Abstract
Efficient hydrolysis of native poly(3-hydroxybutyrate) (nPHB) granules in vitro by soluble PHB depolymerase of Rhodospirillum rubrum requires pretreatment of nPHB with an activator compound present in R. rubrum cells (J. M. Merrick and M. Doudoroff, J. Bacteriol. 88:60-71, 1964). Edman sequencing of the purified activator (17.4 kDa; matrix-assisted laser desorption ionization—time of flight mass spectrometry) revealed identity to a hypothetical protein deduced from a partially sequenced R. rubrum genome. The complete activator gene, apdA (activator of polymer degradation), was cloned from genomic DNA, expressed as a six-His-tagged protein in recombinant Escherichia coli (Mr, 18.3 × 103), and purified. The effect of ApdA on PHB metabolism was studied in vitro and in vivo. In vitro, the activity of the activator could be replaced by trypsin, but recombinant ApdA itself had no protease activity. Comparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the protein patterns of trypsin- and ApdA-treated nPHB granules isolated from different PHB-accumulating bacteria showed that trypsin activated nPHB by removing proteins of the surface layer of nPHB regardless of the origin of nPHB, but ApdA bound to and interacted with the surface layer of nPHB in a nonproteolytic manner, thereby transforming nPHB into an activated form that was accessible to the depolymerase. In vivo, expression of ApdA in E. coli harboring the PHB biosynthetic genes, phaCBA, resulted in significant increases in the number and surface/volume ratio of accumulated PHB granules, which was comparable to the effect of phasin proteins, such as PhaP in Ralstonia eutropha. The amino acid sequence of ApdA was 55% identical to the amino acid sequence of Mms16, a magnetosome-associated protein in magnetotactic Magnetospirillum species. Mms16 was previously reported to be a GTPase with an essential function in magnetosome formation (Y. Okamura, H. Takeyama, and T. Matsunaga, J. Biol. Chem. 276:48183-48188, 2001). However, no GTPase activity of ApdA could be demonstrated. We obtained evidence that Mms16 of Magnetospirillum gryphiswaldense can functionally replace ApdA in R. rubrum. Fusions of apdA and mms16 to gfp or yfp were functionally expressed, and both fusions colocalized with PHB granules after conjugative transfer to R. rubrum. In conclusion, ApdA in vivo is a PHB-bound, phasin-like protein in R. rubrum. The function of Mms16 in magnetotactic bacteria requires further clarification.
Poly(3-hydroxybutyrate) (PHB) is a compound that stores carbon and energy in many bacteria and can account for up to 90% of the cellular dry weight during unbalanced growth (for recent reviews see references 3 and 20). Large quantities of PHB can be isolated by solvent extraction, and due to its thermoplastic properties and biodegradation to water and carbon dioxide PHB has attracted academic and industrial interest over the past two decades. This polymer has been commercialized under the trade name BIOPOL.
Accumulated PHB can be hydrolyzed by the accumulating strain itself during periods of starvation (intracellular PHB hydrolysis by intracellular PHB depolymerases) or by other microorganisms after release of the polymer from the accumulating strain (extracellular PHB hydrolysis by extracellular PHB depolymerases). The differentiation between extra- and intracellular degradation is necessary because PHB can be present in two biophysical conformations. In vivo, the polymer is completely amorphous (native) and is covered by a surface layer that is about one-half the size of a cytoplasmic membrane (1) and consists of proteins (so-called phasins) and phospholipids (6, 22, 34, 38). In Ralstonia eutropha H16 the major phasin protein is PhaP, which is involved in synthesis, morphology, and regulation of PHB synthesis (12, 27, 37, 39, 41, 42). After release of the polymer from the cell (e.g., after cell lysis or solvent extraction) or after removal or damage of the surface layer, the polymer denatures and becomes paracrystalline. For the sake of clarity PHB in its intact intracellular, amorphous form is called native PHB (nPHB), and extracellular, partially crystalline PHB without a surface layer or with a damaged surface layer is called denatured PHB (dPHB). Most enzymes that hydrolyze PHB are specific for one of the two forms (nPHB or dPHB). For example, extracellular PHB depolymerases that are released from PHB-degrading bacteria so that PHB can be used as an exogenous carbon source are able to hydrolyze dPHB. Intracellular PHB depolymerases are necessary for utilization of the previously accumulated PHB by the accumulating strain itself during periods of starvation. They are specific for nPHB and do not hydrolyze dPHB.
About 20 different extracellular dPHB depolymerases (PhaZ) have been characterized during the last decade (for a recent summary see reference 15), and our knowledge concerning intracellular depolymerases has increased only recently (16, 18, 40). For R. eutropha several isoenzymes of intracellular PHB depolymerases (PhaZ1 to PhaZ3) or 3-hydroxybutyrate oligomer hydrolases have been identified (9, 18, 28, 29, 31, 40). Intracellular nPHB depolymerases of R. eutropha are not related to extracellular dPHB depolymerases in terms of their amino acid sequences, but they exhibit significant levels of amino acid similarity to each other and to other putative intracellular PHB depolymerases found in the database (15, 16, 18, 40). None of the PHB depolymerases described previously requires proteins as cofactors. However, Rhodospirillum rubrum appears to be an exception. Hydrolysis of nPHB granules by a partially purified nPHB depolymerase of R. rubrum in vitro was strongly dependent on the presence of a heat-stable factor (activator), but the nature of this compound has never been determined (23, 24). Recently, we were able to purify the activator from soluble cell extracts of R. rubrum (11). In this study we cloned the corresponding activator gene and characterized it by subcellular localization and functional analyses.