Abstract
Inherited forms of cataract are a clinically important and genetically heterogeneous cause of visual impairment that usually present at an early age with or without systemic and/or other ocular abnormalities. Here we have identified a new locus for inherited cataract and high-tension glaucoma with variable anterior segment defects, and characterized an underlying mutation in the gene coding for transient receptor potential cation channel, subfamily M, member-3 (TRPM3, melastatin-2). Genome-wide linkage analysis mapped the ocular disease locus to the pericentric region of human chromosome 9. Whole exome and custom-target next-generation sequencing detected a heterozygous A-to-G transition in exon-3 of TRPM3 that co-segregated with disease. As a consequence of alternative splicing this missense mutation was predicted to result in the substitution of isoleucine-to-methionine at codon 65 (c.195A>G; p.I65 M) of TRPM3 transcript variant 9, and at codon 8 (c.24A>G; p.I8 M) of a novel TRPM3 transcript variant expressed in human lens. In both transcript variants the I-to-M substitution was predicted in silico to exert damaging effects on protein function. Furthermore, transient expression studies of a recombinant TRPM3-GFP reporter product predicted that the I-to-M substitution introduced an alternative translation start-site located 89 codons upstream from the native initiator methionine found in eight other TRPM3 transcript variants (1–8). Collectively, these studies have provided the first evidence that TRPM3 is associated with inherited ocular disease in humans, and further provide support for the important role of this cation channel in normal eye development.
Introduction
Mendelian forms of cataract constitute a clinically and genetically heterogeneous disorder of the ocular lens that occurs with an estimated prevalence of 1–3 cases per 10,000 births [1], [2]. Typically, inherited cataract presents with an early onset ranging from birth (congenital) or infancy (infantile) into the fourth decade and, under slit-lamp examination, exhibits considerable phenotypic variation with respect to the location, shape, density and progression of opacities within the lens [3]. Congenital and infantile forms of cataract are a clinically important cause of impaired form-vision development (deprivation amblyopia) that despite surgical intervention can pose a lifelong risk of progressive visual impairment, notably as a result of glaucoma [4]–[6]. Cataract may be inherited either as an isolated lens phenotype, often with autosomal dominant transmission and high penetrance, or as part of many genetic syndromes and metabolic disorders (OMIM.org) involving systemic abnormalities (e.g. galactosemia, MIM230400) and/or other ocular defects (e.g. anterior segment dysgenesis, MIM107250). So far, genetic studies have identified over 50 genes and loci for inherited cataract, with or without other ocular signs, including those for α-crystallins, β/γ-crystallins, α-connexins and other lens membrane or cytoskeleton proteins, several transcription factors (e.g. HSF4, PITX3, MAF), and an increasing spectrum of functionally diverse genes (e.g. EPHA2, CHMP4B, TDRD7, FYCO1) [2], [7]. Consequently, no clear genotype-phenotype correlation has been observed for inherited cataract rendering both clinical classification and molecular diagnosis challenging.
Transient receptor potential (TRP) channel genes encode a superfamily of variably selective cation influx channels that function as polymodal cellular sensors in diverse physiological processes including the perception of light, temperature, pressure and pain [8]–[10]. The TRP superfamily derives its name from the visually impaired trp-mutant of Drosophila that was found to exhibit a transient photoreceptor response to steady light rather than the sustained electroretinogram observed in wild-type flies [11]. In mammals, 28 TRP channel genes have been identified, and based on sequence similarity they are divided into six sub-families, namely; canonical (TRPC1–7), vanilloid (TRPV1–6), melastatin (TRPM1–8), ankyrin (TRPA1), polycystin (TRPP1–3), and mucolipin (TRPML1–3) [9]. All TRP proteins share a predicted transmembrane topology with six-transmembrane domains including the channel pore-forming loop located between the 5th and 6th domains. The cytosolic N- and C- termini are variable in length and contain different combinations of functional sub-domains including; N-terminal ankyrin repeats, calmodulin-binding motifs, TRP-boxes, coiled-coil regions, and C-terminal enzyme domains [12]. Functional TRP channels are assembled from tetramers of identical (homomeric) or similar (heteromeric) TRP subunits, and exhibit highly variable permeability to Ca2+ and several other monovalent and divalent cations [9]. TRP channels exhibit wide tissue distribution in both plasma-membranes and intracellular membranes particularly from brain and kidney [13]–[14].
So far mutations in at least ten TRP genes spanning all sub-families have been associated with over a dozen human genetic diseases, referred to as ‘channelopathies’ [15]–[17]. These include skeletal dysplasias, sensory neuropathies and spinal muscular atrophies (TRPV4, MIM 605427), kidney diseases (TRPC6, MIM 603652; TRPP1, MIM 601313), mucolipidosis type-IV (TRPML1, MIM 605248), familial episodic pain syndrome (TRPA1, MIM 604775), and Olmsted syndrome (TRPV3, MIM 607066). Within the melastatin sub-family, mutations have been implicated in Guamanian amyotropic lateral sclerosis-parkinsonism/dementia (TRPM7, MIM 605692), progressive familial heart block type-1B (TRPM4; MIM 604559), hypomagnesemia with secondary hypocalcemia (TRPM6; MIM 602014), and congenital stationary night blindness (CSNB) in humans (TRPM1, MIM 603576) and in horses homozygous for Leopard Complex (LP) coat spotting [18]–[22]. TRPM3 (melastatin-2), which is most phylogenetically conserved with TRPM1 (melastatin-1), has not been unambiguously linked with inherited human disease. Rare deletions involving TRPM3 have been reported in cases of Kabuki syndrome and autism, whereas, common non-coding variants in TRPM3 have been tentatively associated with longevity and elevated levels of low-density lipoprotein cholesterol and triglycerides [23]–[26].
Here we map, identify, and characterize a mutation in TRPM3 associated with inherited cataract and glaucoma linked to human chromosome 9q.
Results
A new locus for cataract and glaucoma maps to human chromosome 9
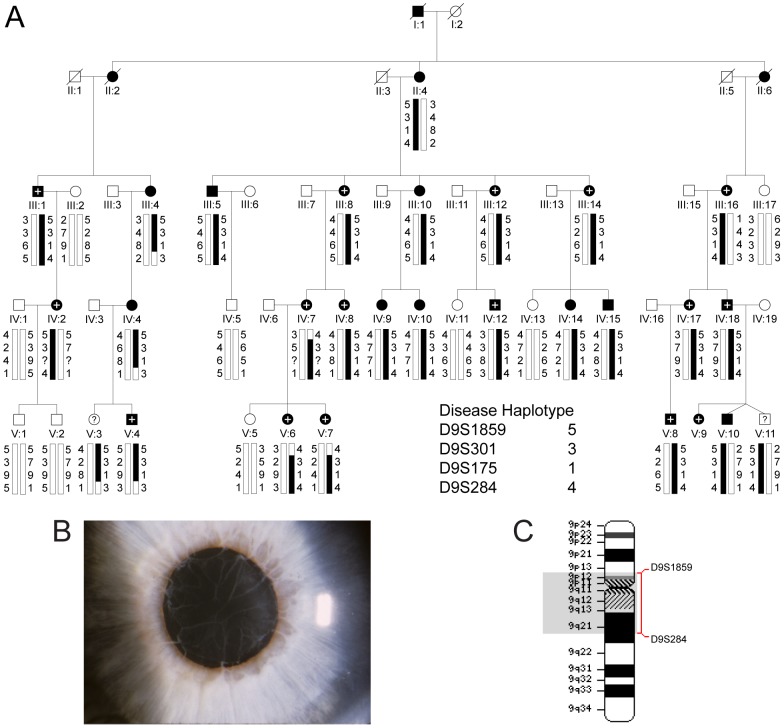
We investigated a 5-generation Caucasian-American pedigree mostly from the mid-west United States segregating cataract and glaucoma with autosomal dominant transmission (Figure 1a; Table S1). Review of ophthalmic records indicated that the cataract was usually bilateral and generally described as congenital, infantile or developmental, with age-at-diagnosis varying from birth (congenital) to 35 years, and age-at-surgery in the first eye ranging from 4 to 40 years. No slit-lamp images of the cataract were available; however, in affected individual IV:17 from the pedigree opacities were described as punctate cortical, and in affected individual V:4 posterior sub-capsular opacities were documented by slit-lamp examination in the medical record. All of the family members with cataract had visually significant lens opacities affecting best corrected visual acuity and underwent lens extraction. Of the 25 relatives with cataract, 15 (60%) were also diagnosed with glaucoma. Glaucoma diagnosis was supported by significantly elevated intraocular pressure (IOP) >30 mm Hg and, in some cases, consistent visual field and/or optic nerve abnormalities. We note that our records of current glaucoma status for two of the undiagnosed individuals (V:10, V:11) are incomplete due to unsuccessful follow-up attempts. Age-at-diagnosis for glaucoma ranged from birth into the fifth decade, and age-at surgery when performed varied from 11-30 years. Glaucoma was variously described in four individuals (III:16, IV:18, V:4, V:9) as congenital, infantile, juvenile, or developmental open-angle by gonioscopic findings. However, in one affected individual (III:1) chronic angle-closure glaucoma was suspected, and in two others (III:14, IV:2) secondary glaucoma was suspected. Typically, cataract was diagnosed before high-tension glaucoma (IOP>30 mm Hg). However, in one case (III:1) glaucoma was diagnosed before cataract, and one individual (III:16) was diagnosed simultaneously with congenital cataract and glaucoma.
