Abstract
Infections involving Staphylococcus aureus are often more severe and difficult to treat when the organism assumes a biofilm mode of growth. The polysaccharide poly-N-acetylglucosamine (PNAG), also known as polysaccharide intercellular adhesin, is synthesized by the products of the intercellular adhesin (ica) locus and plays a key role in biofilm formation. Numerous conditions and exogenous factors influence ica transcription and PNAG synthesis, but the regulatory factors and pathways through which these environmental stimuli act have been only partially characterized. We developed a DNA affinity chromatography system to purify potential regulatory proteins that bind to the ica promoter region. Using this technique, we isolated four proteins, including the staphylococcal gene regulator SarA, a MarR family transcriptional regulator of the teicoplanin-associated locus TcaR, DNA-binding protein II, and topoisomerase IV, that bound to the ica promoter. Site-directed deletion mutagenesis of tcaR indicated that TcaR was a negative regulator of ica transcription, but deletion of tcaR alone did not induce any changes in PNAG production or in adherence to polystyrene. We also investigated the role of IcaR, encoded within the ica locus but divergently transcribed from the biosynthetic genes. As has been shown previously in Staphylococcus epidermidis, we found that IcaR was also a negative regulator of ica transcription in S. aureus. We also demonstrate that mutation of icaR augmented PNAG production and adherence to polystyrene. Transcription of the ica locus, PNAG production, and adherence to polystyrene were further increased in a tcaR icaR double mutant. In summary, TcaR appeared to be a weak negative regulator of transcription of the ica locus, whereas IcaR was a strong negative regulator, and in their absence PNAG production and biofilm formation were enhanced.
Staphylococcus aureus is an exceptionally adaptable organism and responds rapidly to changes in its environment by modulating the expression of appropriate genes. For example, molecular regulatory processes, which at present are not well understood, enable S. aureus to switch from a free-floating and relatively independent existence to an adherent, community-based biofilm mode of growth. Biofilm formation is an important aspect of endocarditis, osteomyelitis, and corneal and medical device infections related to S. aureus (10, 16, 17).
Critical to S. aureus biofilm formation is the production of a poly-N-acetylglucosamine (PNAG) polysaccharide, of which a low-molecular-weight variant referred to as the polysaccharide intercellular adhesin has been studied by some groups (7, 18). The intercellular adhesin (ica) locus, originally identified in Staphylococcus epidermidis (11) and later found in S. aureus (5, 19), encodes the genes involved in PNAG production. The four proteins implicated in PNAG synthesis, IcaA, IcaD, IcaB, and IcaC, are translated from a single transcript. The icaR gene is transcribed divergently from the other genes and encodes a regulatory protein that binds to the ica promoter just upstream of the initiation codon for the IcaA protein (4, 5, 12), although the exact mechanism by which it regulates icaADCB transcription and PNAG synthesis is not known.
In a previous report, we described the use of an isogenic pair of S. aureus strains, MN8 and MN8m, for an initial characterization of transcriptional regulation of the ica locus (12). S. aureus strain MN8 is a clinical isolate that normally produces only low levels of PNAG and a weak biofilm in vitro, which is augmented in the presence of glucose. This strain underwent a spontaneous mutation during a chemostat culture which led it to constitutively hyperproduce PNAG (19). A 5-nucleotide deletion within the ica promoter region of the mutant strain (MN8m) is responsible for the phenotype and confers the PNAG-overproducing phenotype upon other S. aureus strains (12). This report also demonstrated that IcaR binds to the ica promoter but not in the region of the deletion mutation.
In an effort to isolate potential regulatory factors with high affinity for the region of the ica promoter in which the deletion occurs, we developed a DNA affinity chromatography system. This technique revealed a number of proteins that bind this region of the ica promoter, including two nonspecific DNA binding proteins, DNA-binding protein II and topoisomerase IV, the global regulatory factor SarA, previously identified as a regulator of ica (1, 27), and TcaR, a MarR family protein originally described as a putative transcriptional regulator of the teicoplanin-associated locus (tca) (2). Site-directed mutagenesis was used to delete tcaR from MN8, MN8m, and an unrelated strain, and the effects on ica transcription, PNAG production, and adherence to polystyrene were analyzed. We found that ica transcription was augmented approximately fivefold in the absence of TcaR but that PNAG production and adherence were not significantly increased.
In order to compare the role of TcaR with that of IcaR, we deleted icaR and found that deletion of icaR augmented ica transcription approximately 100-fold, and even though it had only a moderate impact on adherence, it increased PNAG production by more than 10-fold. When both tcaR and icaR were deleted, ica transcription increased approximately 500-fold with respect to the parent strains, the amount of PNAG elaborated was 100-fold greater, and adherence was significantly increased with respect to both the parent strains and the icaR single knockouts.
MATERIALS AND METHODS
Staphylococcal strains and media.
S. aureus MN8 is a clinical isolate provided by Patrick Schlievert, Minneapolis, Minn. Strain MN8m is a spontaneous mutant isolated from a chemostat culture of strain MN8 (19). Strain NCTC 10833 (ATCC 25904) is a clumping factor-positive variant of a throat swab isolate. Partial deletion of the ica locus to produce strains 10833 Δica::tet and MN8 Δica::tet was performed as described by Cramton et al. (5). The strains were grown at 37°C on tryptic soy agar plates containing the appropriate antibiotic. Liquid cultures were either in tryptic soy broth (TSB) lacking glucose and containing (per liter) 17 g of peptone from casein, 3 g of peptone from soymeal, 5 g of NaCl, and 2.5 g of K2HPO4 or in TSB-1% glucose.
Plasmids, primers, and cloning.