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. A mutant of R. rubrum resistant to 600 μg of streptomycin per ml and 100 μg of rifampin per ml was isolated by two rounds of repeated transfers to media containing increasing amounts of streptomycin and rifampin. The doubly resistant mutant (R. rubrum SmRif) was used as a recipient for mating experiments. R. rubrum was grown photoheterotrophically (∼1,000 lx, 29 to 30°C) in PYI medium, which contained (per liter) 10 g of peptone, 5 g of NaCl, 5 g of yeast extract, 0.2 g of MgSO4 · 7H2O, 5 ml of a phosphate solution (7 g of Na2HPO4 per liter, 8 g of KH2PO4 per liter), and 1 ml of SL8 (containing [per 1,000 ml of double-distilled water] 5.2 g of Na2-EDTA, 1.5 g of FeSO4 · 4H2O, 70 mg of ZnCl2, 0.1 g of MnCl2 · 4H2O, 62 mg of H3BO3, 190 mg of CoCl2 · 6H2O, 17 mg of CuCl2 · 2H2O, 24 mg of NiCl2 · 6H2O, and 36 mg of Na2MoO4 · 2H2O; pH 6.5). The bacteria produced no PHB granules or only a few PHB granules in this medium. For PHB production, bacteria in a PYI medium culture were transferred (0.05 volume) to glass tubes that were filled almost completely with acetate mineral salts medium (MSM), which contained (per liter) 1.6 g of KH2PO4, 2 g of K2HPO4, 0.2 g of yeast extract, 0.4 g of MgSO4 · 7H2O, 0.4 g of NaCl, 0.4 g of NH4Cl, 8 μg of vitamin B12, 1 ml of SL7, 50 mg of CaCl2 · 2H2O, and 5 mg of Fe(III) citrate, as well as 20 mM sodium acetate. The tubes were incubated in the light at 29 to 30°C. For conjugation experiments R. rubrum SmRif and Escherichia coli S17-1 containing the appropriate plasmids were grown in PYI medium and Luria-Bertani medium, respectively. A mixture of 500 μl of fresh PYI medium, 250 μl of the donor culture, and 250 μl of the recipient culture were spotted onto a PYI agar plate and incubated at 30°C overnight. The bacteria were resuspended in PYI medium and diluted with PYI medium (1:10, 1:100), and 100-μl portions of each dilution were plated on selection agar (PYI agar supplemented with 600 μg of streptomycin per ml, 100 μg of rifampin per ml, and 20 μg of kanamycin per ml). Red colonies of R. rubrum transconjugants appeared in the second week of incubation under air at 30°C. Selected colonies were isolated by repeated transfers on selective medium and were grown in liquid cultures in PYI medium or acetate-containing MSM.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Escherichia coli DH5α | Cloning strain | |
| E. coli S17-1 | Mobilizing strain | 33 |
| E. coli HMS174 | Host for phaP and/or phaCBA | 17 |
| Ralstonia eutropha H16 | Source of PHB | DSMZ428 |
| Bacillus megaterium | Source of PHB | DSMZ32 |
| Bacillus cereus | Source of PHB | DSMZ31 |
| Chromobacterium violaceum | Source of PHASCL | DSMZ30191 |
| Rhodospirillum rubrum S1 | Wild type | DSMZ467 |
| R. rubrum Sm | Spontaneous streptomycin-resistant mutant of S1 (≥600 μg/ml), Smr | This study |
| R. rubrum SmRif | Spontaneous rifampin-resistant mutant of Sm (≥100 μg/ml), Smr Rifr | This study |
| Magnetospirillum gryphiswaldense MSR-1 | Wild type | DSMZ6361 |
| pJoe4036 | Rhamnose-inducible vector for His tagging | 35 |
| pSN2224 | apd in pJoe4036 | This study |
| pEYFP-N1 | eyfp | BD Clontech |
| pEGFP | egfp | BD Clontech |
| pEYFP | eyfp | This study |
| pBBR1-MCS2 | Broad-host-range vector | 19 |
| pJM9238 | phaCBA | 17 |
| pBluescriptKS::phaP | phaP | 17 |
| pSN2338 | Activator-eyfp fusion with lac promotor | This study |
| pSN2343 | Activator-eyfp fusion | This study |
| pSN2372 | Activator-eyfp fusion in pBRR-MCS2 | This study |
| pSN2389 | Activator-eyfp fusion in pBRR-MCS2 with lac promotor | This study |
| pCS11 | mms16-egfp fusion in pBBR1-MCS2 | This study |
DSMZ, Deutsche Sammlung für Mikroorganismen and Zellkulturen; PHASCL, short-chain-length PHA.
Magnetospirillum gryphiswaldense MSR-1 was grown under microaerobic conditions (0.25 × 105 mPa of O2) on LSM medium containing 100 μM ferric citrate in an oxystat fermentor as described previously (13). Bacteria grown under these conditions contained multiple PHB granules per cell, as revealed by electron microscopy (13).
Microscopic analysis.
R. rubrum transconjugants or recombinant E. coli strains in liquid cultures were immobilized on glass by mixing 20 μl of a bacterial suspension with ∼40 μl of 1% agarose at 50 to 60°C and were covered immediately with a coverslip. Bacteria were visualized with a Zeiss Axioplan fluorescence microscope operated in the phase-contrast or fluorescence mode by using F41-007 Cy3 and F41-54 Cy2 filters for analysis of the PHB granules (Nile red stained) and for green fluorescent protein (GFP)-yellow fluorescent protein (YFP) analysis, respectively. If necessary, bacteria were stained with the fluorescent dye Nile red by addition of 0.05 to 0.1 volume of a Nile red solution (10 μg/ml of acetone). Pictures were taken with a digital camera (Coolsnap) and processed with the Metaview/Metamorph software (Visitron Systems).
Isolation of nPHB and dPHB granules.
Semicrystalline dPHB was isolated from accumulating cells of R. eutropha H16 by sodium hypochlorite digestion and subsequent solvent extraction of the dried polymer with acetone-diethyl ether (2:1). nPHB granules with an intact surface layer were prepared from crude extracts (French press) of PHB-rich cells of R. eutropha H16 or from recombinant E. coli harboring the PHB biosynthetic genes phaCBA of R. eutropha with or without phaP (17) by two sodium phosphate-buffered glycerol density gradient centrifugation steps as described previously (10). nPHB granules were isolated from PHB-accumulating cells of Chromobacterium violaceum, Bacillus megaterium, Bacillus cereus, and R. rubum by the same procedure. The content of PHB in lyophilized cells was determined by gas chromatography after conversion of PHB into the 3-hydroxymethylester by methanolysis and by using benzoate methylester as an internal standard.
Assay of R. rubrum PHB depolymerase activity and activator activity (ApdA).
The activity of nPHB depolymerase was assayed by photospectroscopy at 650 nm by using a microtiter plate reader (KC4; Bio-tek Instruments, Inc.), 5- to 20-μl samples in 200-μl assay mixtures at 40°C, and purified soluble nPHB depolymerase of R. rubrum (0.001 to 0.01 mg/ml) (unpublished data) (Fig. 1). Each assay mixture contained 100 mM Tris-HCl (pH 9.0), 1 mM MgCl2, and 500 μg of nPHB granules purified as described above per ml. nPHB granules were activated for 10 min at 40°C in the presence of trypsin (0.6 μg/ml/test; SERVA, Heidelberg, Germany). Depending on the batch of PHB and on the source of trypsin, significant differences in the resulting depolymerase activity were observed. For activator assays trypsin was replaced by the R. rubrum activator by using quantities sufficient to obtain a rate of nPHB hydrolysis comparable to the rate of hydrolysis of trypsin-activated nPHB granules. If necessary, the depolymerase and activator were diluted with buffer or water. It was not possible to measure activator activity quantitatively. It turned out that there was no linear correlation between the amount of activator and the velocity or degree of hydrolysis of nPHB granules. Therefore, experiments were routinely performed with three different concentrations of the activator (and a constant PHB depolymerase concentration).
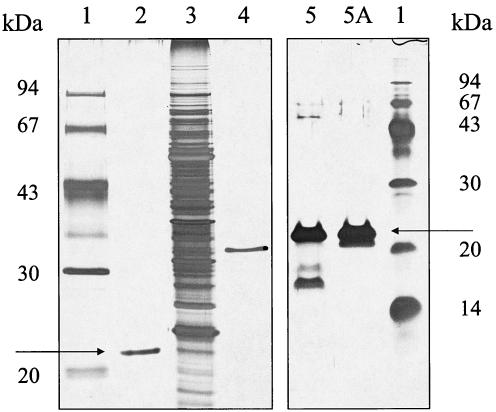
FIG. 1.
Reducing, silver-stained SDS-PAGE analysis of purified ApdA and PHB depolymerase of R. rubrum. Lanes 1, molecular mass standard proteins; lane 2, purified His-tagged recombinant ApdA; lane 3, soluble crude extract of R. rubrum; lane 4, purified His-tagged soluble R. rubrum PHB depolymerase; lanes 5 and 5A, two fractions from the last purification step for wild-type ApdA (11). The arrows indicate the position of ApdA.
GTPase assay.