Figure 1. Linkage analysis of autosomal dominant cataract and glaucoma co-segregating in a 5-generation family.
(A) Pedigree and haplotype analysis showing segregation of markers across chromosome 9cen, listed in descending order from the short-arm telomere (9p-tel). Affected males (filled squares) and females (filled circles) that were diagnosed with cataract and glaucoma are indicated with a + symbol. (B) Ocular phenotype of affected individual V:4 presenting with high-tension glaucoma and persistent pupillary membrane characterized by fibrous strands extending from the iris border across the pupil. (C) Partial ideogram of chromosome 9 showing cytogenetic location of the ocular disease locus. Note, cytogenetic band regions p11.1, q11 and much of q12 are gene-poor centromeric regions, flanked by gene-rich regions in bands p11.2–p13.3 on the short arm and q13–q21.31 on the long arm.
In addition to cataract and glaucoma several other ocular findings were reported in certain family members (Table S1). Bilateral persistent pupillary membrane (PPM) was diagnosed in affected male V:4 (Fig. 1b) and his older sister (V:3). However, the latter has not been diagnosed with cataract or glaucoma at age 24 years raising the possibility of incomplete penetrance. Retinal detachment was reported in at least three affected individuals (IV:2, IV:18, V:9), and in one case (V:10) retinopathy of prematurity (ROP) was documented. However, the ROP is likely to be a coincidental finding. Finally, in one individual (V:9), who was unavailable for genetic testing, the ocular spectrum included suspected infantile glaucoma, bilateral Haab's striae (horizontal breaks in Descemet's membrane), anterior segment dysgenesis, megalocornea, mild corectopia, and PPM. However, intra-ocular pressure (IOP) and fundus (disc) were normal, and cataract was absent at eight years of age. Overall, while early-onset cataract and atypical high-tension glaucoma were commonly observed in this family, other ocular abnormalities that particularly affect the anterior segment may co-exist.
For genome-wide linkage analysis 24 consenting members of the family (19 affected and 5 unaffected) were genotyped by means of a linkage panel comprising 6090 SNP markers uniformly spaced at an average genetic distance of 0.58 cM. Using parametric two-point analysis we detected significant evidence of linkage at SNP markers rs1333342 (pLOD = 4.53), rs735914 (pLOD = 3.72), and rs713478 (pLOD = 3.45) located on chromosome 9q (Figure S1a). Similarly, parametric multipoint analysis detected strong evidence of linkage between SNP markers rs2031197-(∼333.4 kb)-rs1333342 (pLOD = 3.31) on 9q (Figure S1b). Haplotype analysis of the pedigree further identified a common disease interval spanning the pericentric region (9cen), rs987187-(∼46 Mb)-rs2378592 (chr9: 38,375,215-84,495,227 bp) that co-segregated with the disease in all affected relatives (Table S2).
To validate the SNP disease interval from the genome-wide linkage scan, all 36 relatives were re-genotyped with microsatellite markers. We obtained strong confirmation of linkage to 9q (Table 1) using markers D9S175 (Zmax = 8.05, θmax = 0), D9S301 (Zmax = 7.59, θmax = 0), and D9S1876 (Zmax = 6.42, θmax = 0). Haplotype analysis detected an affected female (III:4) and three of her descendents (IV:4, V:3, V:4) who were recombinant at D9S284, and three affected females (IV:7, V:6, V:7) who were recombinant at D9S1859 (Figure 1a). However, no further obligate recombinants were detected at seven intervening markers (Table 1), placing the disease locus in the reduced physical interval D9S1859-(∼41.5 Mb)-D9S284.
Table 1. Two-point Lod scores (Z) for linkage between the ocular disease locus and markers on chromosome 9 listed in physical distance (Mb) from the short-arm telomere (9p-tel).
| Marker (recombinant) | Mb | Z at recombination fraction (θ) = | |||||||
| 0.00 | 0.05 | 0.10 | 0.20 | 0.30 | 0.40 | ZMax | ΘMax | ||
| D9S1804 | 35.96 | -∞ | 3.98 | 3.79 | 3.00 | 1.99 | 0.88 | 3.98 | 0.05 |
| D9S1859 (IV:7, V:6, V:7) | 36.63 | -∞ | 4.11 | 3.94 | 3.17 | 2.14 | 0.99 | 4.11 | 0.05 |
| D9S1874 | 37.22 | 6.76 | 6.13 | 5.47 | 4.08 | 2.62 | 1.17 | 6.76 | 0.00 |
| D9S2148 | 38.30 | 5.18 | 4.76 | 4.27 | 3.19 | 1.99 | 0.81 | 5.18 | 0.00 |
| rs2073478 (V:6) | 38.40 | -∞ | 0.16 | 0.32 | 0.34 | 0.26 | 0.14 | 0.36 | 0.16 |
| TRPM3 (c.195A>G) | 73.48 | 9.51 | 8.75 | 7.96 | 6.23 | 4.30 | 2.14 | 9.51 | 0.00 |
| D9S301 | 73.80 | 7.59 | 6.91 | 6.19 | 4.67 | 3.04 | 1.35 | 7.59 | 0.00 |
| D9S1876 | 75.23 | 6.42 | 5.86 | 5.28 | 4.07 | 2.78 | 1.44 | 6.42 | 0.00 |
| D9S175 | 77.95 | 8.05 | 7.37 | 6.65 | 5.12 | 3.48 | 1.73 | 8.05 | 0.00 |
| D9S284 (III:4) | 78.09 | -∞ | 6.37 | 5.94 | 4.68 | 3.20 | 1.60 | 6.44 | 0.03 |
| D9S276 | 78.51 | -∞ | 4.45 | 4.19 | 3.28 | 2.14 | 0.89 | 4.46 | 0.04 |
| D9S1122 | 79.69 | -∞ | 2.39 | 2.69 | 2.52 | 1.93 | 1.05 | 2.71 | 0.12 |
| D9S1123 | 80.42 | -∞ | 2.84 | 2.77 | 2.29 | 1.63 | 0.87 | 2.84 | 0.06 |
To further refine the disease interval we have genotyped the existing critical recombinant individuals (III:4, IV:4, IV:7, V:3, V:4, V:6, V:7) with additional SNP markers located within the boundaries of the microsatellite interval using conventional Sanger sequencing. Critical affected individual V:6 was also found to be recombinant at SNP marker rs2073478 (G/T), which is located within ALDH1B1 approximately 1.77 Mb centromeric to D9S1859 (Table 1). However, no further recombinants were detected at more centromeric SNP markers indicating that the disease locus lay in the refined physical interval rs2073478-(∼39.7 Mb)-D9S284 (chr9: 38,396,065-78,086,081 bp).
Our disease interval spans the centromere and excludes the adjacent locus for autosomal recessive adult-onset progressive pulverulent cataract (CTPL1/CAAR) that lies in the interval D9S1123-(∼10 Mb)-D9S257 on 9q [27], [28]. In addition the gene coding for Tudor domain containing-7 (TDRD7, Gene ID: 23424), which has recently been associated with autosomal recessive cataract [29], lies approximately 20 Mb telomeric (chr9: 100,174,302-100,258,407 bp) to the disease interval and was excluded as the causative gene.