Plasmid purifications were performed with the QIAprep spin miniprep kit (Qiagen, Valencia, Calif.). Primers were custom synthesized by Qiagen Operon (Alameda, Calif.). Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs (Beverly, Mass.). The tcaR gene was amplified with primer pair tcaRF (TTTCTTCAAAAATATATTTAGTAGCGAATACAC) and tcaRR (AAGGATAAGATTATTGATAACGCAATAAC) and ligated to the expression plasmid pCRT7-NT (Invitrogen, Carlsbad, Calif.). The recombinant proteins were expressed in Escherichia coli BL21/pLysS cells according to the manufacturer's instructions. The pCRT7-NT vector adds a tag of six histidine residues and an Xpress epitope to the amino terminus of the protein. The IcaR and TcaR proteins and the vector expression control (the histidine tag and Xpress epitope alone) were sequentially purified with Probond nickel affinity chromatography resin (Invitrogen) and the strong cation exchange resin HiTrap SP XL (Amersham). The lac operon from Staphylococcus xylosus was amplified by PCR with lacHfwd (GAGTGAGTGCTCATTGCTTG) and lacHrev (GCCCTAGTTGACTATCATTAG) and cloned into the EcoRV site of the temperature-sensitive plasmid pBT9 (3). The resulting plasmid was amplified with the primer pair lacHmutfwd (GGACATGTTCCGACACTCGG) and lacHmutrev (TGATCTTCAAGATAGGTTCCATC) to delete the KpnI site within the lacH gene and religated to create pLacH.
All plasmid constructs were initially transformed into the restriction-deficient S. aureus strain RN4220 according to the method of Lee (15). Constructs were transferred to other strains of S. aureus by transduction with phage 80 (13). In order to create icaR and tcaR deletion mutants, the region surrounding the gene was amplified by PCR from the total DNA obtained from strain 10833 with one of the primer pairs icaRdelfwd and icardelrev (GGGGGTACCGGAAACCTTTTCGTTTTCATTGTGC) or tcaRdelfwd (GGGGGTACCCTTCAGTAACATCTACCGTTTCAGAATTAC) and tcaRdelrev (GGGGGTACCGTGATATGGTGTATGACTTCCGACACCATC) and cloned into pCR-TOPO (Invitrogen). The resulting plasmid was amplified with icaR-xhofwd (CTCGAGCATCAAGTGTTGTACCGTCATACC) and icaR-xhorev (CTCGAGTGGAGCAGTGGAAGAAAGTAAAAGTC) or tcaR-xhofwd (CTCGAGGTAAATGTTTTACCATAATTATTTCTCCC) and tcaR-xhorev (CTCGAGGAAGATATTGAAAATGTAAGGCAAGTATTAGAAG), phosphorylated with T4 kinase, and religated to produce a construct (pIcaRdel or pTcaRdel) with an XhoI restriction site in place of the coding region.
The erythromycin resistance cassette was amplified from Tn917 by PCR with ermfwd (GGGGGTACCCTCGAGGCCTACGAGAAATTTGTATCG) and ermrev (GGGGGTACCCGTGTAACTTTCCAAATTTACAAAAGCG), digested with XhoI, and ligated into the XhoI site of pIcaRdel-TOPO and pTcaRdel-TOPO. The entire insert was then subcloned into the KpnI site of pLacH to produce pIcaR::erm-placH and pTcaR::erm-placH, which were transformed into RN4220 by electroporation.
Transformants were cultured overnight at 30°C in the presence of 10 μg of erythromycin/ml and 10 μg of chloramphenicol/ml, diluted 1:1,000, and subcultured overnight at 42°C without antibiotics. The cultures were diluted 1:10, plated on Luria-Bertani (LB) agar plates containing 10 μg of erythromycin/ml and 200 μg of Bluo-gal (Invitrogen)/ml, and grown for 48 h at 42°C. White colonies were picked and screened for the double-crossover events, initially by PCR and then by DNA sequencing, which was carried out by the Microbiology Core Facility at Harvard Medical School (Boston, Mass.). In order to create the icaR tcaR double knockout, the tetracycline resistance cassette was excised from plasmid pTX15 (22) with NheI and ClaI. The ends were filled in with Klenow, and the fragment was ligated to XhoI-digested, blunt-ended pTcaR::erm-placH to produce pTcaR::tet-pLacH. This plasmid was used to replace tcaR in RN4220 with the tetracycline resistance marker, and the mutation was transduced to MN8 Δica::erm and 10833 Δica::erm.
DNA affinity chromatography.
Four liters of TSB was inoculated with 40 ml of an overnight culture of S. aureus MN8. Cultures were well aerated and grown at 37°C for 5 h (A600 = 0.9). Cells were lysed essentially as described by Fournier et al. (8); they were first washed in 200 ml of buffer A (20 mM Tris-HCl, 50 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mM EDTA, 5% glycerol), frozen for 2 h at −80°C, and lysed in 50 ml of buffer A containing 100-μg/ml lysostaphin and EDTA-free complete protease inhibitors (Boehringer Mannheim, Indianapolis, Ind.) for 3.5 h on ice; 30 ml of buffer A-1.3 M KCl was added, and the incubation was continued on ice for 30 min. Lysates were cleared by centrifugation, precipitated with 50% ammonium sulfate at 4°C for 1 h, resuspended in 2 ml of phosphate-buffered saline (PBS), and dialyzed through a 3,500-molecular-weight-cutoff membrane (Pierce, Rockford, Ill.) overnight against 4 liters of 10 mM HEPES-1 mM MgCl2-0.5 mM DTT at 4°C. Then 2 nmol of double-stranded MID-ICA probe (CTATGTTACAGGAAAATTAAGTTGCAATTACAAATATTTCCGTTTAATTATAA annealed to its reverse complement) was biotinylated at one end and immobilized on 10 mg of streptavidin-coated M-280 Dynabeads (Dynal, Oslo, Norway) according to the manufacturer's instructions. The beads were equilibrated with binding buffer 1 (BB1; 10 mM HEPES, 60 mM KCl, 4 mM MgCl2, 0.1 mM EDTA, 0.1 mg of bovine serum albumin per ml, and 0.25 mM DTT); 1.5 ml of cell-free lysate (approximately 20 mg of protein) was combined with 6 ml of BB1 and 1.5 mg of sonicated salmon sperm DNA and chilled on ice for 10 min. The lysate mixture was added to the beads and incubated on ice for 10 min. The beads were washed once with BB1-200 μg of sonicated salmon sperm DNA per ml and twice with BB1 without bovine serum albumin or sonicated salmon sperm DNA. DNA-binding proteins were eluted with 1 ml of 10 mM HEPES-0.25 M KCl. The eluate was concentrated and desalted with Microcon YM-3.5 centrifugal concentrators (Millipore). The concentrated sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with NuPAGE 4 to 15% gradient gels and MOPS (morpholineethanesulfonic acid) buffer (Invitrogen). The gels were stained with Coomassie blue, and protein bands were excised and submitted to the Molecular Biology Core Facility (Dana-Farber Cancer Institute, Boston, Mass.) for sequencing by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectral analysis.