Five microliters of an appropriate dilution of the purified activator was added to 200 μl of 2 mM GTP in 100 mM Tris-HCl (pH 9) containing 5 mM MgCl2. After incubation for 15 min at 40°C, the sample was diluted with buffer A (80 mM potassium phosphate buffer [pH 6]—methanol, 77:23) containing 5 mM tetrabutylammonium hydrogen sulfate, and 20-μl aliquots were assayed isocratically on a C8-Gromsil 100 high-performance liquid chromatography column (5 μm; 125 by 4 mm) operated at a flow rate of 0.7 ml/min with UV detection at 260 nm. Samples without activator and samples with pure GTP, pure GDP, or mixtures of GDP and GTP (in assay buffer) served as controls. GTP and GDP eluted at 2.78 and 2.45 min, respectively.
Purification of the activator and cloning of its gene.
The activator was purified from soluble cell extracts of a photoheterotrophically grown R. rubrum culture by hydrophobic interaction chromatography (Octyl-Sepharose), chromatofocusing (MonoP), phenol-chloroform extraction, gel filtration (Superdex S75PC), and reverse-phase high-performance liquid chromatography (C18-ChromSil ODS 2FE). Forty of 45 N-terminal amino acids could be identified by Edman degradation of the purified activator and were identical to the DNA-deduced sequence of a hypothetical protein (ZP00014946) that has been identified in a genome sequencing project. Two synthetic oligonucleotides, 5′-ATCGATACATATGGCCAAGCAACCCGAGACC (forward) and 5′-TAGGATCCCTTCTGGGTGGTCGCGGCGCGC (reverse), were used as primers for PCR-based (Pfx polymerase-Taq polymerase, 1:0.75) cloning of the activator gene (apdA) in frame into the NdeI-BamHI-digested rhamnose-inducible His-tagged vector pJOE4036, yielding pSN2224. The correct cloning was confirmed by DNA sequencing of both strands. The activator was purified from recombinant E. coli in one step by metal chelate affinity chromatography by using Ni-NTR agarose and was stored in aliquots at −20°C. Reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis after silver staining confirmed that the purified recombinant activator was almost pure (>90%).
EYFP fusion analysis of ApdA.
A C-terminal fusion between apdA and a derivative of the GFP, the enhanced yellow fluorescent protein (EYFP) (BD Clontech), was constructed. EYFP has a slightly modified fluorescence emission maximum but can be detected by the same filter as GFP. The apdA gene, including 190 bp of the upstream region, was PCR amplified from genomic DNA of R. rubrum by using the synthetic oligonucleotides 5′-GGCGACAGCGGCGGGA (forward) and 5′-ATTGGATCCAACTTCTGGGTGGTCGC (reverse) as the primers. The SalI/BamHI-digested purified product containing 171 bp of the upstream region, including the putative native promoter of apdA, was ligated into SalI/BamHI-opened vector pEYFP and transformed into E. coli JM109. Plasmid pEYFP was constructed by ligating the BamHI/NotI fragment of plasmid pEYFP-N1 (BD Clontech) into the BamHI/NotI-opened vector pECFP (BD Clontech), resulting in pEYFP. Correct cloning of the resulting plasmid (pSN2343) was confirmed by DNA sequencing. Recombinant plasmids were isolated, and the fusion was cut with PaeI, filled with Klenow DNA polymerase, heat inactivated, and digested with SpeI. The isolated and modified fusion was cloned into SmaI/SpeI-opened broad-host-range vector pBBR1MCS2, yielding pSN2372, and transformed into E. coli S17-1. Alternatively, pSN2343 was digested with PvuII/SpeI, and the resulting fusion fragment, including the lac promoter of the parent vector, was cloned into SmaI/SpeI-opened pBBR1MCS2, yielding pSN2389, and transformed into E. coli S17-1. The plasmids constructed were transferred to R. rubrum SmRif by conjugation. Isolated transconjugants were grown in PYI medium and in acetate-containing MSM supplemented with 20 μg of kanamycin per ml and were assayed for PHB formation and EYFP fluorescence.
Cloning of the mms16 gene of M. gryphiswaldense MSR-1 and construction of a C-terminal EGFP fusion.
A gene with a high level of similarity (70% at the protein level) to apdA of R. rubrum was identified in the preliminary genome assembly of M. gryphiswaldense MSR-1 (7) and was designated mms16 (accession no. BX571783) because of its high level of identity (80% at the protein level) to mms16 of Magnetospirillum sp. strain AMB-1. The mms16 gene of M. gryphiswaldense was amplified from genomic DNA by using primer MPPF2 containing a KpnI recognition site (5′-ATATATGGTACCGTGCGTATACGCAAGTATCTATC) and primer MPPRV1containing a BamHI site (5′-ATATATGGATCCACTTCTTCGAGGCCTT-GACGAAC). The KpnI-BamHI-digested PCR product was ligated into the corresponding restrictions sites of the pEGFP-N2 vector (BD Clontech) to generate plasmid pCS1B. Proper insertion of the in-frame mms16-egfp fusion was verified by sequence analysis of the pCS1B insert. The HindIII-XbaI fragment harboring this construct was excised from plasmid pCS1B and subsequently ligated into the HindIII-XbaI sites of pBBR1MCS2 to generate plasmid pCS11. Plasmids used in XbaI digests were previously propagated and isolated from E. coli strain INV110 (Invitrogen).
RESULTS
Bioanalytical characterization of the R. rubrum purified activator.
Recently, we purified and biochemically characterized a protein with the ability to stimulate in vitro hydrolysis of nPHB granules by soluble PHB depolymerase of R. rubrum; this protein was designated an activator (11, 23). Apparent molecular masses of 22 and 17.421 ± 0.1 kDa were determined for the purified activator by SDS-PAGE (Fig. 1) and matrix-assisted laser desorption ionization—time of flight analysis, respectively. The 22-kDa (17.4-kDa) protein was subjected to automated Edman degradation. Forty of 45 amino acids were identified, as follows: NH2-KQPETFLDFDFTRYFADLKVPGVDVESLIASQKRNLXXXTNXXKL. The amino acid sequence obtained by Edman degradation was compared with the protein database, and 100% agreement with positions 3 to 38, 42, 43, 46, and 47 of a hypothetical protein (accession no. ZP00014946) that has been proposed by an R. rubrum genome sequencing project was found. The total molecular mass of the predicted protein was 17.527 kDa, which is close to the value obtained by matrix-assisted laser desorption ionization analysis. We concluded that the gene identified in the database codes for the activator protein. The ability of the purified activator to activate nPHB granules in vitro was not affected by many physical and chemical stresses, such as heat (121°C, autoclaving), pH 1 to 12, dryness, phenol-chloroform, and many other stresses, as described recently (11).
Heterologous expression of the activator in recombinant E. coli and characterization of the activation mechanism in vitro.
The gene coding for the activator protein, apdA (activator of polymer degradation), was PCR amplified as a C-terminal six-His-tagged fusion protein (calculated Mr, 18.349 × 103) from genomic DNA of R. rubrum by using the sequence information in the database. The PCR product was cloned into the vector pJOE4036 under control of a rhamnose-inducible promoter and was transformed into E. coli DH5α. Recombinant ApdA was expressed in E. coli and was purified from rhamnose-induced cells by affinity chromatography on Ni-NTR agarose (Fig. 1). ApdA migrated at an apparent molecular mass of 22 to 23 kDa, similar to ApdA purified from wild-type R. rubrum.