Exome and custom-target sequencing identifies a novel missense mutation in TRPM3
According to the NCBI Map Viewer database (Build 37.3) the refined disease interval contained over 270 positional candidate genes, none of which had been previously associated with eye disease in humans. In order to scan genes in the interval for mutations we undertook whole-exome and custom-target next-generation sequencing of an affected individual (III:16) and an affected-parent-child-unaffected-sibling trio (IV:7, V:5, V:6), respectively, from the pedigree. For exome sequencing (III:16), 96.2% of total paired-end reads (133,419,338) were mapped to the genome, and 78% of mapped reads were present in the captured exome, of which 95.5% achieved a coverage ≥5X. Similarly, for custom-target sequencing (IV:7, V:5, V:6), >98% of total reads (average 22,175,122) were mapped to the target interval on chromosome 9, and >61% of mapped reads were present in the captured target of which >98% achieved a coverage of ≥5X with no unexpected gaps.
Combined, both strategies identified the same list of four heterozygous, exonic variants (three non-synonymous and one frame-shift deletion) within the disease interval in all three index affected relatives (III:16, IV:7, V:6) that were not present in the unaffected relative (V:5) (Table 2). Of these variants, three had known reference sequence (rs) numbers cited in the dbSNP or 1000Genomes databases with no known clinical significance reported. The minor allele frequency (MAF) of two of the non-synonymous variants, rs143826416 (FAM75A3) and rs41310055 (GDA), were 43.4% and 1.7%, respectively. Both variants were predicted to be benign using PolyPhen-2, and rs41310055 (GDA) was also present on the Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS). No MAF was retrieved for the frame-shift variant, rs200487787 (CNTNAP3B), and it was not present on the EVS. However, Sanger sequencing confirmed that the frame-shift variant did not co-segregate with disease in the pedigree (data not shown). Taken overall, these observations effectively exclude three of the four variants as causative for disease.
Table 2. Sequencing variants detected in affected but not unaffected members of the family within the ocular disease interval on chromosome 9 (Figure 1C).
| Chr | Position (bp) | Gene Region | Gene Symbol | Nucleotide Variant | Protein Variant | Affected + Variant | Unaffected + Variant | Translation Impact | SIFT Prediction (Score) | PolyPhen-2 Prediction (Score) | dbSNP ID | MAF (%) | Found on EVS |
| 9 | 43,62,5382 | Exonic | FAM75A3 | c.3305C>T | p.P1102L | 3 | 0 | Nonsynonymous | Tolerated (0.06) | Benign (0.395) | rs143826416 | 43.4 | No |
| 9 | 43,844,265 | Exonic | CNTNAP3B | c.1599delG | p.L534fs* | 3 | 0 | Frameshift | rs200487787 | None | No | ||
| 9 | 73,479,410 | Exonic; 5′UTR | TRPM3 | c.195A>G | p.I65M | 3 | 0 | Nonsynonymous | Intolerant (0.00) | Probably Damaging (1.000) | Novel | None | No |
| 9 | 74,865,703 | Exonic; 3′UTR | GDA | c.1399C>T | p.P467S | 3 | 0 | Nonsynonymous | Intolerant (0.00) | Benign (0.000) | rs41310055 | 1.7 | Yes |
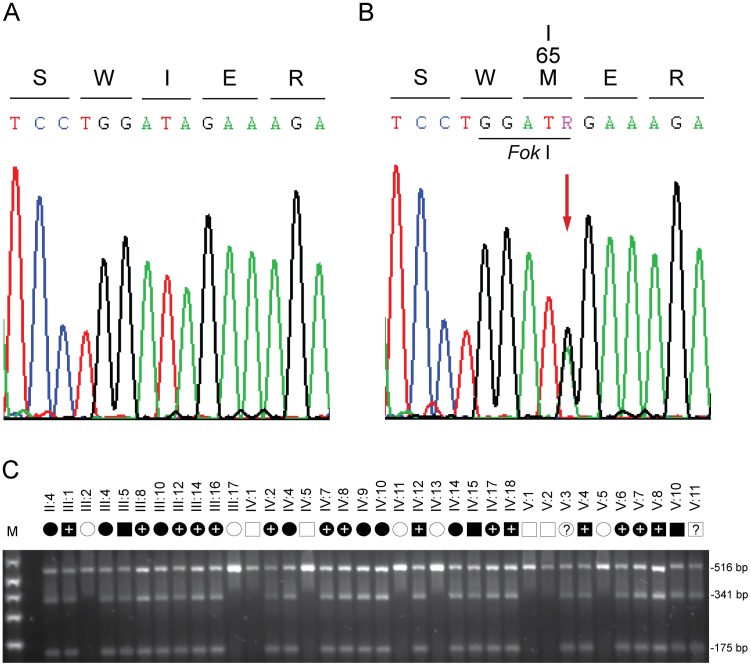
The remaining non-synonymous variant was novel, and comprised a heterozygous A>G transition that introduced a Fok 1 restriction site located in exon-3 of the gene coding for transient receptor potential cation channel subfamily M, member-3 (TRPM3). This missense change occurred at nucleotide position 195 from the first base (A) of the translation start codon in the cDNA sequence for TRPM3 transcript variant-9 (c.195A>G). Sanger sequencing confirmed the presence of the c.195A>G change in exon-3 from the three index affected relatives (Figure 2b), and excluded mutations in other exons and splice-sites of TRPM3. Allele-specific PCR amplification and Fok 1 restriction fragment length analysis confirmed that the c.195A>G change co-segregated with affected but not unaffected relatives across the family (Figure 2c). Moreover, when we tested the c.195A>G change as a bi-allelic marker with a notional frequency of 1% in a two-point LOD score analysis we obtained highly significant confirmation of linkage to TRPM3 (Zmax = 9.51, θmax = 0.0) (Table 1). Finally, this variant was not detected in a panel of 192 normal unrelated Caucasian individuals (384 chromosomes) using allele-specific Fok 1 restriction fragment analysis described in Figure 2c (data not shown). Combined, our sequence and genotype data strongly suggest that the c.195A>G transition constituted a causative mutation and not a benign variant in linkage disequilibrium with the disease.
Figure 2. Mutation analysis of TRPM3.
(A) Sanger sequence trace of the wild-type allele showing translation of isoleucine (I) at codon 65 (ATA) of variant 1 (isoform k), and at codon 8 of a novel lens variant. (B) Sequence trace of the mutant allele showing the heterozygous A-to-G transition (denoted R by the International Union of Pure and Applied Chemistry [IUPAC] code) that is predicted to result in the missense substitution of methionine (ATG) for isoleucine at codon 65 (c.195A>G, p.I65 M) of isoform k, and at codon 8 of a lens abundant isoform (c.24A>G, p.I8 M). (C) Restriction fragment length analysis showing gain of a Fok I site (5′GGATG[9/13]) that co-segregated with affected individuals heterozygous for the A-to-G transition (175 bp).
TRPM3 generates a novel transcript in human lens, and gains an alternative translation start-site in other TRPM3 transcript variants
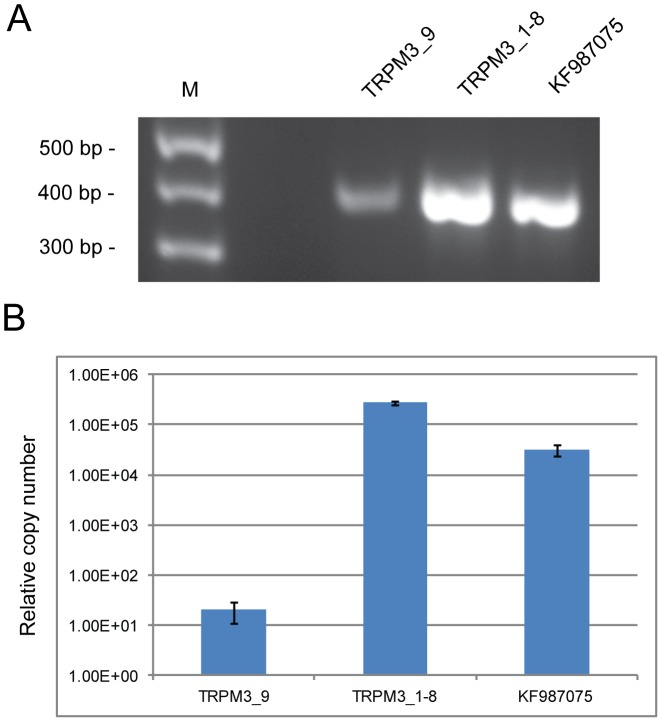
The reference sequence for TRPM3 (Gene ID: 80036) comprises 28 exons that generate at least nine transcript variants (1–9) encoding nine protein isoforms (a–h, k) by means of alternative splicing (Figure 3, Table S3). In order to profile TRPM3 transcript variants expressed in the human lens we undertook reverse transcript PCR (RT-PCR) and 5-prime rapid amplification of cDNA ends (5′-RACE). In postmortem lens RNA we detected strong evidence for multiple TRPM3 reference transcripts (Figure 4). These included transcript variant-9, which encodes protein isoform-k, and some or all of transcript variants 1–8, which encode protein isoforms a-h (Table S3). In addition, we detected abundant levels of a novel transcript variant (GenBank accession number: KF987075) that substituted exon-1 of variant-9 with an expressed sequence tag (EST, GenBank accession number: BM712132) previously found in human lens ([30], Figure 4, Figure S2).
