Abstract
Genetic diversity of T. gondii is a concern of many studies, due to the biological and epidemiological diversity of this parasite. The present study examined sequence variation in rhoptry protein 17 (ROP17) gene among T. gondii isolates from different hosts and geographical regions. The rop17 gene was amplified and sequenced from 10 T. gondii strains, and phylogenetic relationship among these T. gondii strains was reconstructed using maximum parsimony (MP), neighbor-joining (NJ), and maximum likelihood (ML) analyses. The partial rop17 gene sequences were 1375 bp in length and A+T contents varied from 49.45% to 50.11% among all examined T. gondii strains. Sequence analysis identified 33 variable nucleotide positions (2.1%), 16 of which were identified as transitions. Phylogeny reconstruction based on rop17 gene data revealed two major clusters which could readily distinguish Type I and Type II strains. Analyses of sequence variations in nucleotides and amino acids among these strains revealed high ratio of nonsynonymous to synonymous polymorphisms (>1), indicating that rop17 shows signs of positive selection. This study demonstrated the existence of slightly high sequence variability in the rop17 gene sequences among T. gondii strains from different hosts and geographical regions, suggesting that rop17 gene may represent a new genetic marker for population genetic studies of T. gondii isolates.
1. Introduction
Toxoplasma gondii can infect all warm-blooded vertebrates, including mammals and birds [1–3]. Genetic diversity of T. gondii is widespread due to the biological and epidemiological diversity of this parasite. T. gondii isolates can be clustered into six major clades [4], and genetic diversity of T. gondii is especially common in South America [4]. Utilizing 11 genetic markers, T. gondii isolates in North America and Europe are grouped into four major clonal lineage types (I, II, III, and 12) [5, 6] using PCR-RFLP.
Rhoptry kinases are involved in mediating pathogenesis of T. gondii [7], and they are also master regulators that manipulate the host inflammatory responses [8, 9]. T. gondii rhoptry protein 17 (ROP17), a member of the ROP2 subfamily [10], was predicted to have a cellular localization on the parasitophorous vacuole membrane (PVM), which may participate in the manipulation of the host signalling pathways [9]. Previous studies have shown the existence of sequence variation in some ROP genes, such as rop7, rop9, rop13, and rop38 [11–14]. However, it is yet to be known whether sequence diversity exists in rop17 gene of T. gondii. The objective of the present study was to examine sequence variation in rop17 gene among T. gondii strains representing different genotypes and host and origins.
2. Materials and Methods
2.1. T. gondii Isolates
Ten T. gondii strains collected from different hosts and locations were used for analysis in this study (Table 1). These strains have been genotyped and their genomic DNA has been prepared as described previously [15–17].
Table 1.
Details of Toxoplasma gondii strains used in the present study.
| Strain | Host | Geographical origin | Genotype∗ |
|---|---|---|---|
| GT1 | Goat | United States | Reference, Type I |
| PTG | Sheep | United States | Reference, Type II, ToxoDB number 1 |
| CTG | Cat | United States | Reference, Type III, ToxoDB number 2 |
| TgCatBr5 | Cat | Brazil | Reference, ToxoDB number 19 |
| TgCatBr64 | Cat | Brazil | Reference, ToxoDB number 111 |
| TgCgCa1 | Cougar | Canada | Reference, ToxoDB number 66 |
| TgToucan (TgRsCr1) | Toucan | Costa Rica | Reference, ToxoDB number 52 |
| TgPLh | Pig | China | Type I, ToxoDB number 10 |
| QHO | Sheep | China | Type II, ToxoDB number 1 |
| TgWtdSc40 | White-tailed deer | United States | Type 12, ToxoDB number 5 |
2.2. Amplification of rop17 Genes and Sequencing
The rop17 gene was amplified by PCR. Two primers were designed based on the rop17 sequence of T. gondii RH strain available in GenBank (accession number: KC997178): ROP17F, 5′-AGGACAACACTAGGTAGCGAGAACC-3′, and ROP17R, 5′-TGGCGAAGTCAAGAGACGACGCAG-3′. Each reaction was performed in a total volume of 25 μL containing 12.5 μL Premix Taq (TaKaRa, Dalian, China), ROP17F (20 pmol) 0.25 μL, ROP17R (20 pmol) 0.25 μL, template DNA (200 ng) 2 μL, and ddH2O 8 μL, and the reaction conditions were 94°C for 5 min, then 35 cycles of 30 sec at 94°C, 30 sec at 55°C, and 1 min 20 s at 72°C, and a final extension at 72°C for 10 min. All the PCR products were then cloned into pMD18-T vector (TaKaRa, China) after purification using the DNA purification kit (TIANGEN, China) and then sequenced by Songon Biotech Co., Ltd. (Shanghai, China).
