Abstract
flaA1 and wbpB are conserved genes with unknown biological function in Helicobacter pylori. Since both genes are predicted to be involved in lipopolysaccharide (LPS) biosynthesis, flagellum assembly, or protein glycosylation, they could play an important role in the pathogenesis of H. pylori. To determine their biological role, both genes were disrupted in strain NCTC 11637. Both mutants exhibited altered LPS, with loss of most O-antigen and core modification, and increased sensitivity to sodium dodecyl sulfate compared to wild-type bacteria. These defects could be complemented in a gene-specific manner. Also, flaA1 could complement these defects in the wbpB mutant, suggesting a potential redundancy of the reductase activity encoded by both genes. Both mutants were nonmotile, although the wbpB mutant still produced flagella. The defect in the flagellum functionality of this mutant was not due to a defect in flagellin glycosylation since flagellins from wild-type strain NCTC 11637 were shown not to be glycosylated. The flaA1 mutant produced flagellins but no flagellum. Overall, the similar phenotypes observed for both mutants and the complementation of the wbpB mutant by flaA1 suggest that both genes belong to the same biosynthesis pathway. The data also suggest that flaA1 and wbpB are at the interface between several pathways that govern the expression of different virulence factors. We propose that FlaA1 and WbpB synthesize sugar derivatives dedicated to the glycosylation of proteins which are involved in LPS and flagellum production and that glycosylation regulates the activity of these proteins.
Helicobacter pylori is a spiral-shaped gram-negative microaerophilic bacterium that was first isolated from the human stomach in 1984 (42). It is estimated that 70% of the worldwide population is infected by this bacterium. Most infections are asymptomatic, but they can also lead to gastric ulcers and cancers (26, 51, 66, 69). The relationship between colonization by H. pylori and the onset of the disease is not fully understood. Hence, it is important to identify essential bacterial virulence factors and elucidate their contribution to disease development.
Several virulence factors contribute to the stringent host and tissue specificities exhibited by H. pylori (37). Among them, urease helps neutralize the acidic pH surrounding the bacteria and allows their survival in the gastric environment (21, 45). In addition, the spiral shape and unipolar flagella of H. pylori confer on the bacterium a corkscrew motion that enhances motility in the viscous gastric mucus (32, 33, 63) and is essential for host colonization (22, 23, 35). Lipopolysaccharide (LPS) is also important for the virulence of H. pylori since strains lacking the O antigen are significantly impaired in their capacity to colonize the murine stomach (40). The H. pylori O antigen is composed of N-acetyl-d-glucosamine, l-fucose, and d-galactose (4, 5, 46), which form structural motifs that are identical to human blood group antigens Lewis X, Y, and b (4, 5, 46-48). This host mimicry might allow the bacteria to evade human immune defenses and to establish long-term host colonization (54, 61).
This study focuses on two genes of unknown biological function in H. pylori: flaA1 (HP0840) and wbpB (HP0679). They exhibit significant homologies to LPS (9), capsule (8, 57, 58), and/or flagellar biosynthetic genes found in medically relevant bacteria. For example, FlaA1 shows 52% homology to the C-terminal half of WbpM, which is essential for LPS synthesis in Pseudomonas aeruginosa (9). The sequence homologies result in functional equivalence between these proteins, since flaA1 can support O-antigen biosynthesis in a wbpM mutant (15). Additional homologues identified in Campylobacter jejuni (PglF and CJ1293) (25, 64) and Caulobacter crescentus (FlmA) (38) are involved in protein glycosylation and/or influence flagellum production. In contrast to FlaA1, which seems widely distributed in the bacterial world, WbpB has only two homologues in bacterial genomes. One (WbpBPa, 63% homology) is found in the LPS biosynthetic cluster of P. aeruginosa serotype O5 (9), and the other (WlbA, 52% homology, N-terminal half only) is found in the LPS biosynthetic cluster of Bordetella pertussis (1). The homologies described above strongly suggest that flaA1 and wbpB might also be involved in LPS and flagellum biosynthesis in H. pylori. Interestingly, in bacteria where a homologue for each gene is found, these homologues are found together within a cluster of genes that are dedicated to the same biological function, suggesting a potential functional link for the H. pylori genes despite their presence in distinct areas of the chromosome.