Figure 3. Schematic showing the gene structure of TRPM3.
(A) Exon organization, splice variants and protein domains. Exons are indicated by numbered boxes (1–28), and codon numbers are shown below each coding exon. Translation start and stop sites are denoted by ATG and asterisks, respectively. Transcript variants are numbered (1–9), and corresponding protein isoforms are indicated by letters (a–h, k). Exons subject to alternative splicing (1, 2, 8, 15 and 17) are indicated by plus and minus symbols below each exon. (−/−) indicates that the exon is absent from both the transcript variant and protein isoform. (+/−) indicates that the exon is present in the transcript variant but is not translated. Exon 2, exon 8 and exon 15 are skipped in transcript variant 9 (isoform k). In transcript variants 1–8 (isoforms a–h) exon 2, exon 3 and all of exon 4 except the last 3 bases (ATG) are non-coding. EST denotes an expressed sequence tag (BM712132) that replaces exon 1 of variant 9, and exon 2 of variants 1–8, joining directly to exon 3 in a novel lens abundant transcript variant (KF987075). Arrows indicate the orientation and exon location of PCR primers used to amplify and sequence the 5′-ends of TRPM3 transcripts. The micro-RNA gene, MIR204, is located in intron 8. The predicted I-to-M substitution (red) identified in the family studied here is located in a putative calmodulin-binding (CaM) motif [31] near the N-terminus of isoform k and the novel lens isoform. The approximate locations of conserved protein domains are indicated: (pfam00520) ion-transport domain; (TRP1–2) transient receptor potential box 1 and 2; (C–C) coiled-coil domain, and (ICF) amino-acid sequence indispensible for channel function [45]. Note that variants 7 and 8 encode short isoforms h and c, respectively, lacking the conserved domains above. These short cytoplasmic isoforms may play a role in regulation of the full-length transmembrane isoforms [64]. (B) Amino acid sequence alignment of the N-terminal calmodulin-binding motif of human TRPM3 isoform-k, a novel human lens isoform, and homologs from other species showing conservation of isoleucine at the site of the predicted methionine substitution (red). Divergent amino-acid residues are shaded grey.
Figure 4. Alternative splicing of TRPM3 transcripts in the human lens.
(A) RT-PCR showing presence of transcripts for variant 9 (exon 1–5, 411 bp), variants 1–8 (exon 2–5, 414 bp) and a novel lens variant, KF987075 (EST - exon 5, 403 bp). (B) Quantitative RT-PCR showing relative copy numbers of TRPM3 variant 9 (2.06E+01, SD 9.46E+00), variants 1–8 combined (2.76E+05, SD 2.15E+04), and lens variant KF987075 (3.17E+04, SD 8.07E+03) standardized against RPL19. TRPM3 primers used for amplification were as follows; RT1F - RT5R for variant 9, RT2F - RT5R for variants 1–8, and ESTF - RT5R for lens variant KF987075 (Table S5).
The c.195A>G missense change in TRPM3 transcript variant-9 identified above occurred at the third base position of codon 65 (ATA>ATG), and was predicted to result in the conservative substitution of isoleucine-to-methionine (p. I65 M) in TRPM3 isoform-k at the level of protein translation. Notably in the lens transcript variant, which encoded a truncated N-terminal domain compared with isoform-k, the missense change occurred at codon 8 (c.24A>G) shifting the predicted amino-acid substitution accordingly (p.I8M). The predicted I-to-M substitution represented a relatively conservative amino acid change with the non-polar side-group of isoleucine (-C4H9) replaced by the non-polar sulfur-containing side-group of methionine (-C2H4-S-CH3). However, cross-species amino-acid alignment of TRPM3 confirmed that the substituted isoleucine residue is evolutionarily conserved in bony vertebrates, and is located within a putative calmodulin-binding motif near the N-terminus (Figure 3, [31]). Further, in silico missense mutation prediction analysis using the SIFT and PolyPhen-2 algorithms were consistent with damaging effects on protein function (Table 2).
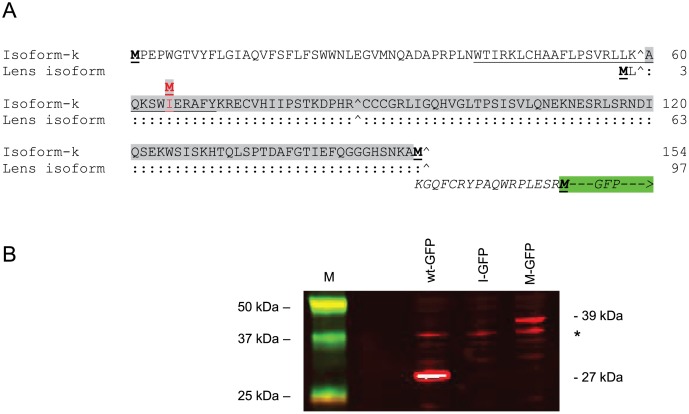
In human TRPM3 transcript variant-9, the translation start codon is located in exon-1, and the c.195A>G (p.I65M) change in exon-3 (Figure 3). Similarly, in the lens transcript variant the start codon is located in the EST and the c.24A>G change (p.I8M) is located in exon-3. By contrast, in human TRPM3 transcript variants 1–8 the translation start-codon is located at the end of exon-4 (Figure 3) effectively placing the predicted p.I65M and p.I8M substitutions in the 5′-untranslated region (5′-UTR) in-frame with the native start-codon lying downstream. Interestingly, the predicted I-to-M substitution is embedded in a partially consensus Kozak sequence raising the possibility that it functions as a novel translation start-site (Figure 5a, Figure S3). In order to test this possibility we sub-cloned part of the 5′-coding sequence from variant-9 (codons 60–153) into a GFP fusion vector such that the I-to-M substitution was in-frame with the consensus translation start-site for GFP located downstream (Figure 5a, Figure S3). Following transient expression of the mutant (methionine) sequence in HEK293T cells and immunoblot analysis with GFP antibody, we detected a soluble antigen (Mr ∼39 kDa) that was ∼12 kDa larger than the native GFP (∼27 kDa, Figure 5b). This larger antigen was consistent with translation of a TRPM3-GFP fusion protein starting from the I-to-M substitution site. By contrast, the ‘wild-type’ (isoleucine) sequence did not produce a detectable GFP-fusion product (Figure 5b) most likely because several out-of-frame translation start-codons were present in the TRPM3 sequence between the I-to-M substitution site and the translation start-site for GFP (Figure S3). These data predict that, in heterozygous individuals, the I-to-M change introduces an alternative translation start-site in transcript variants 1–8, which is able to compete with the endogenous start-sites potentially resulting in the addition of 89 amino acid residues to the N-terminal domain of protein isoforms a–h.
Figure 5. Transient expression of a TRPM3-GFP fusion product in HEK293T cells.