Electrophoretic mobility shift.
The 198-bp probe was generated by PCR with genomic DNA from strain MN8 as a template, obtained from either S. aureus MN8 to yield the wild-type probe or S. aureus MN8m to yield the mutant probe with the primer pair icaFWD (ATTGCGTTATCAATAATCTTATCCTTC) and icaREV (TTGCAATTTCTTTACCTACCTTTC). The nonspecific competitor used in gel shift assays was also a 198-bp PCR product representing a sequence from the icaA gene and was amplified from MN8 total DNA with primer pair icaA-FWD (CCTGTATTTATGTCTATTTACTGG) and icaA-Rev (CTTCTCGTATTTGAGTGCAAG). The PCR products were purified with the Qiagen PCR purification kit. For gel shift analysis, the double-stranded probes were labeled with [γ-32P]ATP with T4 kinase. Gel shift assays were performed essentially as described by Fournier et al. (8). A 20-μl binding reaction containing 5 to 25 ng (14 to 70 nM) of purified recombinant TcaR, 1 μg of sonicated salmon sperm DNA, and 1 μg of poly(dI-dC) in BB1 was incubated at 21°C for 10 min before 20,000 cpm of radiolabeled probe (final concentration of probe, 5 nM) was added. The reaction was incubated for an additional 15 min, loaded onto a 5% nondenaturing polyacrylamide gel, and electrophoresed in chilled 1× Tris-borate-EDTA (TBE) at 360 V for 1 h. Gels were dried and exposed to radiographic film overnight at −80°C.
DNase I footprint analysis.
Footprinting was carried out essentially as described by Sandaltzopoulos and Becker, with the modifications described by Jefferson et al. (12, 26); 10 pmol of icaREV was end labeled with [γ-32P]ATP with T4 kinase. The labeled oligonucleotide was precipitated with ethanol and used in a 50-μl PCR with 10 pmol of 5′-biotinylated icaFWD and genomic DNA from MN8 or MN8m as a template for the amplification of the 198-bp probe representing the ica promoter region. The biotinylated, radiolabeled PCR product was immobilized on 500 μg of Dynabeads according to the manufacturer's instructions. The magnetic beads were resuspended in 100 μl of binding buffer 2 [BB2; 10 mM Tris-Cl, 5 mM MgCl2, 2 mM DTT, 50 μg of bovine serum albumin per ml, 2 μg of poly(dI-dC) per ml, 0.5 μg of sonicated salmon sperm DNA per ml, 100 mM KCl]; 10 μl of oligonucleotide-coated beads, 100 ng of purified recombinant TcaR (112 nM) in Tris buffer, and 25 μl of BB2 were combined and incubated for 10 min at 21°C. Then 5 μl of a DNase I solution (10 mM Tris-Cl, 5 mM MgCl2, 2 mM DTT, 10 mM CaCl2, 100 mM KCl, 2.5 U of DNase I per ml) was added, and the reaction was carried out for 2 min at 21°C before the addition of 50 μl of 2× stop buffer (4 M NaCl, 100 mM EDTA). The beads were washed in 1× stop buffer, resuspended in 95% formamide-6 μg of bromophenol blue per ml, heated to 76°C for 5 min, and loaded on a 6% prerun sequencing gel at 55 W for 25 min. The gel was dried at 80°C and analyzed by autoradiography.
RNA slot blot analysis.
S. aureus cultures were grown in TSB-1% glucose at 37°C for 16 h. RNA was extracted from 109 cells with the RNeasy Protect miniprep kit and RNase-free DNase kit (Qiagen) as described in the manufacturer's instructions except that 0.1 mg of lysostaphin per ml was used in place of lysozyme to lyse the cells. RNA concentrations were determined by absorbance at 260 nm, and 10 μg of each sample was immobilized on a nylon membrane. The single icaADBC transcript was detected with a DNA probe specific for icaAD that was generated by PCR (icaAfwd, GTATTTATGTCTATTTACTGGATTGTCGGTTC, and icaBrev, TCCGGCAATATGATCAAGATACTCAACA) and labeled with the ECL direct nucleic acid labeling and detection system (Amersham). Blots were reprobed with a probe specific for the housekeeping gene for DNA gyrase B, which was also generated by PCR (gyrBFwd, TTATGGTGCTGGGCAAATACAAG, and gyrBRev, ACCTTCGAATTGAGGATCACC) to ensure equal loading.
Adherence assay.
Microtiter well adherence assays for PNAG production were performed essentially as described by Valle et al. with minor modifications (27). Cultures were grown overnight in tryptic soy broth (TSB)-1% glucose, diluted to an optical density at 650 nm (OD650) of 0.2, and aliquoted into 96-well polystyrene flat-bottomed microtiter plates (Corning). The plates were centrifuged at 3,000 rpm for 10 min and incubated at 37°C for 1 h. The wells were emptied, washed twice with phosphate-buffered saline (PBS), dried at ambient temperature, stained for 30 s with safranin, and washed under gently running tap water. The stained bacteria were assessed qualitatively for relative adherence by visual inspection and quantitatively by resuspending the adherent cells in 200 μl of PBS by gentle sonication and measuring the OD405 with an enzyme-linked immunosorbent assay reader. Statistical analysis was performed on the OD405 values with Tukey's multiple-comparison test. Calculations were carried out with Prism software.
PNAG slot blot analysis.