Both genes encode sugar-nucleotide-modifying enzymes. FlaA1 is a UDP-GlcNAc C-6 dehydratase/C-4 reductase (15, 16), and WbpB is predicted to be an oxidoreductase. Consistent with the potential functional link between flaA1 and wbpB mentioned above, WbpB might be involved in the reduction of the UDP-4-keto-6-deoxy-GlcNAc intermediate generated by FlaA1. Interestingly, neither the 4-keto intermediate nor its reduced derivative are present in the LPS of H. pylori (4, 5, 46). In addition, all genes involved in the biosynthesis of the precursors necessary for LPS assembly have been identified (30, 36, 44) and are distinct from flaA1 and wbpB. Consequently, although it is expected that flaA1 and wbpB might affect LPS synthesis based on sequence homologies, such an effect is not anticipated to occur directly via production of LPS sugar precursors.
The sequence homologies described above also suggested that flaA1 and wbpB might be involved in protein glycosylation. This is relevant since flagellin glycosylation has been detected in H. pylori (34, 60) and it is possible that additional proteins—other than the flagellins—are glycosylated, as is the case in the closely related C. jejuni (64, 72).
Using a gene disruption strategy, we investigated the biological function of flaA1 and wbpB in H. pylori. In light of their homologies to LPS biosynthetic and flagellum modification genes found in other bacteria, the study was focused on the role of both genes on LPS biosynthesis and outer membrane barrier function, as well as on their role in flagellum synthesis and function.
MATERIALS AND METHODS
Bacterial strains and common procedures.
The H. pylori strains used in this study were 26695, J99, NCTC 11637, and SS1. Unless stated otherwise, all strains were grown for 36 h in brain heart infusion-yeast extract (0.25%)-agar media (BHI-YE) supplemented with 0.05% sodium pyruvate, 10% fetal calf serum, 5 μg of trimethoprim per ml, 4 μg of amphotericin B per ml, and 10 μg of vancomycin per ml. When necessary, selection with 5 μg of kanamycin per ml or 4 μg of chloramphenicol per ml was applied. Incubations were performed at 37°C under microaerophilic conditions using sealed jars and gas packs (CampyGen; Oxoid).
Cloning experiments were performed with Escherichia coli DH5α unless stated otherwise, using 30 μg of kanamycin per ml, 34 μg of chloramphenicol per ml, or 100 μg of ampicillin per ml when necessary. Each construct was checked by restriction analysis and sequencing. All kits were used as specified by their manufacturer. DNA sequencing was performed at the Robarts Institute Sequencing Facility (London, Canada).
Cloning and sequencing of the flaA1 and wbpB genes from strains SS1 and NCTC 11637.
wbpB and flaA1 were amplified from chromosomal DNA using Pwo (Roche) DNA polymerase and primers HPWB5 (GCTCTCCATGGGTATGCTTTTTGCGATGATTG) and HPWB3 (AAGCAGGATCCTCAAGCCAATTTGACAGACG) for wbpB and Flatop (ACTGTACATGTCAATGCCAAATCATCAAAAC) and Flabot (AAGCTGGATCCTCATAATAATTTCAACAAA) for flaA1. Both PCR products were cloned in Topo-PCR2.1 (Invitrogen) using Top10F′ cells. The constructs were sequenced on both strands using M13 forward and reverse primers.
Cloning and sequencing of the flagellin genes, flaA and flaB, from strain NCTC 11637.
flaA and flaB were amplified from chromosomal DNA using primers based on the sequences of HP0601 and HP0115 from strain 26695. Note that flaA (HP0601, encoding the flagellin) is distinct from flaA1 (HP0840, encoding the enzyme under study). The primers were HPFlaAP3 (GCTCTCCATGGCTTTTCAGGTCAATAC) and HPFlaAP5 (AAGAAGATCTCCTAAGTTAAAAGCCTTAAG) for flaA and HP0115P1 (GCTCTCCATGGGCATGAGTFTTAGGATAAATAC) and HP0115P2 (AAGAGGATCCTTATTGTAAAAGCCTTAAGA) for flaB. The PCR was performed using an Expand long-range template (Roche) with annealing at 46°C. Both PCR products were cloned in the pET23a derivative (13, 49) with an N-terminal histidine tag using NcoI and BglII for flaA and NcoI and BamHI for flaB. The constructs were sequenced using T7 promoter primer and primers HPFlagAP3 (GTGAATGATGTAACTTTAGAG) and HPFlagBP3 (CTTATAATGTCATGGCTACC).
