop 2 corresponding to residues E156 to D161 and Q194 to T210 in the E. coli are involved in DNA binding (23). Both loop sequences are completely conserved in B. burgdorferi, except for an E156D substitution. Individual RecA monomers interact with each other and single-stranded DNA, and filaments formed in this manner can participate in filament-filament interactions. These protein-protein interactions are required for all RecA functions (23). The residues that have been implicated in these interactions in E. coli RecA are highly conserved in the spirochetal proteins.
Although complementation with B. burgdorferi RecA conferred resistance to both UV irradiation and MMC exposure, the extent of complementation was different. Whereas complete repair of MMC-induced damage was observed in the complemented mutants, the level of survival after UV irradiation was only 10% of that observed in mutants complemented with the homologous gene. Essentially similar results were obtained by Putteet-Driver et al. (33), although only a moderate increase in survival following MMC treatment was observed. It is likely that the difference between the observed responses in the two studies was due to differences in the experimental design. UV exposure causes mostly intrastrand thymine dimers in DNA, the repair of which involves nucleotide excision. This pathway is induced by a rapid, transient, RecA-mediated SOS response that removes the damage and quickly returns the cell to its normal preirradiation status (9). In contrast, growth in the presence of MMC induces interstrand cross-links in the DNA, resulting in a more extended SOS response that may involve the differential expression of more than 1,000 genes (22). Thus, although SOS induction is the common outcome of both UV and MMC exposure in E. coli, the extent of the response is greater in MMC-treated cells. Therefore, it is possible that the rapid response after UV exposure cannot be fully provided by B. burgdorferi RecA in E. coli, whereas the longer-lived response to MMC can be fully complemented.
DNA damage-induced alterations in RecA structure promote an increased protein docking function which enhances the normally slow autocatalytic activity of LexA (25, 40). Rapid degradation of intracellular LexA by this coprotease activity of RecA in E. coli results in a 12-fold increase in recA expression and the coordinate induction of over 30 genes repressed by LexA (24). This is referred to as the SOS response (34). In E. coli, this response is induced by the persistence of single-stranded DNA (ssDNA), an indication of replisome inhibition at frequent DNA lesions (16, 24). This response appears to controlled by competition for ssDNA between single-stranded binding protein (SSB) and RecA. Under normal in vivo conditions, SSB prevents RecA from polymerizing onto ssDNA; however, when replication forks are stalled due to DNA damage, SSB assists RecA binding to ssDNA (24). The functional form of RecA is a polymerized filament around an ssDNA molecule that presents a LexA docking site deep in its groove (19, 55). Binding of LexA to this site promotes rapid autocleavage, which results in coordinate activation of the genes in the SOS regulon.
The B. burgdorferi genome does not encode an orthologue of LexA (13), and a scan of the regions upstream of recA and other genes that comprise the SOS regulon in E. coli did not reveal any consensus LexA binding sites in B. burgdorferi. Despite this, the B. burgdorferi RecA protein was able to facilitate DNA repair in E. coli, indicating that it could interact with E. coli LexA. Furthermore, B. burgdorferi RecA was also competent for induction of λ phage, which requires RecA-mediated λ cI repressor autocleavage. Indeed, λ phage induction mediated by RecA was enhanced >50-fold compared with that observed in E. coli DH5α complemented with the homologous RecA protein. This suggests that the B. burgdorferi protein harbors an enhanced coprotease activity. The presence of either of two point mutations, E39K and Q174K, in E. coli RecA results in increased coprotease activity (53). Interestingly, B. burgdorferi RecA contains both K39 and K174 in the wild-type sequence. These naturally occurring amino acid changes could explain the enhanced coprotease activity observed, as first suggested by Dew-Jager et al. (10).
The lack of a LexA protein has also been previously reported in Bacillus fragilis (17), Thiobacillus ferrooxidans (35), Porphyromonas gingivalis (12), and Acinetobacter calcoaceticus (18). In radiation-resistant Deinococcus radiodurans LexA is present, but cleavage is not enhanced by recA expression (30). The possible presence of an alternate protein(s) in B. burgdorferi that may provide LexA-like regulatory functions remains an open question.