2.3. Sequence Analysis and Reconstruction of Phylogenetic Relationships
The rop17 gene sequences of different T. gondii strains were aligned using Multiple Sequence Alignment Program, Clustal X 1.83 [18], and the sequence differences were determined according to Chilton et al. [19] and Zhao et al. [20]. Phylogenetic reconstruction was based on the rop17 gene sequences determined in the present study plus the corresponding sequences of strains TgC7, PRU, and RH available in GenBank (accession numbers: KC997176, KC997177, and KC997178) using three inference methods, namely, neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP), using the sequence of Neospora caninum (NCLIV_027930) as the outgroup. All the analyses were conducted following previous studies [20, 21]. Phylograms were drawn using the Tree View program version 1.65 [22].
3. Results and Discussion
The length of the rop17 genes from all examined T. gondii isolates was 1375 bp and A+T contents varied from 49.45% to 50.11%. The alignment of 10 rop17 sequences plus the corresponding sequences of the RH, PRU, and TgC7 strains available in GenBank revealed nucleotide polymorphisms at 33 positions, with an intraspecific variation of 0–2.1%. The genetic diversity in rop17 gene was higher than our previous studies for PLP1 [23], ROP7 [11], eIF4A [24], and MIC13 [25] genes and the whole genome, secretome, and kinome of T. gondii [8]. 16 variable positions were identified as transitions and the rest variable nucleotides were classified as transversions, and no deletions were detected in the 13 rop17 gene sequences.
Phylogeny reconstruction using MP, NJ, and ML analyses revealed two major clusters (Figure 1(a)). Topologies of all trees based on nucleotide sequences inferred by three different methods were similar, with only the small difference of bootstrap values. The classical genotypes II and III and atypical Type 12 strain were clustered in one clade. The subtree of NJ analysis further showed that genotype III (strain CTG) was separated from other strains which were supported by bootstrap analysis, and the atypical Type 12 (TgWtSc40 strain) was closely related to classical genotype II (strain PRU) (Figure 1(b)). T. gondii genotype II is one of the parental lineage of Type 12 based on the analysis of the inheritance of multilocus genotypes [6, 26]. The somewhat close relationship between Type II and Type 12 strains coincided with analyses of UPRT and SAG1 loci [6]. All the strains belonging to genotype I in this study were clustered together, including strain TgPLh and typical strains GT1 and RH. Atypical strains TgCat1, TgToucan, TgCatBr64, and TgCatBr5 were phylogenetically clustered more closely with Type I strains. Of these, TgCatBr64 and TgCatBr5 strains which originated from cats in Brazil were grouped together. Based on the limited T. gondii strains examined in the present study, the rop17 gene sequences could distinguish the major clonal lineages, but not all, showing the weak differentiation of T. gondii genotypes compared to analyses using GRA5, GRA6, and AK gene sequences as genetic markers [27–29]. Further validation of the rop17 gene sequences as genetic marker is warranted by sampling more T. gondii strains from wider geographical locations and more hosts.
Figure 1.
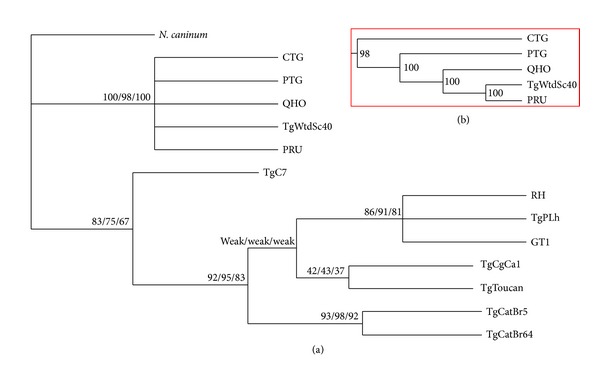
Phylogenetic analysis of 13 Toxoplasma gondii strains based on analysis of the rop17 gene sequences. The tree was built by neighbor-joining (NJ), maximum likelihood (ML), and maximum parsimony (MP) analyses. The numbers at notes indicate bootstrap values resulting from different analyses in the order MP/NJ/ML. (a) The much higher genetic divergence in rop17 revealed two major clusters (denoted by I and II). (b) Subtree in the red box showing results of analysis using neighbor-joining (NJ).