PNAG slot blots were performed essentially as described previously (6) with minor modifications. Bacteria were grown overnight in TSB-1% glucose, and 109 cells were pelleted, washed once with PBS, resuspended in 50 μl of 5 N NaOH, and vortexed for 30 s. The extracts were cleared by centrifugation, and diluted 1:5 in Tris-buffered saline (TBS), and 100 μl of extract from S. aureus strain MN8 or 10833 or 1 μl from strain MN8m was serially diluted in TBS and immobilized on nitrocellulose with a vacuum manifold. Blots were blocked for 1 h in 5% skim milk, probed with 1:50,000-diluted rabbit antiserum specific for PNAG (18) for 2 h at 21°C, washed, and probed with 1:10,000-diluted goat anti-rabbit immunoglobulin-horseradish peroxidase conjugate for 1 h at 21°C. Bands were visualized with the enhanced chemiluminescence kit (Amersham).
RESULTS
DNA affinity chromatography.
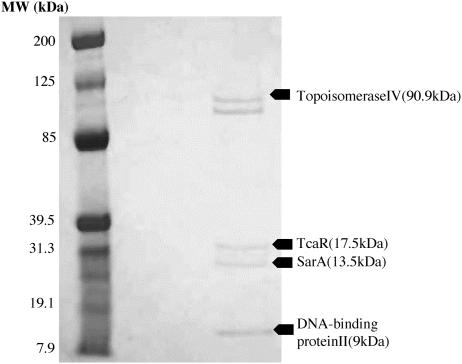
S. aureus strain MN8m was originally isolated as a spontaneous PNAG-hyperproducing mutant of strain MN8. A deletion of a 5-nucleotide TATTT sequence located in the center of the ica promoter is sufficient for the PNAG-overproducing phenotype associated with strain MN8m, but the molecular basis for this effect has not been elucidated (12). IcaR binds to a distinct region of the ica promoter, and binding is unaffected by the deletion mutation. We therefore sought to isolate potential regulatory factors that bind to the region of the ica promoter in which the deletion mutation occurs. We designed a DNA affinity chromatography system to isolate proteins with high affinity for a 53-bp oligonucleotide representing the region of the ica promoter surrounding the TATTT motif. The oligonucleotide, MID-ICA, spans the ica promoter sequence from 131 to 79 bases upstream from the icaA start site. MID-ICA was biotinylated at one end, immobilized on streptavidin-coated magnetic particles, and used to purify specific DNA-binding proteins from an S. aureus lysate. The purified proteins were separated by SDS-PAGE and stained with Coomassie blue, as shown in Fig. 1. Bands were carefully excised and analyzed by MALDI-TOF mass spectral analysis.
FIG. 1.
SDS-PAGE analysis of proteins isolated by DNA affinity chromatography. The left lane contains standard proteins, and the right lane contains proteins isolated by their affinity for an oligonucleotide representing the sequence from the center of the ica promoter (MID-ICA). The gel was stained with Coomassie blue. The band intensities in this figure were digitally enhanced by using Adobe PhotoShop for the purpose of clarity. The indicated bands were excised and sequenced by MALDI-TOF mass spectral analysis. The proteins predicted by the sequence obtained are indicated.
Mass spectral analysis identified two nonspecific DNA binding proteins, DNA-binding protein II and topoisomerase IV, and SarA, which has been described previously as an ica regulator (1, 27). Studies with these proteins were not pursued further. The approximately 90-kDa protein that migrated slightly faster than topoisomerase IV was not successfully sequenced. Mass spectral analysis also indicated that an approximately 20-kDa protein isolated from the DNA affinity beads was most likely TcaR, a protein that displays significant homology to the MarR family of regulatory factors. The tcaR gene is part of the three-cistron teicoplanin-associated locus (tca), so named for the discovery that teicoplanin resistance increases in S. aureus when the locus is deleted or interrupted (2).
Affinity of TcaR for the ica promoter.
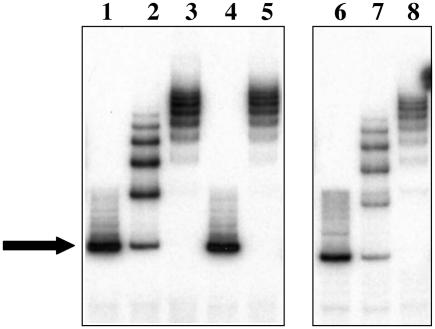
In order to determine whether or not TcaR binds specifically to the ica promoter, we expressed histidine-tagged TcaR in E. coli and combined purified recombinant TcaR with a 198-bp probe representing the sequence of the entire wild-type ica promoter from strain MN8 or the mutant ica promoter from strain MN8m in electrophoretic mobility shift assays. The recombinant TcaR protein induced shifts in both the wild-type (lanes 2 and 3) and mutant (lanes 7 and 8) probes, producing a ladder-like pattern of migration (Fig. 2). The TcaR-induced shifts were lost when a 100-fold excess of unlabeled specific probe was added to the mixture but were unaffected by a 100-fold excess of a nonspecific, control probe (a 198-bp fragment of the icaA gene). An expression control (purified lysate from E. coli containing the empty expression vector) failed to shift either probe (data not shown). These results indicate that TcaR binds to the ica promoter specifically and that binding is not dependent upon the 5-bp TATTT motif.
FIG. 2.
Electrophoretic mobility shift assay indicates that TcaR binds to the ica promoter. Lanes 1 to 5 show mobility shift analysis of the probe representing the wild-type strain MN8 ica promoter. Additions to this probe were as follows: lane 1, nothing (free probe only); lane 2, 5 ng (14 nM) of purified recombinant TcaR; lane 3, 25 ng (70 nM) of TcaR; lane 4, a 100-fold excess of specific competitor (unlabeled wild-type probe) and 25 ng (70 nM) of TcaR; and lane 5, 100-fold excess nonspecific competitor (198 bp sequence from icaA) and 25 ng (70 nM) of TcaR. Lanes 6 to 8 show mobility shift analyses of the probe representing the ica promoter of the mucoid strain MN8m (MUC). Additions to this probe were as follows: lane 6, nothing (free probe only); lane 7, 5 ng (14 nM) of TcaR; and lane 8, 25 ng (70 nM) of TcaR. The solid arrow indicates the free probe. TcaR induced a number of shifts in both the wild-type and mutant probes.