The experiment whose results are shown in Fig. 4 demonstrated that B. burgdorferi cannot effectively repair UV-induced damage. Furthermore, several findings suggest that the RecA function in B. burgdorferi is different from the role that this protein plays in E. coli. RecA-mediated DNA repair is exerted through its cleavage of LexA. As described above, no lexA orthologue has been identified in B. burgdorferi. Additionally, UV irradiation of E. coli results increased expression of recA, but such induction does not occur in B. burgdorferi (Fig. 5). Thus, the role that RecA plays in normal B. burgdorferi biology remains to be clarified.
Little is known about the natural mechanism(s) of gene transfer, homologous recombination, and DNA repair in B. burgdorferi. Evidence suggests that vls, erp, and ospC loci undergo sequence variation that may require homologous recombination (45, 51, 57). Additionally, evolution of the B. burgdorferi genome by recombination between the linear chromosome and plasmids, as well as genetic exchange among various plasmids, has been suggested (6). Dykhuizen and Baranton estimated recombination frequencies for many B. burgdorferi isolates and concluded that this organism is mostly clonal and that there is limited genetic exchange between individual isolates (11). They hypothesized that B. burgdorferi contains a genetic transfer system that may permit the recombination of small (<1-kb) DNA fragments into its genome.
The results of the present study and those of Putteet-Driver et al. (33) demonstrate that B. burgdorferi RecA possesses both intermolecular and intramolecular recombination activities. It is, therefore, reasonable to suggest that this is the primary role of recA in B. burgdorferi. It is noteworthy that B. burgdorferi and T. pallidum both contain a reduced number of genes involved in recombination and DNA repair compared to E. coli. Interestingly, B. burgdorferi contains genes encoding only the RecBCD recombinational pathway, whereas only the RecF pathway genes are evident in T. pallidum (13, 14, 46). In E. coli, recombination is mediated primarily via proteins of the RecBCD pathway, and the function of the RecF pathway becomes evident only in recBCD mutants (23, 41, 42). The possible role that RecA may play in recombination in B. burgdorferi is currently under investigation.
Acknowledgments
We thank Guiqing Wang and Radha Iyer for many helpful discussions and with assistance in preparation of the manuscript and Adrienne Putteet-Driver, J. Zhong, and Alan Barbour for sharing their manuscript prior to publication.
This study is supported by in part by NIH grants AR41511 and AI45801 (to I.S.).
REFERENCES
- 1.Alekshun, M., M. Kashlev, and I. Schwartz. 1997. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene 186:227-235. [DOI] [PubMed] [Google Scholar]
- 2.Anguita, J., V. Thomas, S. Samanta, R. Persinski, C. Hernanz, S. W. Barthold, and E. Fikrig. 2001. Borrelia burgdorferi-induced inflammation facilitates spirochete adaptation and variable major protein-like sequence locus recombination. J. Immunol. 167:3383-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., D. R. Walker, and E. V. Koonin. 1999. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 27:1223-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber, W. 1983. A beginner's guide to lambda biology, p. 381-395. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 5.Boursaux-Eude, C., D. Margarita, J. Belfaiza, I. G. Old, and I. Saint Girons. 1998. Homologues of helicase genes priA and ruvAB of Borrelia burgdorferi, the Lyme borreliosis agent. Res. Microbiol. 149:235-245. [DOI] [PubMed] [Google Scholar]
- 6.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 7.Cloud, J. L., R. T. Marconi, C. H. Eggers, C. F. Garon, K. Tilly, and D. S. Samuels. 1997. Cloning and expression of the Borrelia burgdorferi lon gene. Gene 194:137-141. [DOI] [PubMed] [Google Scholar]
- 8.Concepcion, M. B., and D. R. Nelson. 2003. Expression of spoT in Borrelia burgdorferi during serum starvation. J. Bacteriol. 185:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Courcelle, J., and P. C. Hanawalt. 2001. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc. Natl. Acad. Sci. 98:8196-8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dew-Jager, K., W. Q. Yu, and W. M. Huang. 1995. The recA gene of Borrelia burgdorferi. Gene 167:137-140. [DOI] [PubMed] [Google Scholar]