The analyses of sequence variations in nucleotides and amino acids among different strains showed high ratio of nonsynonymous to synonymous polymorphisms (>1), suggesting that T. gondii rop17 shows signs of positive selection, although more isolates will be required to determine whether rop17 gene is under selection at the population level. Under the immunized stresses of host cells, the positive selection occurring in rop17 gene may increase stress resistance. Ongoing positive selection is also found in several polymorphic dense granule (GRA) antigens [30, 31] and some other ROPs [8].
4. Conclusion
In summary, the present study demonstrated the existence of slightly high sequence variability in the rop17 gene sequences among T. gondii strains from different hosts and regions, which may be explored as a new genetic marker for population genetic studies of T. gondii isolates, and contributed to discovery of the new strategies for vaccination, treatment, or diagnosis.
Acknowledgments
Project support was provided by National Natural Science Foundation of China (Grant nos. 31228022, 31172316, and 31230073) and the Science Fund for Creative Research Groups of Gansu Province (Grant no. 1210RJIA006). Associate Professor Chunlei Su at the Department of Microbiology, the University of Tennessee, Knoxville, USA, is gratefully thanked for providing reference T. gondii strains.
Conflict of Interests
The authors declare that there is no conflict of interests in this paper.
References
- 1.Dubey JP, editor. Toxoplasmosis of Animals and Humans. Boca Raton, Fla, USA: CRC Publishing; 2010. [Google Scholar]
- 2.Huang S, Cong W, Zhou P, et al. First report of genotyping of Toxoplasma gondii isolates from wild birds in China. Journal of Parasitology. 2012;98(3):681–682. doi: 10.1645/GE-3038.1. [DOI] [PubMed] [Google Scholar]
- 3.Tian Y-M, Dai F-Y, Huang S-Y, et al. First report of Toxoplasma gondii seroprevalence in peafowls in Yunnan Province, Southwestern China. Parasites & Vectors. 2012;5(1, article 205) doi: 10.1186/1756-3305-5-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su C, Khan A, Zhou P, et al. Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(15):5844–5849. doi: 10.1073/pnas.1203190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su C, Zhang X, Dubey JP. Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. International Journal for Parasitology. 2006;36(7):841–848. doi: 10.1016/j.ijpara.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Khan A, Dubey JP, Su C, Ajioka JW, Rosenthal BM, Sibley LD. Genetic analyses of atypical Toxoplasma gondii strains reveal a fourth clonal lineage in North America. International Journal for Parasitology. 2011;41(6):645–655. doi: 10.1016/j.ijpara.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sibley LD, Qiu W, Fentress S, Taylor SJ, Khan A, Hui R. Forward genetics in Toxoplasma gondii reveals a family of rhoptry kinases that mediates pathogenesis. Eukaryotic Cell. 2009;8(8):1085–1093. doi: 10.1128/EC.00107-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peixoto L, Chen F, Harb OS, et al. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host and Microbe. 2010;8(2):208–218. doi: 10.1016/j.chom.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melo MB, Jensen KDC, Saeij JPJ. Toxoplasma gondii effectors are master regulators of the inflammatory response. Trends in Parasitology. 2011;27(11):487–495. doi: 10.1016/j.pt.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laliberté J, Carruthers VB. Host cell manipulation by the human pathogen Toxoplasma gondii . Cellular and Molecular Life Sciences. 2008;65(12):1900–1915. doi: 10.1007/s00018-008-7556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Lu P, Xu MJ, Ren D. Sequence variation in TgROP7 gene among Toxoplasma gondii isolates from different hosts and geographical regions. African Journal of Biotechnology. 2012;11(25):6658–6661. [Google Scholar]
- 12.Wang PY, Lu P, Xu MJ, Li J. Genetic diversity among Toxoplasma gondii isolates from different hosts and geographical locations revealed by analysis of ROP13 gene sequences. African Journal of Biotechnology. 2012;11(25):6662–6665. [Google Scholar]