Production of anti-flagellin A antibody.
Flagellin A was overexpressed from the pET23 construct in BL21(DE3)pLys cells with induction by 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37°C. The protein was purified by metal chelation as described previously (13) in the presence of 6 M guanidine-HCl. The protein was dialyzed in 50 mM ammonium bicarbonate in the presence of 1% sodium dodecyl sulfate (SDS), subjected to microelution from SDS-polyacrylamide gel electrophoresis (PAGE) gels (Bio-Rad Electro-Eluter), and redialyzed in the same buffer. The dialyzate was lyophilized, resuspended in 1.7% saline, and used to immunize rabbits after 1:1 (vol/vol) dilution with incomplete Freund's adjuvant. The immunization schedule and procedure for adsorption of the collected serum against an E. coli BL21(DE3)pLys extract were as reported previously (14).
To further enhance the specificity of the serum toward flagellins, the serum was passed through a Blue Sepharose 6 Fast Flow column (Pharmacia Biotech) and purified onto a flagellin A affinity column that was made by coupling purified overexpressed flagellin A to CNBr-activated Sepharose (Pharmacia Biotech) in the presence of guanidine-HCl. After being washed with 1 column volume (CV) of 150 mM glycine (pH 8.9)-350 mM NaCl and after acid shock with 1 CV of 100 mM glycine (pH 3.2), flagellin-specific antibodies were eluted with 1 CV of 1 M ammonium hydroxide and immediately neutralized with 0.09 CV of 0.2 M acetic acid. The recovered antibodies were further adsorbed against a lysate from a H. pylori FlaA knockout mutant (see below) as described previously (14).
Preparation of the knockout and complementation constructs. (i) flaA1 knockout construct.
flaA1 (HP0840) and its potential promoter were PCR amplified from genomic DNA using Taq DNA polymerase (Invitrogen) and primers FlaTop2 (GCGAGCGCGAATCTTTAT) and FlaA1Top (ATAGAACCGCTCACGAGC). The PCR product was cloned into Topo-PCR2.1 to generate TopoFla and was subsequently subcloned into the BamHI and ApaI sites of pBluescript II SK. The construct was digested with NcoI and blunted using T4 DNA polymerase. The kanamycin resistance cassette and its promoter were cut out from pHel3 (27) using SmaI and ligated into the blunted flaA1-pBluescript fragment to generate the flaA1 knockout construct.
(ii) wbpB knockout construct.
wbpB (HP0679) and its potential promoter were PCR amplified from genomic DNA using primers WbpBup (AACAGAGCCCACGAACGA) and WbpBdown (ATCACGCTTGCGATTGGC) and Taq DNA polymerase. The PCR product was cloned into Topo-PCR2.1 to generate TopoWbpB and subcloned into the EcoRI site of pBluescript II SK to generate pBSWbpB. The construct was digested with NsiI and blunted using T4 DNA polymerase. The kanamycin resistance cassette was cut from pHel3 (27) using SmaI and ligated into the blunted pBSWbpB fragment to generate the wbpB knockout construct.
(iii) flaA knockout construct.
Inverse PCR amplification of the flaA/pET23 construct was performed with primers HPFlagAP3 (GTGAATGATGTAACTTTAGAG) and HPFlagAP4 (GAACGATGTCAGATTGAATC) and Expand long-range template polymerase (Roche). The PCR product was blunted and dephosphorylated before being ligated to the SmaI-extracted kanamycin cassette as described above.
(iv) Complementation constructs.
The EcoRI fragment containing either gene with its promoter was cut out from TopoFla or TopoWbpB, blunted with T4 DNA polymerase, and cloned into the EcoRV-cut pHel2 shuttle vector (27) to generate the complementation constructs pHel2-FlaA1 and pHel2-WbpB.
Southern blotting.
Southern blotting was performed using the digoxigenin-labeling method with detection with anti-digoxigenin-alkaline phosphatase-CSPD (disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate) substrate as specified by the manufacturer (Roche). Chromosomal DNA was extracted from H. pylori using DNAzol (Invitrogen) and cut with HaeII/HindIII (flaA1 mutant) or MaeIII (wbpB mutant).