- 11.Dykhuizen, D. E., and G. Baranton. 2001. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 12.Fletcher, H. M., R. M. Morgan, and F. L. Macrina. 1997. Nucleotide sequence of the Porphyromonas gingivalis W83 recA homolog and construction of a recA-deficient mutant. Infect. Immun. 65:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 14.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed]
- 15.Ge, Y., I. G. Old, I. Saint Girons, and N. W. Charon. 1997. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus sigma70 promoter. J. Bacteriol. 179:2289-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gigliani, F., C. Ciotta, M. F. Del Grosso, and P. A. Battaglia. 1993. pR plasmid replication provides evidence that single-stranded DNA induces the SOS system in vivo. Mol. Gen. Genet. 238:333-338. [DOI] [PubMed] [Google Scholar]
- 17.Goodman, H. J., J. R. Parker, J. A. Southern, and D. R. Woods. 1987. Cloning and expression in Escherichia coli of a recA-like gene from Bacteroides fragilis. Gene 58:265-271. [DOI] [PubMed] [Google Scholar]
- 18.Gregg-Jolly, L. A., and L. N. Ornston. 1994. Properties of Acinetobacter calcoaceticus recA and its contribution to intracellular gene conversion. Mol. Microbiol. 12:985-992. [DOI] [PubMed] [Google Scholar]
- 19.Horii, T., T. Ogawa, and H. Ogawa. 1981. Nucleotide sequence of the lexA gene of E. coli. Cell 23:689-697. [DOI] [PubMed] [Google Scholar]
- 20.Hubner, A., A. T. Revel, D. M. Nolen, K. E. Hagman, and M. V. Norgard. 2003. Expression of a luxS gene is not required for Borrelia burgdorferi infection of mice via needle inoculation. Infect. Immun. 71:2892-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlin, S., and L. Brocchieri. 1996. Evolutionary conservation of RecA genes in relation to protein structure and function. J. Bacteriol. 178:1881-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khil, P. P., and R. D. Camerini-Otero. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol. Microbiol. 44:89-105. [DOI] [PubMed] [Google Scholar]
- 23.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavery, P. E., and S. C. Kowalczykowski. 1992. A postsynaptic role for single-stranded DNA-binding protein in RecA protein-promoted DNA strand exchange. J. Biol. Chem. 267:9315-9320. [PubMed] [Google Scholar]
- 26.Lin, B., S. A. Short, M. Eskildsen, M. S. Klempner, and L. T. Hu. 2001. Functional testing of putative oligopeptide permease (Opp) proteins of Borrelia burgdorferi: a complementation model in opp− Escherichia coli. Biochim. Biophys. Acta 1499:222-231. [DOI] [PubMed] [Google Scholar]
- 27.Liveris, D., G. Wang, G. Girao, D. W. Byrne, J. Nowakowski, D. McKenna, R. Nadelman, G. P. Wormser, and I. Schwartz. 2002. Quantitative detection of Borrelia burgdorferi in 2-millimeter skin samples of erythema migrans lesions: correlation of results with clinical and laboratory findings. J. Clin. Microbiol. 40:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics, p. 150-156 and 268-275. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Miller, R. V., and T. A. Kokjohn. 1990. General microbiology of recA: environmental and evolutionary significance. Annu. Rev. Microbiol. 44:365-394. [DOI] [PubMed] [Google Scholar]
- 30.Narumi, I., K. Satoh, M. Kikuchi, T. Funayama, T. Yanagisawa, Y. Kobayashi, H. Watanabe, and K. Yamamoto. 2001. The LexA protein from Deinococcus radiodurans is not involved in RecA induction following gamma irradiation. J. Bacteriol. 183:6951-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohnishi, J., J. Piesman, and A. M. de Silva. 2001. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl. Acad. Sci. 98:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. K. Howell, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 33.Putteet-Driver, A. D., J. Zhong, and A. G. Barbour. 2004. Transgenic expression of RecA of the spirochetes Borrelia burgdorferi and Borrelia hermsii in Escherichia coli revealed differences in DNA repair and recombination phenotypes. J. Bacteriol. 186:2266-2274. [DOI] [PMC free article] [PubMed]