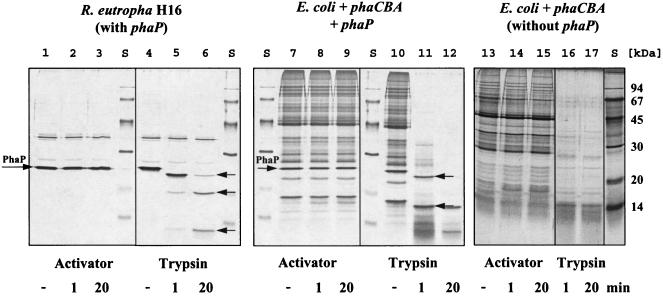
Purified recombinant ApdA was able to activate nPHB granules in a manner comparable to the manner of wild-type ApdA, as first described by Merrick and Doudoroff (23). Figure 2A shows the hydrolysis of nPHB granules by R. rubrum PHB depolymerase after trypsin activation and after activation with purified ApdA. Both types of activated granules were rapidly hydrolyzed by soluble PHB depolymerase of R. rubrum. However, when a trypsin inhibitor (ovomucoid) was added, only the reaction with trypsin was inhibited; the reaction with ApdA was not inhibited (11, 23). Similar results were obtained if nPHB granules isolated from C. violaceum, B. megaterium, B. cereus, or recombinant E. coli harboring the PHB biosynthetic genes (phaCBA) and the major phasin gene (phaP) were used instead of nPHB granules isolated from R. eutropha. nPHB granules isolated from R. rubrum were not suited very well for ApdA analysis because it was not possible to isolate ApdA-free granules. Therefore, nPHB granules of R. rubrum did not require additional ApdA for activation in vitro. However, nPHB granules of R. rubrum responded to trypsin activation by an increased rate of hydrolysis after addition of soluble PHB depolymerase. nPHB granules from recombinant E. coli without phaP were easily activated by trypsin but required a higher concentration of ApdA and a lag phase of 5 to 10 min before activation became visible by subsequent hydrolysis by soluble PHB depolymerase. From these results we concluded that trypsin and ApdA activate nPHB by different mechanisms. To obtain further experimental evidence for this conclusion, the SDS-PAGE protein profiles of nPHB granules purified from R. eutropha H16, recombinant E. coli with phaCBA, and E. coli with phaCBA and phaP before and after activation by trypsin or ApdA were compared. The profiles of nPHB granules showed that there was significant proteolytic digestion of the major phasin protein PhaP and of other granule-associated proteins protein after 1 min of trypsin treatment and that the digestion increased after 20 min of trypsin treatment (Fig. 3). In contrast, no digestion was observed with the profiles of ApdA-treated nPHB granules. This is in agreement with the absence of any protease activity of ApdA either with a natural substrate (casein) or with an artificial substrate (N-α-benzoyl-l-arginine-nitranilide) (11). Apparently, the activation effect of ApdA was not caused by proteolysis. We assume that ApdA interacted with the surface layer of nPHB granules in a nonproteolytic manner.
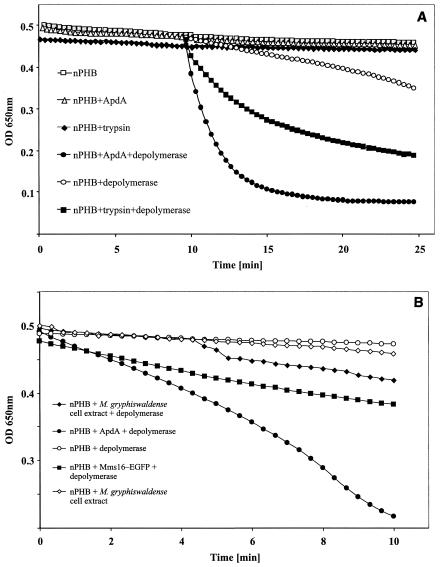
FIG. 2.
In vitro hydrolysis of nPHB granules. (A) nPHB granules from R. eutropha H16 were incubated for 10 min at 40°C (activation) before the hydrolysis reaction was started by addition of soluble PHB depolymerase. In some experiments the hydrolysis reaction for trypsin-activated nPHB was as fast as the ApdA-activated reaction. (B) nPHB granules from R. eutropha H16 with different additions (no preincubation) were hydrolyzed by addition of soluble PHB depolymerase at 40°C. Note the lag time compared to the data shown in panel A. Reactions with depolymerase or activating compounds alone served as controls. Note the different scales in panels A and B. OD 650nm, optical density at 650 nm.
FIG. 3.
Influence of trypsin and purified activator on protein profiles of isolated nPHB granules. A reducing SDS-PAGE analysis of nPHB granules purified from R. eutropha (lanes 1 to 6) (11), from E. coli with phaCBA and phaP (lanes7 to 12), and from E. coli with phaCBA but not phaP (lanes 13 to 17) was performed, and the gels were stained with silver. nPHB granules were treated with ApdA or trypsin. The time of activation (1 or 20 min) is indicated at the bottom. A minus sign indicates that neither trypsin nor activator was added. Lanes S contained a molecular mass standard. The arrows indicate the positions of PhaP and degradation products of phasin proteins. nPHB granules after the treatments were heated in the presence of denaturation-loading buffer and centrifuged. Each supernatant was loaded on an SDS-PAGE gel.
ApdA has significant homology to magnetosome-associated proteins of magnetotactic bacteria and to phasin proteins of PHB-accumulating bacteria.
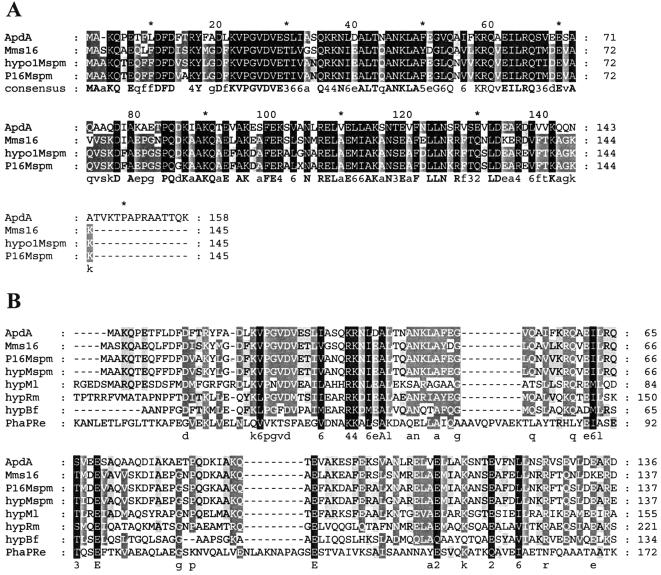
Blast searches of the amino acid sequence of ApdA (accession no. AAO00724) resulted in identification of several closely related sequences. The highest levels of similarity (54 to 55% identity) were found with magnetosome-associated proteins of several magnetotactic bacteria, including Magnetospirillum magnetotacticum (accession no. ZP00056330), Magnetospirillum sp. strain AMB-1 (Mms16; accession no. BAB47588) (26), and M. gryphiswaldense (accession no. BX571783) (7). Moderate levels of similarity (≤40% identity) were found with hypothetical proteins of Mesorhizobium loti (40% identity; accession no. NP102799), Ralstonia metallidurans (34%; accession no. ZO00023358), Burkholderia fungorum (29%; accession no. ZP00031181), and R. eutropha (26%; accession no. CAA59734). Low levels of similarity (<25%) were found with PHB-granule-bound proteins, the so-called phasin proteins of several PHB-accumulating bacteria, such as Azotobacter sp. (24%; accession no. CAD42757), Azotobacter vinelandii (24%; accession no. ZP00088789), and others. Phasins are relatively small proteins (Mr, ≤20 × 103) that are localized in the surface layer of PHB granules and are involved in regulation of PHB synthesis and in determination of the size of PHB granules (34, 37, 38). An alignment of the amino acid sequence of ApdA with the most closely related sequences in the database (August 2003) is shown in Fig. 4. ApdA also corresponded to a highly conserved protein family (COG5490.1) in the conserved domain database. This family comprises several PHB granule-associated proteins, such as PhaP (R. eutropha; accession no. S57610), Gap11 and Gap20 (Methylobacterium extorquens AM1; accession no. AF442748 and AF442749), several hypothetical proteins from M. magnetotacticum, and many other proteins whose functions are unknown.