(A) N-terminal amino-acid sequence of TRPM3 isoform k (NP_001007472), and a lens abundant isoform (KF987075). The sequence coding for amino-acids 60-153 (exon 3) of isoform k (codons 3–96 of the lens isoform) that was inserted into the GFP fusion vector is shaded grey. The p.I65M substitution in isoform k (p.I8M in the lens isoform) is shown in red. Translation initiator methionine residues are shown in bold and underlined. The translation start-site for isoform k is located 57 codons upstream of that for the lens isoform, and 153 codons upstream of that for isoforms 1–8. Italics denote the 17 amino-acid sequence linking the TRPM3 sequence in-frame to GFP (green box). The GFP translation start-site is located in-frame 106 codons downstream from the I-to-M substitution in isoform k and the lens isoform. (:) denotes identical amino-acids, (∧) indicates intron boundaries between exons 1, 3 and 4 (exon 2 is skipped). The CaM binding domain is underlined. (B) Immunoblot analysis of transfected HEK293T cell lysates showing presence of the predicted TRPM3-GFP fusion product (∼39 kDa) that is ∼12 kDa larger than native GFP (27 kDa). (*) indicates non-specific cross-reaction with the HEK293T cell lysate.
Discussion
In cases where early-onset cataract and glaucoma are co-inherited, suspicion is first directed toward genes encoding ocular transcription factors, notably PAX6, FOXE3, PITX3, and MAF that control development of anterior ocular structures including the cornea, iris, ciliary-body, iridocorneal angle drainage system (trabecular meshwork and Schlemms canal) and lens [32]–[34]. Following exclusion of these and many other candidate genes we have identified TRPM3 on human chromosome 9q as a novel gene for autosomal dominant cataract and high-tension glaucoma. The A-to-G mutation discovered in transcript variant-9 (c.195A>G) was predicted to result in a missense substitution (p.I65M) in TRPM3 isoform-k that was located within a putative calmodulin binding motif. However, because of extensive alternative splicing of TRPM3 transcripts, the A-to-G mutation was also predicted to cause a missense substitution (c.24A>G; p.I8M) in a novel TRPM3 transcript abundant in human lens. In both transcript variants, the I-to-M substitution was predicted in silico to exert damaging effects on protein function. Moreover, the I-to-M substitution was predicted in vitro to function as an alternate translation initiation-site in TRPM3 transcript variants 1–8 resulting in aberrant N-terminal extension of TRPM3 isoforms a–h. Structure-function analyses of TRPM channels indicates that the N-terminal and C-terminal domains form a large cytoplasmic complex that functions in membrane localization, tetrameric assembly, and ligand interactions [35], [36]. Conceivably, abnormal extension of the N-terminal domain may reduce overall protein stability and compromise some or all of these functions. While we cannot exclude loss-of-function defects, these observations raise the likelihood that the I-to-M substitution triggers deleterious gain-of-function effects leading to lens opacities, high-tension glaucoma, and variable anterior segment defects.
In mice, both Trpm3 and its intronically hosted, non-coding micro-RNA gene (Mir204) are co-expressed in ocular tissues, particularly the ciliary-body, lens epithelium, and retinal pigment epithelium [37]–[39]. Recently, the paired-box transcription factor gene, Pax6, has been shown to co-regulate Trpm3 and Mir204 during mouse eye development, and an evolutionarily conserved mechanism has been described in medaka fish [40]–[42]. Up-regulation of miR-204 results in suppression of several target genes involved in neurogenesis, cell-motility, and epithelial-to-mesenchymal transition (EMT), consistent with a role in determining and/or maintaining epithelial cell fate [43], [44]. By contrast, the precise function(s) of Trpm3 up-regulation in ocular tissues remains unclear. TRPM3 has been shown to function in vitro as a spontaneous, steroid-activated, Ca2+-permeable channel, with selectivity for other cations influenced by alternative splicing [45], [46]. In vivo, TRPM3 channel activity has been found to facilitate diverse cellular processes including, insulin release by pancreatic β-cells [47], mechano-sensing in vascular smooth muscle cells [48], and thermo-sensing by dorsal root ganglia neurons [49]. Recently, Trpm3-null mice were found to display an attenuated pupillary light reflex (iris constriction) under bright light and dim light conditions [50]. The abundant expression of Trpm3 in the ciliary body coupled with the requirement for sustained Ca2+ influx during ciliary muscle contraction, support the notion that Trpm3 functions in regulating pupil constriction [50]. In addition to pupil response, changes in the contractile state of the ciliary muscle are known to modulate aqueous humor outflow via the trabecular meshwork [51]. Further, the ciliary epithelium (pigmented and non-pigmented) co-operate in the production of aqueous humor [52]. These observations raise the possibility that Trpm3 expression in the ciliary body may influence intra-ocular pressure. Outside the ciliary body, the abundant expression of Trpm3 in the lens epithelium and retinal pigment epithelium suggests that this cation channel may also participate in the maintenance of intracellular Ca2+ essential for preserving lens transparency, and in the regulation of Ca2+ concentration fluxes in the sub-retinal space that accompany light/dark transitions [53], [54].
Based on its Pax6-driven ocular expression pattern, Trpm3/Mir204 represents an intriguing locus for eye disease. In mice, disruption of Trpm3 results in a mild pupilary phenotype [50], whereas, Pax6-deficiency results in a severe eye phenotype characterized by aphakia, microphthalmia and anophthalmia [55]. The divergent nature of these eye phenotypes reflects, in part, contrasting loss-of-function effects associated with Mir204, Trpm3 and Pax6. In Trpm3-null mice the gene-targeting site is located in exon-17 [49], well downstream from Mir204 located in intron-8, and would not be expected to interfere with ocular expression of the miR-204 transcript. By contrast, early ocular expression of both Mir204 and Trpm3 is lost in Pax6-null mice [40]. Further, ocular defects resulting from Trpm3 loss-of-function may be partially rescued because of functional redundancy with other TRP(M) channels expressed in the eye. For example, Trpm1, which shares ∼57% amino acid identity with Trpm3 at the protein level, is also expressed in the ciliary body [56]. Conversely, the severe ocular defects resulting from Pax6-deficiency cannot be functionally rescued by other ocular transcription factors.
In humans, rare deletions involving TRPM3 have been reported in one case of Kabuki syndrome (KS, OMIM 147920), and in a nuclear family with two cases of autism [23], [24]. In the autism family, a paternal deletion of exons 1–9 that included MIR204 was shared by two affected sons and an unaffected daughter [24]. However, no rare variants were found in the maternal copy of TRPM3/MIR204 in the affected sons. Since ocular abnormalities were not reported in the autism family, and we do not have any evidence of autism in the family studied here, the role of TRPM3 in autism remains unclear. By contrast, ocular abnormalities have been found in about 30–50% of KS cases [57]. Typical ocular findings include ptosis, strabismus and/or blue sclerae, whereas, less common findings include cataract, Peters' anomaly, corneal opacities and colobomas. Again, we have no evidence of KS in the family studied here, and the importance of TRPM3 in the etiology of KS is further obscured since the majority of KS patients have been found to harbor mutations in MLL2 on chromosome 12q [58]. Interestingly, PAX6 mutations primarily result in a variable pan-ocular phenotype that includes aniridia (iris hypoplasia), cataract, glaucoma, corneal clouding, foveal dysplasia, and optic nerve hypoplasia [59], [60]. In addition to cataract and glaucoma, the TRPM3 mutation (I-to-M) discovered here was associated with a mild iris anomaly in three affected family members (V:3, V:4, V9), and in one case (V:9) anterior segment dysgenesis was also documented (Figure 1b, Table S1). Such overlap in the ocular phenotype associated with TRPM3 and PAX6 mutations is consistent with the close functional synergy between these genes during eye development. Our data predict that the I-to-M coding mutation in TRPM3 affects the N-terminal translation of multiple TRPM3 isoforms (Figure 5) potentially resulting in deleterious gain-of-function effects that manifest differently to loss-of-function effects in Trpm3-null mice. In addition, the I-to-M mutation is located upstream of the human MIR204 homolog hosted in intron-8 of TRPM3 (Figure 3a) and, although it is unlikely, we cannot rule out a deleterious effect on the ocular expression of MIR204. Further insights regarding the genotype-phenotype complexity associated with TRPM3 await the discovery of additional disease causing mutations associated with this cation entry channel.
Materials and Methods
Ethics statement
Ethical approval for this study was obtained from the Washington University Human Research Protection Office (HRPO), and written informed consent was provided by all participants prior to enrollment in accordance with the tenets of the Declaration of Helsinki, and Health Insurance Portability and Accountability Act (HIPAA) regulations.