- 34.Radman, M. 1975. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis, p. 355-367. In P. S. R. Hanawalt (ed.), Molecular mechanisms for repair of DNA. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 35.Ramesar, R. S., V. Abratt, D. R. Woods, and D. E. Rawlings. 1989. Nucleotide sequence and expression of a cloned Thiobacillus ferrooxidans recA gene in Escherichia coli. Gene 78:1-8. [DOI] [PubMed] [Google Scholar]
- 36.Roberts, J. W., C. W. Roberts, and N. L. Craig. 1978. Escherichia coli recA gene product inactivates phage lambda repressor. Proc. Natl. Acad. Sci. 75:4714-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosa, P. A., T. Schwan, and D. Hogan. 1992. Recombination between genes encoding major outer surface proteins A and B of Borrelia burgdorferi. Mol. Microbiol. 6:3031-3040. [DOI] [PubMed] [Google Scholar]
- 38.Sartakova, M., E. Dobrikova, and F. C. Cabello. 2000. Development of an extrachromosomal cloning vector system for use in Borrelia burgdorferi. Proc. Natl. Acad. Sci. 97:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sassanfar, M., and J. W. Roberts. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol. 212:79-96. [DOI] [PubMed] [Google Scholar]
- 40.Slilaty, S. N., and J. W. Little. 1987. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc. Natl. Acad. Sci. 84:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, G. R. 1988. Homologous recombination in procaryotes. Microbiol. Rev. 52:1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, G. R. 1989. Homologous recombination in E. coli: multiple pathways for multiple reasons. Cell 58:807-809. [DOI] [PubMed] [Google Scholar]
- 43.Stamm, L. V., E. A. Parrish, and F. C. Gherardini. 1991. Cloning of the recA gene from a free-living leptospire and distribution of RecA-like protein among spirochetes. Appl. Environ. Microbiol. 57:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson, B., and K. Babb. 2002. LuxS-mediated quorum sensing in Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 70:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869-1879. [DOI] [PubMed] [Google Scholar]
- 46.Subramanian, G., E. V. Koonin, and L. Aravind. 2000. Comparative genome analysis of the pathogenic spirochetes Borrelia burgdorferi and Treponema pallidum. Infect. Immun. 68:1633-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sung, S. Y., J. V. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect. Immun. 68:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tchamedeu Kameni, A. P., E. Couture-Tosi, I. Saint-Girons, and M. Picardeau. 2002. Inactivation of the spirochete recA gene results in a mutant with low viability and irregular nucleoid morphology. J. Bacteriol. 184:452-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tilly, K., J. Fuhrman, J. Campbell, and D. S. Samuels. 1996. Isolation of Borrelia burgdorferi genes encoding homologues of DNA-binding protein HU and ribosomal protein S20. Microbiology 142:2471-2479. [DOI] [PubMed] [Google Scholar]
- 50.Tilly, K., R. Hauser, J. Campbell, and G. J. Ostheimer. 1993. Isolation of dnaJ, dnaK, and grpE homologues from Borrelia burgdorferi and complementation of Escherichia coli mutants. Mol. Microbiol. 7:359-369. [DOI] [PubMed] [Google Scholar]
- 51.Wang, G., A. P. van Dam, and J. Dankert. 1999. Evidence for frequent ospC gene transfer between Borrelia valaisiana sp. nov. and other Lyme disease spirochetes. FEMS Microbiol. Lett. 177:289-296. [DOI] [PubMed] [Google Scholar]
- 52.Wang, I. N., D. E. Dykhuizen, W. Qiu, J. J. Dunn, E. M. Bosler, and B. J. Luft. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, W. B., E. S. Tessman, and I. Tessman. 1988. Activation of protease-constitutive RecA proteins of Escherichia coli by rRNA and tRNA. J. Bacteriol. 170:4823-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Yu, X., and E. H. Egelman. 1993. The LexA repressor binds within the deep helical groove of the activated RecA filament. J. Mol. Biol. 231:29-40. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, J. R., and S. J. Norris. 1998. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect. Immun. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]