In order to localize the TcaR binding site(s) within the ica promoter, we performed DNase I footprint analysis. DNase I footprinting resulted in a diffuse pattern of protection over approximately 120 bases in the center of the ica promoter, indicating that TcaR protects multiple sites within the ica promoter (data not shown). The pattern of protection was similar within the wild-type and mutant ica promoter sequences. This result, coupled with the observation of a ladder effect in the gel shift assays, could indicate the presence of multiple TcaR recognition sites within the ica promoter sequence or that TcaR oligomerizes once it is bound to the promoter region.
Effect of TcaR on ica transcription.
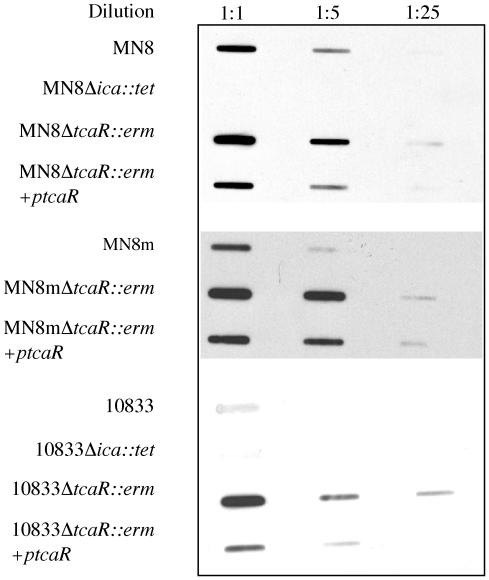
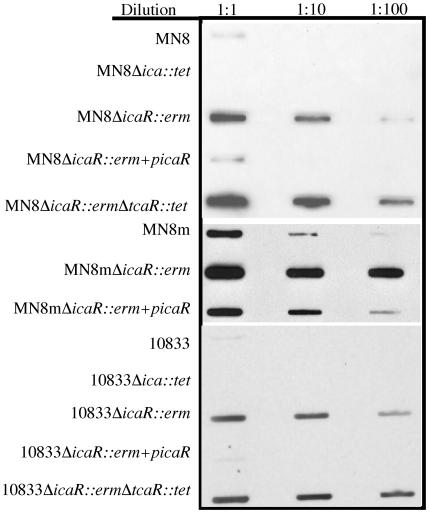
The specificity with which TcaR bound to the ica promoter suggested that the protein may have a regulatory function in ica transcription. In order to characterize the effect of TcaR on ica transcription and PNAG synthesis, we replaced the tcaR gene in the RN4220 chromosome by allelic replacement with an erythromycin resistance cassette and transduced the mutation to S. aureus strains MN8, MN8m, and 10833. The transcriptional activity of the ica locus was characterized in the resulting strains, MN8 ΔtcaR::erm, MN8m ΔtcaR::erm, and 10833 ΔtcaR::erm. We constructed a probe that binds to icaAD mRNA and used it to detect ica transcript levels in bacterial extracts by Northern slot blot analysis. The icaADBC genes are transcribed as a single transcript, so icaAD mRNA is representative of the transcription of all four genes. All blots were reprobed with a probe specific for the housekeeping gene DNA gyrase B to ensure equal loading of RNA (data not shown). Transcription of the ica locus was previously shown to be elevated in the PNAG hyperproducing strain MN8m, so only 2 μg of total RNA from the MN8m strains was blotted onto nylon membranes, whereas 10 μg of total RNA from the weakly PNAG-producing strains 10833 and MN8 was analyzed. Northern analysis indicated an approximately fivefold increase in icaAD transcript level in all three strains in the absence of tcaR (Fig. 3). This increase was lost when the mutants were complemented with a plasmid carrying tcaR under the control of an inducible promoter (ptcaR), indicating that the phenotype was due to the lack of tcaR. Thus, TcaR appears to be a negative regulator of ica transcription.
FIG. 3.
Slot blot analysis of RNA indicates that deletion of tcaR augments icaADBC transcription in strains MN8, MN8m, and 10833. Total RNA was isolated from the indicated strains, serial dilutions of the RNA were made, and 10, 2, or 0.4 μg of MN8 and 10833 RNA or 2, 0.4, or 0.08 μg of MN8m RNA was immobilized on nylon membranes with a slot blot apparatus. The blots were probed with a single-stranded DNA probe specific for the icaAD transcript labeled with horseradish peroxidase. A chemiluminescent substrate was added, and the blots were exposed to autoradiographic film. Blots representative of three separate experiments are shown.
Effect of TcaR on PNAG production.
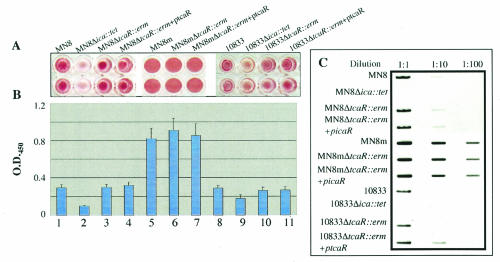
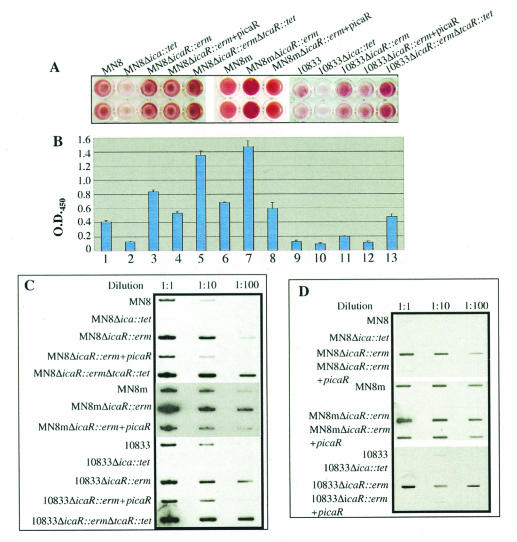
We hypothesized that because ica transcription was elevated in the tcaR deletion mutants, PNAG synthesis and biofilm formation would also be augmented in these strains. The ability of these strains to form a biofilm was measured by a microtiter well adherence assay. The strains were grown in TSB-1% glucose overnight and then diluted and allowed to adhere to polystyrene microtiter wells. Adherence was assessed by visual inspection of the dry, stained biofilms and by optical density measurement of resuspended, stained bacteria. The MN8 Δica::tet and 10833 Δica::tet mutants, which lack the icaADBC genes and are therefore unable to synthesize PNAG, were used as controls in the adherence assay, and both produced very little biofilm. As shown in Fig. 4A and B, deletion of tcaR did not have a significant effect on the ability of the strains to adhere (P > 0.05, Tukey's multiple comparison test).