Family participants
A 5-generation Caucasian-American pedigree segregating cataract and glaucoma was ascertained through medical records in the Department of Ophthalmology and Visual Sciences at Washington University School of Medicine (Figure 1a). Retrospective review of medical records limited our diagnoses of cataract and glaucoma to various lens opacities requiring surgical extraction for visual rehabilitation, and significant elevation of intraocular pressure (IOP>30 mm Hg) associated with or at high risk for optic nerve damage and/or visual field loss, respectively. Blood samples were obtained from 36 family members including 25 affected, seven unaffected, two undiagnosed and two spouses (Table S1). Leukocyte genomic DNA was purified using the Gentra Puregene Blood kit (Qiagen, Valencia, CA), and quantified by absorbance at 260 nm (NanoDrop 2000, Wilmington, DE).
SNP genotyping and linkage analysis
For genome-wide linkage analysis, genotyping was performed by means of the HumanLinkage-12 genotyping beadchip and the Infinium-II whole-genome amplification and single-base extension assay (Illumina, San Diego CA), in the Genome Technology Access Center (GTAC) at Washington University. Two-point and multipoint parametric linkage analysis was performed with Superlink (v. 1.6) and GeneHunter (v. 2.1r5), respectively, from the easyLINKAGE Plus (v. 5.08) package of programs (http://genetik.charite.de/hoffmann/easyLINKAGE/). SNP marker allele frequencies used for linkage analysis were those calculated for Caucasians by the HapMap project (http://www.hapmap.org/). A frequency of 0.001 and a penetrance of 100% were assumed for the disease allele. Multipoint computation was performed with GeneHunter in sets of 100 markers.
Microsatellite genotyping and linkage analysis
Microsatellite markers from the National Center for Biotechnology Information (NCBI) combined Généthon, Marshfield, and deCODE genetic linkage maps (http://www.ncbi.nlm.nih.gov/genome/guide/human/) were genotyped by means of a 4200 DNA analyzer running Gene ImagIR software (Li-Cor, Lincoln, NE) as described [61]. Pedigree and haploptype data were managed using Cyrillic (v. 2.1) software (FamilyGenetix Ltd., Reading, UK), and two-point LOD scores (Z) calculated using the MLINK sub-program from the LINKAGE (5.1) package of programs (http://linkage.rockefeller.edu/soft/). Marker allele frequencies were assumed to be equal. A frequency of 0.01% and a penetrance of 100% were assumed for the disease allele.
Whole exome and custom-target capture
Whole exome capture was achieved using the SureSelect Human All Exon 50 Mb Kit (Agilent Technologies), and custom-target capture was performed with the SureSelect Target Enrichment Kit (Agilent) and oligo-probes designed using the web-based eArray application (http://www.genomics.agilent.com/en/Custom-Design-Tools/eArray/?cid=AG-PT-122&tabId=AG-PR-1047), according to manufacturer's instructions. Briefly, genomic DNA (3 ug) was fragmented (150–200 bp) by acoustic shearing, ligated to adapter primers, and PCR-amplified. Following denaturation (95°C, 5 min), amplified DNA-fragment libraries (∼500 ng) were hybridized in solution under high stringency (65°C, 24 hr) with biotinylated RNA capture probes (∼120 bp). Resulting DNA/RNA hybrids were recovered by streptavidin-coated magnetic bead separation (Dynal, Invitrogen) and captured DNA eluted (NaOH) prior to next-generation sequencing.
Next-generation sequencing
Solid-phase (flow-cell) massively parallel sequencing was performed on a HiSeq2000 System (Illumina) using the Illumina Multiplexing Sample Preparation Oligo-nucleotide Kit, and the HiSeq 2000 Paired-End Cluster Generation Kit according to the manufacturers instructions. Briefly, hybrid-capture libraries were amplified to add indexing (identifying) tags and sequencing primers then subject to paired-end (2×101 bp read length), multiplex sequencing-by-synthesis using fluorescent reversible (3′-blocked) terminators. Raw sequence data was aligned to the reference genome by NovoalignMPI (www.novocraft.com), and sequence variants called using the Sequence Alignment/Map format (SAMtools) and Picard programs (http://samtools.sourceforge.net/), and further annotated using SeattleSeq (http://snp.gs.washington.edu/SeattleSeqAnnotation131/). Target coverage and read depth were reviewed by the Integrated Genomics Viewer (IGV, http://www.broadinstitute.org/igv/). Variants were reviewed for presence/absence in dbSNP and 1000_genomes databases, frequency in the Exome Variant Server/database (http://evs.gs.washington.edu/EVS/), and predicted effect on protein function using the PolyPhen2 and SIFT servers [62], [63].
Sanger Sequencing
Genomic sequence for TRPM3 (Gene ID: 80036) was obtained from the Ensembl human genome browser (http://www.ensembl.org/index.html), and gene-specific M13-tailed PCR primers (Table S4) were selected from the NCBI re-sequencing amplicon (RSA) probe database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=probe) or custom designed with Primer Quest (IDT.com) or Exon Primer (UCSC Genome Bioinformatics - http://genome.ucsc.edu). Genomic DNA (2.5 ng/ul, 20 ul reactions), was amplified (35–40 cycles) in a GeneAmp 9700 thermal cycler using AmpliTaq polymerase (Applied Biosystems, Foster City, CA) and gene-specific primers (10 pmol). Resulting PCR amplicons were either enzyme-purified with ExoSAP-IT (USB Corporation, Cleveland, OH) or gel-purified with the QIAquick gel-extraction kit (Qiagen). Purified amplicons were direct cycle-sequenced in both directions with BigDye Terminator Ready Reaction Mix (v3.1) containing M13 forward or reverse sequencing primers then ethanol precipitated and detected by capillary electrophoresis on a 3130xl Genetic Analyzer running Sequence Analysis (v5.2) software (Applied Biosystems), and Chromas (v2.23) software (Technelysium, Tewantin, Queensland, Australia).
RT-PCR
Postmortem human lens total RNA was isolated with Trizol Reagent (Invitrogen), and reverse transcribed into cDNA by random priming with the iScript cDNA Synthesis Kit (Bio-Rad). TRPM3 sequence located between exon 1 and exon 5 was amplified with AmpliTaq polymerase (Applied Biosystems) and TRPM3 gene-specific primers (Table S5). Sequence identity was confirmed by Sanger sequencing as above. For quantization purposes, RT-PCR products were amplified in a 10-fold dilution series (in triplicate) by means of the iQ SYBR green Supermix (Bio-Rad) in a real-time PCR detection system (iQ5, Bio-Rad). Transcript copy number was determined by melt-curve analysis and standardization against a control transcript (RPL19) that was amplified separately in a similar 10-fold dilution series of the same RT-PCR products using RPL19 gene-specific forward (5′-catccgcaagcctgtgac) and reverse (5′-gtgaccttctctggcattcg) primers.
5′-RACE
Rapid amplification of 5′-cDNA ends was achieved using the GeneRacer Kit (Invitrogen). Briefly, total lens RNA (1–5 ug) was dephosphorylated (CIP), decapped (TAP), and ligated to the GeneRacer RNA oligo with T4 RNA ligase then reverse transcribed with AMV-RT and random primers. First-strand cDNA was PCR amplified using the GeneRacer 5′-primer and a reverse TRPM3 gene-specific primer and then re-amplified using nested primers (Table S5) to increase product specificity and yield. Following purification (SNAP column), resulting 5′-RACE products with 3′A-overhangs were sub-cloned into pCR4-TOPO and Sanger sequenced in both directions using flanking T7 and T3 sequencing primers.
GFP fusion product
Generation of a C-terminal TRPM3-GFP fusion product was achieved using the GFP Fusion TOPO TA Expression Kit (Invitrogen) according to the manufacturer's instructions. Briefly, lens total RNA (1 ug) was reverse transcribed into cDNA by random priming with the iScript cDNA Synthesis Kit (Bio-Rad). TRPM3 coding sequence located between codons 60–154 of isoform-k (Figure 5) was PCR amplified using either a wild-type (I65) or mutant (M65) forward primer located in exon-3 paired with a reverse anchor primer located in exon-4 (Table S5). PCR product (283 bp) with non-template 3′-A overhang was ligated to the C-terminal fusion vector (pcDNA3.1/CT-GFP-TOPO) using topoisomerase. Following transformation of competent TOP10 cells, plasmid DNA was purified from overnight cultures by the QIAprep Spin Kit (Qiagen), and Sanger sequenced using the T7 and GFP-reverse sequencing primers to verify correct orientation and reading frame.