FIG. 4.
Amino acid alignment for ApdA, Mms16, and related proteins. (A) ApdA, activator of polymer degradation from R. rubrum (accession no. AAO00724); Mms16, Mms16 protein of M. gryphiswaldense (accession no. BX5171783); hypo1Mspm, hypothetical protein of M. magnetotacticum (accession no. ZP00056330); P16Mspm, magnetic-particle-membrane-specific GTPase P16/magnetosome-associated protein of Magnetospirillum sp. strain AMB-1 (accession no. BAB47588). (B) hypMl, hypothetical protein of M. loti (accession no. BAB48585); hypRm, hypothetical protein of R. metallidurans (accession no. ZO00023358); hypBf, hypothetical protein of B. fungorum (accession no. ZP00031181); PhaPRe, PHA-granule-associated protein/phasin of R. eutropha (accession no. AAC78327). The alignments were constructed with ClustalX, version 1.8.1, by using the Gonnet series as a protein weight matrix (gap opening, 10; extension, 0.2) and GeneDoc (shading in conservation mode). The shading indicates the degree of homology, as follows: black, 100% conserved; dark gray, 80 to 100% conserved; gray, 60 to 80% conserved; no shading, less than 60% conserved. The values in the consensus line are phylogenetic tree scores for conserved residues; a lower value is better (i.e., there is less evolutionary cost).
Mms16 has been described as the major magnetosome-associated protein in Magnetospirillum sp. strain AMB-1 (25, 26). Okamura et al. found evidence of a GTPase activity for Mms16 and assumed that Mms16 has a key function in magnetosome biosynthesis. Since PHB granules and magnetosomes have some things in common, such as the presence of a membranous surface layer (1, 5, 8), we speculated that the ApdA and Mms16 proteins could have analogous unknown functions in synthesis, maintenance, structure, and/or degradation of these bacterial subcellular structures. We therefore attempted to determine the in vivo subcellular localization of these proteins in R. rubrum.
In vivo localization of ApdA in R. rubrum determined by fusion to EYFP.
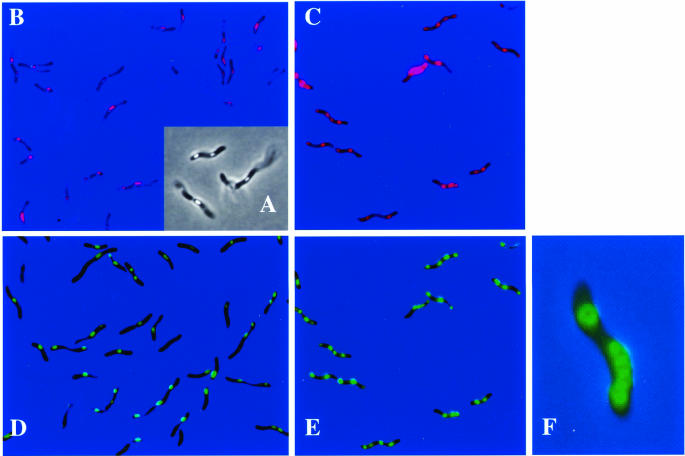
ApdA was isolated from soluble cell extracts of R. rubrum and therefore was first assumed to be localized in the cytoplasm. However, cell extracts of the insoluble fractions also contained significant ApdA activity (data not shown). In order to determine the true localization of ApdA in vivo, a C-terminal fusion of ApdA with the EYFP was constructed and cloned into the broad-host-range vector pBBR1-MCS2 as described in Materials and Methods and then transformed into E. coli S17-1. When recombinant E. coli strains harboring the apdA-eyfp fusion with or without the lac promoter of the parent vector were investigated with the fluorescence microscope, only the strain with the lac promoter showed strong fluorescence in the whole cell and exhibited functional expression of the EYFP portion of the fusion protein in the cytoplasm. The construct with the 5′ upstream region (171 bp) of apdA but without the lac promoter showed no fluorescence. Apparently, the native promoter of apdA was not present in the cloned 5′ upstream region or was not recognized by the E. coli RNA polymerase. Significant ApdA activity was found in soluble cell extracts of the fluorescent E. coli strain. Control cells which harbored only eyfp without ApdA showed strong fluorescence in the cells but had no ApdA activity. We concluded that the apdA-eyfp fusion was functionally expressed in E. coli (from the lac promoter) and that the fusion did not inhibit the activity of ApdA. Both apdA-eyfp fusions (with and without the lac promoter) were transferred to R. rubrum SmRif by conjugation. Selected transconjugants were isolated, were grown photoheterotrophically in PYI medium and acetate-containing MSM, and were analyzed for PHB granule formation and EYFP expression. Since both transconjugants (with and without the lac promoter) showed identical results, only the results obtained with the 5′ upstream region of apdA but without lac are shown below. Almost no PHB was formed in PYI medium, and no EYFP-specific fluorescence was visible. When transconjugants were transferred to acetate-containing MSM and incubated anaerobically in the light, several refractive globular particles per cell became visible within a few hours after growth began as determined by microscopic inspection in the phase-contrast mode, and this indicated that there was synthesis of significant amounts of PHB granules (Fig. 5A). This result was confirmed (i) by gas chromatography-based PHB analysis of lyophilized cells (data not shown) and (ii) by staining of the cells with the PHB-specific dye Nile red. All cells contained several red fluorescent granules (Fig. 5B). The red fluorescent inclusion bodies concomitantly exhibited EYFP fluorescence (Fig. 5D). The results clearly indicate that there was colocalization of ApdA with PHB granules. The EYFP fluorescence did not change significantly during prolonged incubation on acetate-containing MSM, except that the number of PHB granules increased after 24 h (data not shown). We concluded that apdA was expressed from its own promoter during PHB synthesis on acetate-containing MSM and that ApdA is a PHB-granule-bound protein in R. rubrum.
FIG. 5.
Phase-contrast (A) and fluorescence (B to F) microscopic analyses of recombinant R. rubrum cells harboring an apd-eyfp fusion (A, B, and D) or an mms16-egfp fusion, (C and F) in pBBR1-MCS2. Cells were stained with Nile red and were visualized by using a Nile red- and PHB-specific filter (BP546/FT580/LP590) (B and C) or a GFP- and YFP-specific filter (BP450-490/FT510/LP515) (D to F). Bacteria were grown anaerobically in the light in acetate-containing MSM.