FIG. 4.
Adherence assays and slot blot analysis of cell surface extracts indicate that the tcaR gene does not have a significant effect on PNAG production or adherence in S. aureus. (A) Bacteria were grown in TSB-1% glucose overnight. Strains were diluted to an OD650 of 0.2, and the cells in 200 μl of each culture were centrifuged to the bottom of wells in a polystyrene microtiter plate. The wells were washed with PBS, and adherent cells were stained with safranin. (B) After staining, 200 μl of PBS was added to each microtiter well, and the bacteria were homogenized by sonication. Bars reflect the mean OD405 of eight samples, and error bars show the standard deviations. (C) Bacteria were grown in 5 ml of TSB-1% glucose for 16 h. Cultures were centrifuged to pellet the cells. Cell surface extracts were made by resuspending bacteria in 50 μl of 5 N NaOH. Extracts were cleared by centrifugation and diluted with 250 μl of TBS, and serial 10-fold dilutions of 100 μl of MN8 and 10833 extracts or of 1 μl of MN8m extract were immobilized on nitrocellulose membranes. The blots were blocked with 5% milk and probed with PNAG-specific rabbit antiserum and polyclonal goat anti-rabbit immunoglobulin-horseradish peroxidase conjugate. A chemiluminescent substrate was added to the blots, and the blots were exposed to autoradiographic film.
Adherence to plastic is a good relative measure of PNAG production, but other surface proteins and polysaccharides can also play a role in adherence. In order to measure PNAG production more directly, we performed a PNAG slot blot assay with polyclonal antiserum specific for PNAG. Cell surface extracts were prepared from stationary-phase cultures grown in TSB-1% glucose, blotted onto nitrocellulose, and probed with a PNAG-specific polyclonal antiserum (18). As MN8m is a PNAG hyperproducer, extracts from this strain and MN8m ΔtcaR::erm were diluted 100-fold before they were blotted onto nitrocellulose. Again, MN8 Δica::tet and 10833 Δica::tet were used as negative controls and did not react with the PNAG-specific antiserum. There was no obvious difference in the amount of PNAG elaborated by the tcaR mutants with respect to the parent strains (Fig. 4).
We were surprised that despite the increase in ica transcription in the absence of TcaR, there was no discernible effect on PNAG synthesis, and we reasoned that the effects of the tcaR deletion might be manifested under different growth conditions. To test this, cultures were grown under various conditions that have been shown previously to affect biofilm formation, including high osmolarity (4% NaCl), elevated temperature (45°C), and the presence of a subinhibitory concentration of tetracycline (0.05 μg/ml) (24). The tcaR deletant did not exhibit significantly greater PNAG expression than the wild-type strain under any conditions (data not shown). These findings suggests that while TcaR appears to be a negative regulator of ica transcription, an effect on PNAG expression in the tcaR deletant strains was not detected by our methods. The effect of the tcaR deletion might be masked under normal growth conditions either by a posttranscriptional regulatory mechanism or possibly by an additional negative regulator of transcription. We therefore hypothesized that IcaR, a known regulator of ica transcription in S. epidermidis, might obscure the effect of TcaR, and we next sought to compare the phenotype of an icaR deletant with that of the tcaR deletant.
Effect of IcaR on icaADBC transcript levels.
It has been reported that IcaR is a repressor of icaADBC transcription in S. epidermidis, although in this report, deletion of icaR did not affect biofilm formation (4). We previously demonstrated, by DNase I protection and electrophoretic mobility shift analysis, that IcaR binds to the S. aureus ica promoter, but direct evidence that it is a repressor of ica transcription in S. aureus has not yet been presented (12). In order to confirm that IcaR acts as a repressor of ica transcription in S. aureus, we inactivated the icaR gene within three S. aureus strains, MN8, MN8m, and 10833, by allelic replacement with an erythromycin resistance cassette. Total RNA from 16-h TSB-1% glucose cultures of the resulting strains, MN8 ΔicaR::erm, MN8m ΔicaR::erm, and 10833 ΔicaR::erm, was subjected to Northern slot blot analysis with the icaAD-specific probe. All blots were also analyzed with a DNA gyrase-specific probe to ensure equal loading (data not shown). As explained above, only 2 μg of RNA from the MN8m and MN8m ΔicaR::erm strains was analyzed, whereas 10 μg of RNA from the MN8 and 10833 strains was analyzed.
In all three strains, inactivation of icaR increased the level of the icaAD transcript (Fig. 5). This increase was estimated to be at least 100-fold, as measured by densitometric analysis with NIH Image software (data not shown). These results demonstrate that, in S. aureus, IcaR is a repressor of ica transcription and that it is functional even in the PNAG-overproducing strain MN8m. The finding that the negative regulatory effects of both IcaR and TcaR are unaffected by the 5-bp deletion within the ica promoter of MN8m is somewhat surprising, considering that PNAG expression is so high in this strain. This suggests that MN8m may have lost a requirement for a positive regulator or that the 5-bp deletion somehow increases the efficiency of transcriptional initiation by the RNA polymerase complex at this promoter.
FIG. 5.