Cell culture and transfection
HEK293T cells were cultured (37°C, 5% CO2) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2 mM glutamine, 100 IU penicillin, 100 ug streptomycin (Cellgro, Mediatech, Manassas, VA), and 10% bovine serum (Invitrogen). Plasmid DNA (8 ug) was transfected into HEK293T cell monolayers in 60 mm dishes (60–80% confluence) using Lipofectamine 2000 reagent (Invitrogen) in serum-free media (OptiMEM, Invitrogen) for 4 hr and then cultured for 24–48 hr in serum supplemented media. Transfected cells were washed (PBS), detached (EDTA), and centrifuged (1500×g, 5 min).
Immunoblot analysis
Transfected cell pellets were re-suspended (50 ul, 37°C, 10 min) in detergent lysis buffer (1% IGEPAL, 50 mM Tris-HCL, 150 mM NaCl, pH 7.8; Sigma) then centrifuged (10,000×g, 10 min) to pellet cell nuclei. Post-nuclear lysate was removed and soluble protein concentration was determined using the Non-interfering assay (G-Bioscience). Soluble proteins (20 ug) were separated on SDS-PAGE gels (12%) then transferred onto nitrocellulose, incubated with anti-GFP rabbit serum (Invitrogen, dil 1∶1000), and visualized using an Odyssey Infrared Imaging System (Li-Cor Bioscience).
Supporting Information
Genome-wide linkage analysis of ocular disease in the family using SNP markers. (A) Parametric two-point LOD scores (pLOD SPT) indicating linkage to chromosome 9. (B) Parametric multi-point LOD scores (pLOD MPT) confirming linkage to chromosome 9.
(TIF)
N-terminal reading frame of the novel lens abundant TRPM3 transcript (KF987075) detected in Figure 4 . A previously identified lens EST (BM712132) [30] is shaded grey. The predicted translation initiator methionine is shown in bold. Note, the translation initiator methionine for transcript variants 1–8 is located 97 codons downstream (bold underlined). The I8 M mutation site is shown in red. Upstream translation stop-codons, in-frame with the translation start-codons, are shown in italics. Paired PCR primer sequences (Table S5) located in the EST and in exon 10 are underlined. (∧) indicates intron boundaries between the EST and exons 3, 4, 5, 6, 7, and 9 (exon 8 is skipped).
(TIFF)
N-terminal coding sequence of TRPM3 that was used to generate a GFP fusion product ( Figure 5 ). Note the A-to-G missense change (I-to-M substitution) is embedded in a partially consensus Kozak translation start-site (TGG ATG G). Several potential out-of-frame translation start-sites are located between the predicted I-to-M substitution site (red) and the consensus Kozak start-site for GFP (A/GNN ATG G). Vector sequence is shown in italics.
(TIF)
Clinical ocular status of affected individuals in the family.
(XLSX)
SNP haplotype for the ocular disease locus on chromosome 9.
(DOCX)
Transcript variants of TRPM3 derived from alternative splicing.
(XLSX)
PCR primers for Sanger sequencing of TRPM3 exons.
(DOCX)
RT-PCR primers for amplification of TRPM3 cDNA.
(DOCX)
Acknowledgments
We thank family members for participating in this study, Dr. O. Boskovska for help with family ascertainment, and Dr. D. Hodzic for providing HEK293T cells.
Funding Statement
This work was supported by National Institutes of Health/National Eye Institute (NIH/NEI) grants EY012284 (to AS), and EY02687 (Core Grant for Vision Research), and by an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness (RPB). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Trumler AA (2011) Evaluation of pediatric cataracts and systemic disorders. Curr Opin Ophthalmol 22: 365–379. [DOI] [PubMed] [Google Scholar]
- 2. Shiels A, Hejtmancik JF (2013) Genetics of human cataract. Clin Genet 84: 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rahi JS, Dezateux C (2001) Measuring and interpreting the incidence of congenital ocular anomalies: lessons from a national study of congenital cataract in the UK. Invest Ophthalmol Vis Sci 42: 1444–1448. [PubMed] [Google Scholar]
- 4. Amaya L, Taylor D, Russell-Eggitt I, Nischal KK, Lengyel D (2003) The morphology and natural history of childhood cataracts. Surv Ophthalmol 48: 125–44. [DOI] [PubMed] [Google Scholar]
- 5. Zetterstrom C, Lundvall A, Kugelberg M (2005) Cataracts in children. J Cataract Refract Surg 31: 824–840. [DOI] [PubMed] [Google Scholar]
- 6. Lambert SR, Purohit A, Superak HM, Lynn MJ, Beck AD (2013) Long-term risk of glaucoma after congenital cataract surgery. Am J Ophthalmol 156: 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiels A, Bennett TM, Hejtmancik JF (2010) Cat-Map: putting cataract on the map. Mol Vis 16: 2007–2015. [PMC free article] [PubMed] [Google Scholar]
- 8. Clapham DE (2003) TRP channels as cellular sensors. Nature 426: 517–524. [DOI] [PubMed] [Google Scholar]
- 9. Venkatachalam K, Montell C (2007) TRP channels. Annu Rev Biochem 76: 387–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nilius B, Owsianik G (2011) The transient receptor potential family of ion channels. Genome Biol 12: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Montell C, Rubin GM (1989) Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron 2: 1313–1323. [DOI] [PubMed] [Google Scholar]
- 12. Latorre R, Zaelzer C, Brauchi S (2009) Structure-functional intimacies of transient receptor potential channels. Q Rev Biophys 42: 201–246. [DOI] [PubMed] [Google Scholar]
- 13. Kunert-Keil C, Bisping F, Kruger J, Brinkmeier H (2006) Tissue-specific expression of TRP channel genes in the mouse and its variation in three different mouse strains. BMC Genomics 7: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Toro CA, Arias LA, Brauchi S (2011) Sub-cellular distribution and translocation of TRP channels. Curr Pharm Biotechnol 12: 12–23. [DOI] [PubMed] [Google Scholar]
- 15. Nilius B, Owsianik G (2010) Transient receptor potential channelopathies. Pflugers Arch 460: 437–450. [DOI] [PubMed] [Google Scholar]
- 16. Everett KV (2011) Transient receptor potential genes and human inherited disease. Adv Exp Med Biol 704: 1011–1032. [DOI] [PubMed] [Google Scholar]
- 17. Lin Z, Chen Q, Lee M, Cao X, Zhang J, et al. (2012) Exome sequencing reveals mutations in TRPV3 as a cause of Olmsted syndrome. Am J Hum Genet 90: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z, Sergouniotis PI, Michaelides M, Mackay DS, Wright GA, et al. (2009) Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet 85: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Audo I, Kohl S, Leroy BP, Munier FL, Guillonneau X, et al. (2009) TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet 85: 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Genderen MM, Bijveld MM, Claassen YB, Florijn RJ, Pearring JN, et al. (2009) Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet 85: 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bellone RR, Brooks SA, Sandmeyer L, Murphy BA, Forsyth G, et al. (2008) Differential gene expression of TRPM1, the potential cause of congenital stationary night blindness and coat spotting patterns (LP) in the Appaloosa horse (Equus caballus). Genetics 179: 1861–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sandmeyer LS, Bellone RR, Archer S, Bauer BS, Nelson J, et al. (2012) Congenital stationary night blindness is associated with the leopard complex in the Miniature Horse. Vet Ophthalmol 15: 18–22. [DOI] [PubMed] [Google Scholar]
- 23. Kuniba H, Yoshiura K, Kondoh T, Ohashi H, Kurosawa K, et al. (2009) Molecular karyotyping in 17 patients and mutation screening in 41 patients with Kabuki syndrome. J Hum Genet 54: 304–309. [DOI] [PubMed] [Google Scholar]
- 24. Pagnamenta AT, Holt R, Yusuf M, Pinto D, Wing K, et al. (2011) A family with autism and rare copy number variants disrupting the Duchenne/Becker muscular dystrophy gene DMD and TRPM3. J Neurodev Disord 3: 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yashin AI, Wu D, Arbeev KG, Ukraintseva SV (2010) Joint influence of small-effect genetic variants on human longevity. Aging (Albany NY) 2: 612–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park SH, Lee JY, Kim S (2011) A methodology for multivariate phenotype-based genome-wide association studies to mine pleiotropic genes. BMC Syst Biol 5 Suppl 2: S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heon E, Paterson AD, Fraser M, Billingsley G, Priston M, et al. (2001) A progressive autosomal recessive cataract locus maps to chromosome 9q13–q22. Am J Hum Genet 68: 772–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Forshew T, Johnson CA, Khaliq S, Pasha S, Willis C, et al. (2005) Locus heterogeneity in autosomal recessive congenital cataracts: linkage to 9q and germline HSF4 mutations. Hum Genet 117: 452–459. [DOI] [PubMed] [Google Scholar]