Localization of Mms16 in transgene R. rubrum and evidence for ApdA activity of Mms16.
Because of the high level of similarity of the amino acid sequences of ApdA and Mms16, we wondered whether Mms16 could functionally replace ApdA. To address this question, soluble cell extracts of M. gryphiswaldense MSR-1 were prepared and tested for the presence of ApdA activity. As shown in Fig. 2B, low but significant activity was found, and this indicated that functional ApdA activity was present in M. gryphiswaldense MSR-1. We assumed that if Mms16 was responsible for the observed activity, it should be able to functionally replace the ApdA activity of R. rubrum in vitro. A fusion of the M. gryphiswaldense MSR-1 mms16 gene with egfp was constructed and cloned into broad-host-range vector pBBR1-MCS2, resulting in plasmid pCS11. E. coli harboring pCS11 had fluorescence distributed evenly within the cytoplasm, and soluble cell extracts had ApdA activity (Fig. 2B). When pCS11 was transferred into R. rubrum via conjugation, a fluorescence pattern indistinguishable from that of the R. rubrum apdA-eyfp fusion was observed, indicating that there was colocalization of the fusion protein with PHB granules (Fig. 5C and E). Apparently, in R. rubrum, Mms16 has the same subcellular localization and biological activity as ApdA. Inspection of the bacteria after 24 h of growth led to the impression that the fluorescence was not equally distributed in the granules; it appeared that the fluorescence was stronger at the surface of the granules (Fig. 5F) (the effect was much clearer in the original than it appears to be in Fig. 5F). This effect was not visible with Nile red-stained granules when the Cy3 filter was used and was not caused by an image that was out of focus. In order to test whether phasin proteins of PHB-accumulating bacteria (PhaP) also have ApdA activity, cell extracts of R. eutropha expressing PhaP were assayed. However, no activator activity was observed (data not shown). Apparently, PhaP is unable to replace ApdA for activation of nPHB granules for hydrolysis by R. rubrum PHB depolymerase.
ApdA lacks GTPase activity.
A GTPase activity has been described for Mms16, and it was assumed that the GTPase activity was essential for the biological function of Mms16 (26). To test whether ApdA has GTPase activity that might be involved in regulation of PHB degradation, we tested purified ApdA for the presence of GTPase activity. The same conditions that were used for the assay of ApdA and PHB depolymerase activity were used for the GTPase assay, except that the magnesium concentration was raised from 1 to 5 mM. However, no evidence of GTPase activity of ApdA was found even if we used the 80-fold concentration of ApdA that was usually necessary to obtain significant activation of nPHB granules (data not shown). Apparently, ApdA does not have significant GTPase activity in vitro, at least under conditions under which ApdA is active in the in vitro assay.
Effect of ApdA on PHB synthesis in vivo.
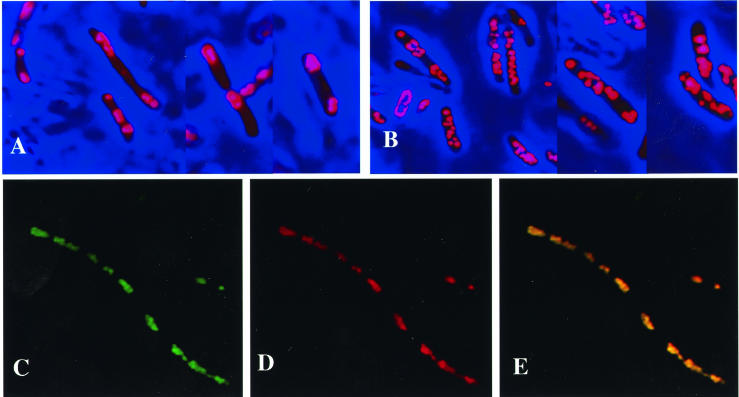
An EYFP fusion analysis of ApdA clearly demonstrated that ApdA was localized at the surface of PHB granules. According to the definition of phasins (34), the R. rubrum activator must be considered a phasin. A comparison of the amino acid sequences revealed low but nevertheless significant similarities to phasin protein PhaP of R. eutropha and other PHB-accumulating bacteria. Several amino acids, including a few hydrophobic residues, two neighboring residues with positive charges, and three glutamate residues, were strictly conserved (Fig. 4). However, PhaP apparently had no ApdA activity (see above). Phasins (PhaP) of other bacteria have been shown to affect the size and number (i.e., the surface/volume ratio) of PHB granules (21, 27, 38, 42). The influence of ApdA on PHB granule formation was analyzed by transformation of the apdA-eyfp fusion (pSN2338) into E. coli HMS174, which already harbored a plasmid with the PHB biosynthetic genes (phaCBA) (17). Recombinant E. coli harboring only the PHB biosynthetic genes (phaCBA) and recombinant E. coli harboring phaCBA and phaP served as controls (Fig. 6A and B). Coexpression of the apdA-eyfp fusion with the PHB biosynthetic genes resulted in formation of very long cells harboring a greatly increased number of PHB granules, but the PHB granules were smaller; this effect was comparable to the effect of PhaP on the surface/volume ratio of PHB granules (Fig. 6) All visible cellular inclusions showed fluorescence of EYFP- and Nile red-specific fluorescence, confirming the localization of ApdA in the PHB-accumulating recombinant E. coli. E. coli expressing the PHB biosynthetic genes alone produced only a few very big PHB granules. Apparently, ApdA has an effect on PHB granule formation comparable to the effect of PhaP in R. eutropha and other PHB-accumulating bacteria if the ApdA activator is overexpressed in E. coli.
FIG. 6.
Fluorescence microscopic analysis of recombinant E. coli with phaCBA but not phaP (A), E. coli with phaCBA and phaP (B), and E. coli with phaCBA and pSN2338 harboring the apd-eyfp fusion (C to E). Cells were stained with Nile red and were visualized by using a Nile red- and PHB-specific filter (BP546/FT580/LP590) (A, B, and D) or a GFP- and YFP-specific filter (BP450-490/FT510/LP515) (C). Colocalization of ApdA-EYFP and Nile red-stained PHB is shown in the fluorescence overlay (E). Bacteria were grown in Luria-Bertani medium, and apdA-eyfp (C, D, and E) or phaP (B) expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM). PHB synthesis was induced byshifting the temperature for 10 min to 42°C. Glucose (1%, wt/vol) was added, and cells were cultivated for another 10 h at 39°C. An image stack was recorded at a primary magnification of ×1,000, and the resulting pictures were deconvoluted by two-dimensional nearest-neighbor analysis.