- 13.Chen J, Fang S, Li Z, Zhou D, Liu G, Zhu X. Sequence variation in ROP9 gene among Toxoplasma gondii strains from different hosts and geographical locations. Journal of Animal and Veterinary Advances. 2012;11(22):4288–4291. [Google Scholar]
- 14.Xu Y, Zhang NZ, Chen J, et al. Toxoplasma gondii rhoptry protein 38 gene: sequence variation among isolates from different hosts and geographical locations. Genetics and Molecular Research. 2014 doi: 10.4238/2014.January.14.3. [DOI] [PubMed] [Google Scholar]
- 15.Zhou P, Zhang H, Lin R, et al. Genetic characterization of Toxoplasma gondii isolates from China. Parasitology International. 2009;58(2):193–195. doi: 10.1016/j.parint.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Zhou P, Nie H, Zhang L, et al. Genetic characterization of toxoplasma gondii isolates from pigs in China. Journal of Parasitology. 2010;96(5):1027–1029. doi: 10.1645/GE-2465.1. [DOI] [PubMed] [Google Scholar]
- 17.Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;137(1):1–11. doi: 10.1017/S0031182009991065. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chilton NB, Gasser RB, Beveridge I. Differences in a ribosomal DNA sequence of morphologically indistinguishable species within the Hypodontus macropi complex (Nematoda: Strongyloidea) International Journal for Parasitology. 1995;25(5):647–651. doi: 10.1016/0020-7519(94)00171-j. [DOI] [PubMed] [Google Scholar]
- 20.Zhao GH, Li J, Song HQ, et al. A specific PCR assay for the identification and differentiation of Schistosoma japonicum geographical isolates in mainland China based on analysis of mitochondrial genome sequences. Infection, Genetics and Evolution. 2012;12(5):1027–1036. doi: 10.1016/j.meegid.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Strimmer K, Von Haeseler A. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Molecular Biology and Evolution. 1996;13(7):964–969. [Google Scholar]
- 22.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 23.Yan HK, Song HQ, Zhou Y, et al. Sequence variation in perforin-like protein 1 gene among six Toxoplasma gondii strains. Journal of Animal and Veterinary Advances. 2011;10(17):2244–2247. [Google Scholar]
- 24.Chen J, Fang SF, Zhou DH, Li ZY, Liu GH, Zhu XQ. Sequence variation in the Toxoplasma gondii eIF4A gene among strains from different hosts and geographical locations. Genetics and Molecular Research. 2014;13(2):3356–3361. doi: 10.4238/2014.April.29.14. [DOI] [PubMed] [Google Scholar]
- 25.Ren D, Zhou DH, Xu MJ, et al. Sequence variation in Toxoplasma gondii MIC13 gene among isolates from different hosts and geographical locations. African Journal of Microbiology Research. 2012;6(6):1333–1337. [Google Scholar]
- 26.Yu L, Shen J, Su C, Sundermann CA. Genetic characterization of Toxoplasma gondii in wildlife from Alabama, USA. Parasitology Research. 2013;112(3):1333–1336. doi: 10.1007/s00436-012-3187-0. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Li Z-Y, Zhou D-H, Liu G-H, Zhu X-Q. Genetic diversity among Toxoplasma gondii strains from different hosts and geographical regions revealed by sequence analysis of GRA5 gene. Parasites and Vectors. 2012;5(1, article 279) doi: 10.1186/1756-3305-5-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fazaeli A, Carter PE, Darde ML, Pennington TH. Molecular typing of Toxoplasma gondii strains by GRA6 gene sequence analysis. International Journal for Parasitology. 2000;30(5):637–642. doi: 10.1016/s0020-7519(00)00036-9. [DOI] [PubMed] [Google Scholar]
- 29.Boughattas S, Ben-Abdallah R, Siala E, et al. Tunisian Toxoplasma gondii strains genotyping by the use of AK69 marker. Parasites and Vectors. 2011;4(1, article 167) doi: 10.1186/1756-3305-4-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs D, Vercammen M, Saman E. Evaluation of recombinant dense granule antigen 7 (GRA7) of Toxoplasma gondii for detection of immunoglobulin G antibodies and analysis of a major antigenic domain. Clinical and Diagnostic Laboratory Immunology. 1999;6(1):24–29. doi: 10.1128/cdli.6.1.24-29.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong J, Grigg ME, Uyetake L, Parmley S, Boothroyd JC. Serotyping of Toxoplasma gondii infections in humans using synthetic peptides. Journal of Infectious Diseases. 2003;187(9):1484–1495. doi: 10.1086/374647. [DOI] [PubMed] [Google Scholar]