Slot blot analysis of RNA indicates that deletion of icaR augments icaADBC transcription in strains MN8, MN8m, and 10833. Total RNA was isolated from the indicated strains, serial dilutions of the RNA were made, and 10, 1, or 0.1 μg of MN8 and 10833 RNA or 2, 0.2, or 2 ng of MN8m RNA was immobilized onto nylon membranes with a slot blot apparatus. The blots were probed with a single-stranded DNA probe specific for the icaAD transcript labeled with horseradish peroxidase. A chemiluminescent substrate was added, and the blots were exposed to autoradiographic film. Blots representative of three separate experiments are shown.
Mutation of icaR augmented ica transcription 100-fold, whereas mutation of tcaR induced only a 5-fold increase in transcription, suggesting that IcaR is a stronger repressor of ica transcription. We therefore reasoned that functional IcaR likely prevents any increase in PNAG production in the tcaR mutant strains. We replaced tcaR with a tetracycline resistance cassette in the icaR knockout strains to produce MN8 ΔicaR::erm ΔtcaR::tet and 10833 ΔicaR::erm ΔtcaR::tet. The double-knockout strains exhibited an approximately 500-fold increase in ica transcription with respect to the wild-type strain, suggesting a synergistic effect of deleting both regulators.
Effect of icaR on PNAG production.
To assess PNAG production in the icaR mutants, the microtiter adherence assay was again used. As shown in Fig. 6A and B, the icaR knockout strains exhibited increased adherence to the polystyrene microtiter wells with respect to their parent strains (P < 0.001, Tukey's multiple-comparison test). There was an approximately twofold increase in the OD405 of the MN8 ΔicaR::erm, MN8m ΔicaR::erm, and 10833 ΔicaR::erm constructs compared to strains MN8, MN8m, and 10833, respectively.
FIG. 6.
Slot blot analysis of cell surface extracts indicates that deletion of the icaR gene augments PNAG production and adherence in S. aureus. (A) Bacteria were grown in TSB-1% glucose overnight. Strains were diluted to an OD650 of 0.2, and the cells in 200 μl of each culture were centrifuged to the bottom of the wells in a polystyrene microtiter plate. The wells were washed with PBS, and adherent cells were stained with safranin. (B) After staining, 200 μl of PBS was added to each microtiter well, and the bacteria were homogenized by sonication. Bars reflect the mean OD405 of eight samples, and error bars show the standard deviations. (C) Bacteria were grown in TSB-1% glucose overnight. Cultures were diluted to an OD650 of 1.0, and 1 ml was centrifuged to pellet the cells. Cell surface extracts were made by resuspending bacteria in 50 μl of 5 N NaOH. Extracts were cleared by centrifugation and diluted with 250 μl of TBS, and serial 10-fold dilutions of 100 μl of MN8 and 10833 extracts or of 1 μl of MN8m extract were immobilized on nitrocellulose membranes. The blots were blocked with 5% milk and probed with PNAG-specific rabbit antiserum and polyclonal goat anti-rabbit immunoglobulin-horseradish peroxidase conjugate. A chemiluminescent substrate was added to the blots, and they were exposed to autoradiographic film.
To confirm that the increase in adherence was a direct result of inactivating icaR, we complemented 10833 ΔicaR::erm, MN8 ΔicaR::erm, and MN8m ΔicaR::erm with a plasmid carrying an inducible copy of the icaR gene (picaR). Complementation of the icaR deletions reduced adherence, confirming that the increase in adherence was a result of deleting icaR (Fig. 6A and B). It is notable that strain MN8m, which normally produces copious quantities of PNAG, exhibited even greater adherence and was more aggregative in the absence of IcaR. On TSA plates, MN8m ΔicaR::erm formed small tight colonies that were even stickier that MN8m colonies, and when grown in TSB, the bacteria were so aggregative that they formed a single compact pellet (data not shown). This suggests that despite the PNAG-hyperproducing phenotype of strain MN8m, IcaR-mediated repression of ica is still effective in this strain.
Adherence of the icaR tcaR double knockout strains MN8 ΔicaR::erm ΔtcaR::tet and 10833 ΔicaR::erm ΔtcaR::tet was significantly greater than that of the single icaR mutant strains (P < 0.001), suggesting that, as would be expected from its role in regulation of ica transcription, TcaR can affect biofilm formation.
We also measured PNAG production in the icaR mutant strains. Figure 6C shows that PNAG synthesis was approximately 10-fold higher in the icaR deletion mutants than in the respective parent strains, substantiating the findings in the adherence assays. Complementation of the icaR gene reduced PNAG expression to wild-type levels (Fig. 6C). Interestingly, deletion of both icaR and tcaR resulted in a 100-fold increase in PNAG elaboration, 10-fold greater than the increase observed in the icaR single knockout. Overall, these results suggest that both IcaR and TcaR repress ica transcription and subsequent PNAG production but that IcaR is a more potent repressor and can override the effects of deleting tcaR. Deleting both tcaR and icaR resulted in a great increase in PNAG production and adherence, but the level of PNAG synthesis in the double knockouts of the wild-type strains was not as high as that associated with strain MN8m, suggesting that other regulatory factors are involved in ica transcriptional control.
When working with the MN8m strains, we noted that while this strain was not highly aggregative in TSB made without glucose, MN8m ΔicaR::erm grew as an aggregated cellular mass even in the absence of glucose. We therefore performed PNAG blots on the strains and icaR deletants grown in TSB without any glucose. The strains were first grown overnight in the absence of glucose, diluted the following day 1:100 in TSB without glucose, and grown to late log phase. Figure 6D shows that deletion of icaR significantly augmented PNAG production in the absence of glucose. In fact, in strains MN8 and 10833, PNAG production was undetectable by our methods in the absence of glucose but was strong in the icaR knockout strains. This suggests that IcaR plays a role in the suppression of PNAG production in the absence of glucose.