DISCUSSION
Biosynthesis and degradation of PHB and other polyhydroxyalkanoates (PHA) have been a focus of academic and industrial interest worldwide during the last two decades. More than 20 extracellular denatured PHA depolymerases and several intracellular nPHB depolymerases have been analyzed at the molecular level (15, 30). Some bacteria apparently have only one depolymerase, while others have several isoenzymes of PHA depolymerases. For example, Paucimonas lemoignei (formerly Pseudomonas lemoignei [14]), the model organism for extracellular PHA degradation, has six extracellular dPHA depolymerases (PhaZ1 to PhaZ6), one extracellular nPHB depolymerase (PhaZ7) (10), and at least one (still undiscovered) intracellular nPHB depolymerase. For R. eutropha, the model organism for PHB synthesis, three intracellular nPHB depolymerases have been identified, which are expressed under different physiological conditions and apparently have different tasks in the cells (41). Previous studies with R. eutropha have shown that PHB is simultaneously synthesized and degraded (2, 36). This observation implies that there is a futile cycle of PHB turnover. One explanation might be that PHB is a buffer of reducing power and carbon comparable to a water reservoir that is continuously filled by a river and is continually used depending on the demand for water. Depending on the nutrition and the cellular demand for carbon and reducing power, the net balance between PHB synthesis and degradation is positive (PHB accumulation) or negative (PHB degradation). Regulated expression of selected isoenzymes under different physiological conditions is one possible way for bacteria to differentially respond to various physiological conditions. However, in R. rubrum the situation appears to be different. This bacterium possesses a soluble PHB depolymerase (PhaZ1) whose activity can be modulated in vitro by the absence or presence of a heat-stable activator compound (ApdA) over a range of about 1 order of magnitude. In addition, R. rubrum apparently has an additional, presumably nonsoluble PHB depolymerase with low activity compared to the activity of PhaZ1 (PhaZ2; accession number AY217774). The R. rubrum activator was first analyzed by Merrick and coworkers 40 ago (23, 24). However, ApdA could not be purified, and the nature of ApdA was obscure because of its unusual heat stability. In a previous study ApdA was purified from soluble cell extracts of R. rubrum (11). Purified ApdA was resistant to various physical and chemical stresses (autoclaving, pH 1 to 12, 3% H2O2, phenol-chloroform, 5 M guanidinium hydrochloride, etc.), and before cloning of apdA whether ApdA is a protein at all was questionable. The sensitivity of purified ApdA to proteases, its N-terminal sequence, and finally its functional expression as a His-tagged fusion in recombinant E. coli in this study confirmed that the activator really is a polypeptide.
Our results showed that ApdA of R. rubrum has two different activities in R. rubrum. First, ApdA is able to activate nPHB granules isolated from different species, including recombinant E. coli, so that the granules can be hydrolyzed by soluble R. rubrum PHB depolymerase in the in vitro test system. Second, ApdA is an nPHB-granule-bound protein (phasin) in vivo and influences the surface/volume ratio of PHB granules in a manner typical of PhaP (R. eutropha) and other phasin proteins of different PHB-accumulating bacteria (21, 27, 37, 41). At present, it is not clear whether the in vitro action of the activator is an artifact of the depolymerase test system or whether the activator is a cellular tool that modulates the activity of the depolymerase according to the cellular demands. nPHB granules isolated from R. rubrum contain ApdA at the polymer surface and therefore do not require activation in vitro. The fact that PhaP-free nPHB granules from recombinant E. coli can be (slowly) activated by ApdA suggests that activation is an in vitro artifact because these granules do not contain the putative target of activation (phasin) or any other granule-bound protein of the wild type (except PHB-synthesizing enzymes). However, nPHB granules containing phasin (PhaP) could be activated significantly faster and with shorter lag times than phasin-free nPHB (11), and this result is in agreement with the assumption that ApdA has a physiological function. Since R. rubrum has phototrophic metabolism, one could imagine that PHB is produced anaerobically in the light and is reutilized during the night aerobically, which would require a fine-tuned regulation mechanism in which ApdA could be a useful tool. To address this question and to come to solid conclusions, construction and genetic analysis of apdA-deficient mutants will be necessary in the future.
Two classes of proteins with different levels of similarity to ApdA were found in a database search. The first class comprised highly similar sequences of a magnetosome-associated protein (Mms16) found in several magnetotactic bacteria, such as M. magnetotacticum, Magnetospirillum sp. strain AMB-1, and M. gryphiswaldense MSR-1 (7), and several nonmagnetotactic bacteria (M. loti, R. metallidurans, B. fungorum). Magnetosomes are intracellular structures of magnetotactic bacteria, and similar inclusions with unknown functions have been found recently in Shewanella putrefaciens (references 4 and 32 and references therein). As far as we know, magnetotactic bacteria are also able to accumulate PHB. The second class of proteins with low levels of similarity to ApdA contained PHB-granule-associated proteins, such as PhaP of R. eutropha (similarity, <25%). Apparently, ApdA is related to proteins that are present in two different forms of bacterial inclusion bodies, namely, magnetosomes (possibly with Mms16) and PHB granules (with phasins). In order to unravel the function of ApdA in R. rubrum, the question of the true localization of ApdA in vivo was answered by EYFP fusion analysis. As clearly shown in Fig. 5, ApdA is a PHB-granule-bound protein (phasin) that is expressed at significant levels during PHB accumulation. No significant fluorescence was detected within the cytoplasm or at the cell membrane. However, large amounts of ApdA were found in the soluble fraction after disruption of the cells. This result demonstrates that in vitro localization experiments based on separated cell fractions can be misleading.
The high level of similarity of ApdA to Mms16 of magnetotactic bacteria raised the question of the functional equivalence of Mms16 and ApdA. In the study of Okamura et al. (26), the authors provided evidence that Mms16 has GTPase activity, and they assumed that this activity is crucially involved in magnetosome formation. A poorly conserved P-loop-like motif (GXXXGK) is present in Mms16. However, we were not able to detect any GTPase activity even if a high concentration of purified ApdA was used. This in agreement with the absence of a GTP/ATP binding motif (P-loop) in the ApdA amino acid sequence. Cell extracts of M. gryphiswaldense and of recombinant E. coli harboring the mms16-egfp fusion clearly showed ApdA activity in PHB hydrolysis in vitro. Moreover, the fusion transformed to R. rubrum was localized in PHB granules and could not be distinguished from the corresponding ApdA-EYFP fusion. In conclusion, Mms16 is able to functionally replace the activator in R. rubrum. Since magnetotactic bacteria accumulate PHB and since we were able to demonstrate the subcellular localization of Mms16 in PHB granules in vivo in recombinant R. rubrum, it appears to be reasonable to assume that Mms16 in vivo might also be a PHB-bound phasin in magnetospirilla. Further studies are required to analyze the expression and localization of Mms16-EGFP in its native background.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft and by a scholarship from the Studienstiftung des Deutschen Volkes to R. Handrick. D. Schultheiss and D. Schüler were supported by the BMBF Biofuture program.
REFERENCES
- 1.Boatman, E. S. 1964. Observations on the fine structure of spheroplasts of Rhodospirillum rubrum. J. Cell Biol. 20:297-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doi, Y., A. Segawa, Y. Kawaguchi, and M. Kunioka. 1990. Cyclic nature of poly(3-hydroxyalkanoate) metabolism in Alcaligenes eutrophus. FEMS Microbiol. Lett. 67:165-170. [DOI] [PubMed] [Google Scholar]
- 3.Doi, Y., and A. Steinbüchel (ed.). 2001. Biopolymers, vol. 3a. Polyesters I—biological systems and biotechnological production. Wiley-VCH, Weinheim, Germany.