- 29. Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, et al. (2011) Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 331: 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bonaldo MF, Lennon G, Soares MB (1996) Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res 6: 791–806. [DOI] [PubMed] [Google Scholar]
- 31. Holakovska B, Grycova L, Jirku M, Sulc M, Bumba L, et al. (2012) Calmodulin and S100A1 protein interact with N terminus of TRPM3 channel. J Biol Chem 287: 16645–16655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gould DB, John SW (2002) Anterior segment dysgenesis and the developmental glaucomas are complex traits. Hum Mol Genet 11: 1185–1193. [DOI] [PubMed] [Google Scholar]
- 33. Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, et al. (2002) Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet 11: 33–42. [DOI] [PubMed] [Google Scholar]
- 34. Reis LM, Semina EV (2011) Genetics of anterior segment dysgenesis disorders. Curr Opin Ophthalmol 22: 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Phelps CB, Gaudet R (2007) The role of the N terminus and transmembrane domain of TRPM8 in channel localization and tetramerization. J Biol Chem 282: 36474–36480. [DOI] [PubMed] [Google Scholar]
- 36. Moiseenkova-Bell V, Wensel TG (2011) Functional and structural studies of TRP channels heterologously expressed in budding yeast. Adv Exp Med Biol 704: 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deo M, Yu JY, Chung KH, Tippens M, Turner DL (2006) Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn 235: 2538–2548. [DOI] [PubMed] [Google Scholar]
- 38. Karali M, Peluso I, Marigo V, Banfi S (2007) Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci 48: 509–515. [DOI] [PubMed] [Google Scholar]
- 39. Gilliam JC, Wensel TG (2011) TRP channel gene expression in the mouse retina. Vision Res 51: 2440–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D, et al. (2013) Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet 9: e1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, et al. (2010) miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci U S A 107: 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xie Q, Yang Y, Huang J, Ninkovic J, Walcher T, et al. (2013) Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS One 8: e54507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, et al. (2010) MicroRNA-204/211 alters epithelial physiology. Faseb J 24: 1552–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y, Li W, Zang X, Chen N, Liu T, et al. (2013) MicroRNA-204-5p regulates epithelial-to-mesenchymal transition during human posterior capsule opacification by targeting SMAD4. Invest Ophthalmol Vis Sci 54: 323–332. [DOI] [PubMed] [Google Scholar]
- 45. Oberwinkler J, Lis A, Giehl KM, Flockerzi V, Philipp SE (2005) Alternative splicing switches the divalent cation selectivity of TRPM3 channels. J Biol Chem 280: 22540–22548. [DOI] [PubMed] [Google Scholar]
- 46. Fruhwald J, Camacho Londono J, Dembla S, Mannebach S, Lis A, et al. (2012) Alternative splicing of a protein domain indispensable for function of transient receptor potential melastatin 3 (TRPM3) ion channels. J Biol Chem 287: 36663–36672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wagner TF, Loch S, Lambert S, Straub I, Mannebach S, et al. (2008) Transient receptor potential M3 channels are ionotropic steroid receptors in pancreatic beta cells. Nat Cell Biol 10: 1421–1430. [DOI] [PubMed] [Google Scholar]
- 48. Naylor J, Li J, Milligan CJ, Zeng F, Sukumar P, et al. (2010) Pregnenolone sulphate- and cholesterol-regulated TRPM3 channels coupled to vascular smooth muscle secretion and contraction. Circ Res 106: 1507–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vriens J, Owsianik G, Hofmann T, Philipp SE, Stab J, et al. (2011) TRPM3 is a nociceptor channel involved in the detection of noxious heat. Neuron 70: 482–494. [DOI] [PubMed] [Google Scholar]
- 50. Hughes S, Pothecary CA, Jagannath A, Foster RG, Hankins MW, et al. (2012) Profound defects in pupillary responses to light in TRPM-channel null mice: a role for TRPM channels in non-image-forming photoreception. Eur J Neurosci 35: 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wiederholt M, Thieme H, Stumpff F (2000) The regulation of trabecular meshwork and ciliary muscle contractility. Prog Retin Eye Res 19: 271–295. [DOI] [PubMed] [Google Scholar]
- 52. Civan MM, Macknight AD (2004) The ins and outs of aqueous humour secretion. Exp Eye Res 78: 625–631. [DOI] [PubMed] [Google Scholar]
- 53. Rhodes JD, Sanderson J (2009) The mechanisms of calcium homeostasis and signaling in the lens. Exp Eye Res 88: 226–234. [DOI] [PubMed] [Google Scholar]
- 54. Wimmers S, Karl MO, Strauss O (2007) Ion channels in the RPE. Prog Retin Eye Res 26: 263–30. [DOI] [PubMed] [Google Scholar]
- 55. Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, et al. (1991) Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354: 522–525. [DOI] [PubMed] [Google Scholar]
- 56. Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, et al. (2008) Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kluijt I, van Dorp DB, Kwee ML, Toutain A, Keppler-Noreuil K, et al. (2000) Kabuki syndrome – report of six cases and review of the literature with emphasis on ocular features. Ophthal Genet 21: 51–61. [PubMed] [Google Scholar]
- 58. Banka S, Veeramachaneni R, Reardon W, Howard E, Bunstone S, et al. (2012) How genetically heterogeneous is Kabuki syndrome?: MLL2 testing in 116 patients, review and analyses of mutation and phenotypic spectrum. Eur J Hum Genet 20: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lee H, Khan R, O'Keefe M (2008) Aniridia: current pathology and management. Acta Ophthalmol 86: 708–715. [DOI] [PubMed] [Google Scholar]
- 60. Hingorani M, Hanson I, van Heyningen V (2012) Aniridia. Eur J Hum Genet 20: 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mackay DS, Andley UP, Shiels A (2003) Cell death triggered by a novel mutation in the alphaA-crystallin gene underlies autosomal dominant cataract linked to chromosome 21q. Eur J Hum Genet 11: 784–793. [DOI] [PubMed] [Google Scholar]
- 62. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, et al. (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40: W452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu XZ, Moebius F, Gill DL, Montell C (2001) Regulation of melastatin, a TRP-related protein, through interaction with a cytoplasmic isoform. Proc Natl Acad Sci USA 98: 10692–10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome-wide linkage analysis of ocular disease in the family using SNP markers. (A) Parametric two-point LOD scores (pLOD SPT) indicating linkage to chromosome 9. (B) Parametric multi-point LOD scores (pLOD MPT) confirming linkage to chromosome 9.
(TIF)
N-terminal reading frame of the novel lens abundant TRPM3 transcript (KF987075) detected in Figure 4 . A previously identified lens EST (BM712132) [30] is shaded grey. The predicted translation initiator methionine is shown in bold. Note, the translation initiator methionine for transcript variants 1–8 is located 97 codons downstream (bold underlined). The I8 M mutation site is shown in red. Upstream translation stop-codons, in-frame with the translation start-codons, are shown in italics. Paired PCR primer sequences (Table S5) located in the EST and in exon 10 are underlined. (∧) indicates intron boundaries between the EST and exons 3, 4, 5, 6, 7, and 9 (exon 8 is skipped).
(TIFF)
N-terminal coding sequence of TRPM3 that was used to generate a GFP fusion product ( Figure 5 ). Note the A-to-G missense change (I-to-M substitution) is embedded in a partially consensus Kozak translation start-site (TGG ATG G). Several potential out-of-frame translation start-sites are located between the predicted I-to-M substitution site (red) and the consensus Kozak start-site for GFP (A/GNN ATG G). Vector sequence is shown in italics.
(TIF)
Clinical ocular status of affected individuals in the family.
(XLSX)
SNP haplotype for the ocular disease locus on chromosome 9.
(DOCX)
Transcript variants of TRPM3 derived from alternative splicing.
(XLSX)
PCR primers for Sanger sequencing of TRPM3 exons.
(DOCX)
RT-PCR primers for amplification of TRPM3 cDNA.
(DOCX)