DISCUSSION
A thorough comprehension of the mechanism by which PNAG elaboration is regulated in S. aureus is an important prerequisite to understanding biofilm formation and could ultimately lead to the development of methods to repress the expression of this important virulence factor. It is becoming increasingly apparent, however, that transcriptional regulation of the genes involved in PNAG synthesis, the ica genes, is quite complex. It has previously been reported that IcaR is a repressor of ica transcription in S. epidermidis, although an influence of IcaR on PNAG production and bacterial adherence or biofilm formation was not demonstrated (4). We have shown here that IcaR is also a repressor of ica transcription in S. aureus and that deletion of icaR results in a substantial increase in PNAG elaboration and bacterial adherence to polystyrene. We also present evidence that glucose impacts ica transcription and PNAG production by counteracting the negative regulatory effect of IcaR. Dobinsky et al. reported that despite elevated transcription of the icaADBC genes, a single strain of S. epidermidis did not form a biofilm or produce detectable amounts of the low-molecular-weight form of PNAG in the absence of glucose (7). In the presence of glucose, the level of icaADBC transcript was very low, but biofilms and the low-molecular-weight form of PNAG were produced. The most intuitive explanation for this dissociation between ica transcription and PNAG production is that glucose is required as a substrate for factors involved in PNAG production. The results presented here indicate that in S. aureus, the glucose-mediated increase in PNAG is due, at least in part, to alleviation of IcaR-mediated repression and a subsequent increase in ica transcription. When icaR was deleted in S. aureus strains MN8, 10833, and MN8m, ica transcription was elevated and PNAG production was greatly increased even in the absence of any exogenous glucose source. This strongly suggests that the positive effect that glucose has on PNAG expression is at least partially attributable to alleviation of IcaR-mediated repression and transcriptional activation of ica. Further studies will be required to determine whether glucose interacts directly with IcaR to alleviate repression of ica transcription or if glucose exerts its effect on IcaR indirectly, through some other regulatory factor.
Interestingly, the MN8m ΔicaR::erm strain, which produced copious quantities of PNAG, was quite growth deficient. Colonies were small, and in broth the bacteria formed a small compact pellet. Presumably the bacteria were so aggregative that they were deprived of the space and nutrients required for replication. This helps to explain why the ica locus is so tightly regulated. Limited PNAG production likely allows the bacteria to form a protective biofilm and to localize in a nutrient-rich environment, but when PNAG is produced unchecked, growth of the cell population is retarded. In addition to its role in biofilm formation, PNAG may act as a “sink” in which excess glucose can be removed from the cell and possibly stored for future use. Alternately, unregulated PNAG production may divert glucose and other substrates into polysaccharide production and deprive the bacterial cells of sufficient nutrients to grow and divide. Overall, if a bacterium synthesizes too much PNAG, it is likely using excessive energy and resources for the production of one factor at the expense of reproduction, indicating one reason why staphylococci need an effective means of tightly regulating PNAG elaboration.
TcaR proved to have a strong interaction with the ica promoter in vitro. Although Northern analysis of tcaR knockout strains suggested that TcaR is a negative regulator of ica transcription, deletion of tcaR did not have a significant impact on PNAG production in strains that were also able to make IcaR. IcaR appeared to be a more potent repressor of ica transcription and was able to mask any phenotypic effects due to the deletion of tcaR. The effects of knocking out both icaR and tcaR on ica transcription, PNAG production, and adherence were synergistic. Overall, both the icaR and tcaR genes encode proteins regulating ica transcription and PNAG production, which, along with sarA, can now be considered part of the regulatory network affecting biofilm formation by S. aureus.
Acknowledgments
We thank Tomàs Maira-Litran for provision of the PNAG-specific antiserum.
This work was supported by NIH grants AI46707 and F32AI51892 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bachi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophys. Acta 1523:135-139. [DOI] [PubMed] [Google Scholar]
- 3.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 4.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramton, S. E., M. Ulrich, F. Götz, and G. Döring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 185:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fournier, B., R. Aras, and D. C. Hooper. 2000. Expression of the multidrug resistance transporter NorA from Staphylococcus aureus is modified by a two-component regulatory system. J. Bacteriol. 182:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galdbart, J. O., J. Allignet, H. S. Tung, C. Ryden, and N. El Solh. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 10.Götz, F., C. Heilmann, and S. E. Cramton. 2000. Molecular basis of catheter associated infections by staphylococci. Adv. Exp. Med. Biol. 485:103-111. [DOI] [PubMed] [Google Scholar]
- 11.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 12.Jefferson, K. K., S. E. Cramton, F. Götz, and G. B. Pier. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889-899. [DOI] [PubMed] [Google Scholar]
- 13.Kasatiya, S. S., and J. N. Baldwin. 1967. Nature of the determinant of tetracycline resistance in Staphylococcus aureus. Can. J. Microbiol. 13:1079-1086. [DOI] [PubMed] [Google Scholar]
- 14.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, J. C. 1993. Methods in molecular biology, vol. 47. Electrotransformation of staphylococci. Humana Press Inc., Totowa, N.J. [DOI] [PubMed]
- 16.Leid, J. G., J. W. Costerton, M. E. Shirtliff, M. S. Gilmore, and M. Engelbert. 2002. Immunology of Staphylococcal biofilm infections in the eye: new tools to study biofilm endophthalmitis. DNA Cell Biol. 21:405-413. [DOI] [PubMed] [Google Scholar]
- 17.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 18.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenney, D., K. L. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Döring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:1523-1527. [DOI] [PubMed] [Google Scholar]
- 21.Peacock, S. J., C. E. Moore, A. Justice, M. Kantzanou, L. Story, K. Mackie, G. O'Neill, and N. P. Day. 2002. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70:4987-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peschel, A., B. Ottenwalder, and F. Götz. 1996. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol. Lett. 137:279-284. [DOI] [PubMed] [Google Scholar]
- 23.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor 32 σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reese, M. G. 2001. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 26:51-56. [DOI] [PubMed] [Google Scholar]
- 26.Sandaltzopoulos, R., and P. B. Becker. 1994. Solid phase DNase I footprinting: quick and versatile. Nucleic Acids Res. 22:1511-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not sigma(B) is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]