- 4.Glasauer, S., S. Langley, and T. J. Beveridge. 2002. Intracellular iron minerals in a dissimilatory iron-reducing bacterium. Science 295:117-119. [DOI] [PubMed] [Google Scholar]
- 5.Gorby, Y. A., T. J. Beveridge, and R. P. Blakemore. 1988. Characterization of the bacterial magnetosome membrane. J. Bacteriol. 170:834-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griebel, R., Z. Smith, and J. M. Merrick. 1968. Metabolism of poly-β-hydroxybutyrate. I. Purification, composition, and properties of native poly-β-hydroxybutyrate granules from Bacillus megaterium. Biochemistry 7:3676-3681. [DOI] [PubMed] [Google Scholar]
- 7.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handrick, R., S. Reinhard, and D. Jendrossek. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J. Bacteriol. 182:5916-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handrick, R., S. Reinhardt, M. L. Focarete, M. Scandola, G. Adamus, M. Kowalczuk, and D. Jendrossek. 2001. A new type of thermoalkalophilic hydrolase of Paucimonas lemoignei with high specificity for amorphous polyesters of short-chain-length hydroxyalkanoic acids. J. Biol. Chem. 276:36215-36224. [DOI] [PubMed] [Google Scholar]
- 11.Handrick, R., U. Technow, T. Reichart, S. Reinhardt, T. Sander, and D. Jendrossek. 2004. The activator of the Rhodospirillum rubrum PHB depolymerase is a polypeptide that is extremely resistant to high temperature (121°C) and other physical or chemical stresses. FEMS Microbiol. Lett. 230:265-274. [DOI] [PubMed] [Google Scholar]
- 12.Hanley, S. Z., D. J. Pappin, D. Rahman, A. J. White, K. M. Elborough, and A. R. Slabas. 1999. Re-evaluation of the primary structure of Ralstonia eutropha phasin and implications for polyhydroxyalkanoic acid granule binding. FEBS Lett. 447:99-105. [DOI] [PubMed] [Google Scholar]
- 13.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 14.Jendrossek, D. 2001. Transfer of [Pseudomanas] lemoignei, a Gram-negative rod with restricted catabolic capacity, to Paucimonas gen. nov. with one species, Paucimonas lemoignei comb. nov. Int. J. Syst. Evol. Microbiol. 51:905-908. [DOI] [PubMed] [Google Scholar]
- 15.Jendrossek, D., and R. Handrick. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56:403-432. [DOI] [PubMed] [Google Scholar]
- 16.Kadouri, D., E. Jurkevitch, and Y. Okon. 2003. Poly-beta-hydroxybutyrate depolymerase (PhaZ) in Azospirillum brasilense and characterization of a phaZ mutant. Arch. Microbiol. 180:309-318. [Online.] [DOI] [PubMed]
- 17.Kidwell, J., H. E. Valentin, and D. Dennis. 1995. Regulated expression of the Alcaligenes eutrophus pha biosynthesis genes in Escherichia coli. Appl. Environ. Microbiol. 61:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, T., M. Shiraki, T. Abe, A. Sugiyama, and T. Saito. 2003. Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase. J. Bacteriol. 185:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 20.Madison, L. L., and G. W. Huisman. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maehara, A., S. Ueda, H. Nakano, and T. Yamane. 1999. Analysis of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 181:2914-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer, F., M. H. Madkour, U. Pieper-Fürst, R. Wieczorek, M. Liebergesell, and A. Steinbüchel. 1996. Electron microscopic observations on the macromolecular organization of the boundary layer of bacterial PHA inclusion bodies. J. Gen. Appl. Microbiol. 42:445-455. [Google Scholar]
- 23.Merrick, J. M., and M. Doudoroff. 1964. Depolymerization of poly-β-hydroxybutyrate by an intracellular enzyme system. J. Bacteriol. 88:60-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merrick, J. M., R. Steger, and D. Dombroski. 1999. Hydrolysis of native poly(hydroxybutyrate) granules (PHB), crystalline PHB, and artificial amorphous PHB granules by intracellular and extracellular depolymerases. Int. J. Biol. Macromol. 25:129-134. [DOI] [PubMed] [Google Scholar]
- 25.Okamura, Y., H. Takeyama, and T. Matsunaga. 2000. Two-dimensional analysis of proteins specific to the bacterial magnetic particle membrane from Magnetospirillum sp. AMB-1. Appl. Biochem. Biotechnol. 84-86:441-446. [DOI] [PubMed] [Google Scholar]
- 26.Okamura, Y., H. Takeyama, and T. Matsunaga. 2001. A magnetosome-specific GTPase from the magnetic bacterium Magnetospirillum magneticum AMB-1. J. Biol. Chem. 276:48183-48188. [DOI] [PubMed] [Google Scholar]
- 27.Potter, M., M. H. Madkour, F. Mayer, and A. Steinbüchel. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148:2413-2426. [DOI] [PubMed] [Google Scholar]
- 28.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[d(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2002. Cloning of an intracellular d(−)-3-hydroxybutyrate-oligomer hydrolase gene from Ralstonia eutropha H16 and identification of the active site serine residue by site-directed mutagenesis. J. Biosci. Bioeng. 94:106-112. [DOI] [PubMed] [Google Scholar]
- 30.Saito, T., and T. Kobayashi. 2001. Intracellular degradation of PHAs, p. 41-83. In A. Doi and A. Steinbüchel (ed.), Polyesters. Wiley-VCH Weinheim, Germany.
- 31.Saito, T., K. Takizawa, and H. Saegusa. 1995. Intracellular poly(3-hydroxybutyrate) depolymerase in Alcaligenes eutrophus. Can. J. Microbiol. 41(Suppl. 1):187-191. [Google Scholar]
- 32.Schüler, D. 2002. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int. Microbiol. 5:209-214. [DOI] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 34.Steinbüchel, A., K. Aerts, W. Babel, C. Föllner, M. Liebergesell, M. H. Madkour, F. Mayer, U. Pieper-Fürst, A. Pries, H. E. Valentin, and R. Wieczorek. 1995. Considerations on the structure and biochemistry of bacterial polyhydroxyalkanoic acid inclusions. Can. J. Microbiol. 41(Suppl. 1):94-105. [DOI] [PubMed] [Google Scholar]
- 35.Stumpp, T., B. Wilms, and J. Altenbuchner. 2000. Ein neues l-Rhamnose induzierbares Expressionssystem für Escherichia coli. Biospektrum 6:33-36. [Google Scholar]
- 36.Taidi, B., D. A. Mansfield, and A. Anderson. 1995. Turnover of poly(3-hydroxybutyrate) (PHB) and its influence on the molecular mass of the polymer accumulated by Alcaligenes eutrophus during batch culture. FEMS Microbiol. Lett. 129:201-206. [Google Scholar]
- 37.Wieczorek, R., A. Pries, A. Steinbüchel, and F. Mayer. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177:2425-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wieczorek, R., A. Steinbüchel, and B. Schmidt. 1996. Occurrence of polyhydroxyalkanoic acid granule-associated proteins related to the Alcaligenes eutrophus H16 GA24 protein in other bacteria. FEMS Microbiol. Lett. 135:23-30. [DOI] [PubMed] [Google Scholar]
- 39.York, G. M., B. H. Junker, J. A. Stubbe, and A. J. Sinskey. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183:4217-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.York, G. M., J. Lupberger, J. Tian, A. G. Lawrence, J. Stubbe, and A. J. Sinskey. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185:3788-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.York, G. M., J. Stubbe, and A. J. Sinskey. 2001. New insight into the role of the PhaP phasin of Ralstonia eutropha in promoting synthesis of polyhydroxybutyrate. J. Bacteriol. 183:2394-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York, G. M., J. Stubbe, and A. J. Sinskey. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J. Bacteriol. 184:59-66. [DOI] [PMC free article] [PubMed] [Google Scholar]