Preparation of knockout mutants and complemented clones.
The knockout mutants were generated by electroporation-mediated allelic exchange (63) with modifications described by McGee et al. (43) and selection on kanamycin. Potential transformants were analyzed for gene integration by PCR and Southern blotting. Complementation constructs were introduced into the mutants by electroporation, with selection onto kanamycin and chloramphenicol. The plasmids were extracted from the complemented strains and subjected to restriction analysis to ensure their integrity. Transformants harboring intact plasmids were selected for further studies.
LPS analysis.
LPS prepared as described previously (28) was analyzed on SDS-PAGE (15% polyacrylamide) or 10 to 20% Tricine gradient (Novex) gels. Detection was performed by silver staining (24) or Western blotting with anti-Lewis Y (Calbiochem) or anti-lipid A monoclonal antibodies (19).
Preparation of soluble cell extracts.
Soluble extracts were obtained by lysing H. pylori cells harvested from one BHI-YE plate in 100 μl of breaking buffer (20 mM sodium phosphate [pH 7.5], 1 mM EDTA) with acid-washed glass beads (Sigma G-4649). After being vortexed three times for 30 s each, the cells were pelletted for 10 min at 12,000 × g at room temperature and the supernatant was used for SDS-PAGE analysis.
Protein deglycosylation.
Enzymatic deglycosylation of flagellum preparations obtained by glycine extraction (50) was performed under denaturing conditions using five different enzymes as provided in the Calbiochem glycoprotein deglycosylation kit (no. 362280).
MS identification of proteins.
Protein bands cut out from Coomassie-stained gels were subjected to in-gel trypsinolysis. The peptides were analyzed by liquid chromatography mass spectrometry (MS) (Q-TOF2) and MALDI-MS at the Dr. Don Rix Protein Identification Facility of the University of Western Ontario.
RT-PCR.
RNA extracted using the RNeasy kit (Qiagen) was treated with RNase-free DNase I (Invitrogen) and subjected to reverse transcription (RT) using Superscript II RNase H− reverse transcriptase (Invitrogen) and random hexanucleotide primers (Roche). PCR (15 cycles) was subsequently performed with Taq DNA polymerase at an annealing temperature of 52°C and elongation time of 45 s, using gene-specific primers. The primers were Fla363 (GCTATCAGTCAGGTTATCGC) and Fla550 (ACGGCACCACGCTCCCAC) to amplify a 187-bp fragment of flaA1, HPWB8 (CAGAACACATGGGAGTAGC) and HPWB7 (GCCGTCCGAGCGCCAATTTGACAGACGC) to amplify a 171-bp fragment of wbpB, and HP1045P1 (GTCATTATCTATATGCCCAT) and HP1045P2 (CTGGCTTGAGCATGTAAGG) to amplify a 200-bp fragment from HP1045. The PCR products were analyzed on 2% agarose gels with ethidium bromide staining.
SDS, bile salts, and novobiocin sensitivity assays.
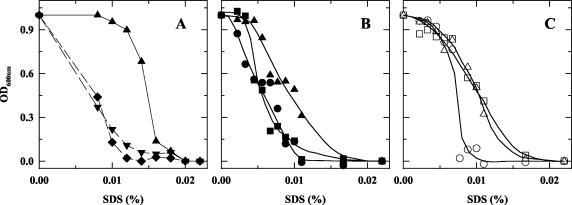
Harvested bacteria were diluted to an optical density at 600 nm (OD600) of 1 in BHI-YE. A 30-μl volume of cell suspension was added to 175 μl of BHI-YE supplemented with the appropriate antibiotics and/or detergents, with concentrations up to 15 μg/ml for novobiocin, 0.02% for SDS, and 0.18 mg/ml for bile salts (50% sodium cholate, 50% sodium deoxycholate (Sigma) (29). The assays were performed three times with independent cultures in 96-well plates incubated for 4 to 5 days with agitation at 37°C under microaerophilic conditions. On each plate, each strain was tested in duplicate. A representative example of the SDS-sensitivity experiment is provided in Fig. 6.
FIG. 6.
SDS sensitivity of the wild-type, mutant, and complemented strains. To compare wild-type and mutant strains, the assay was performed in the presence of kanamycin. Resistance to kanamycin was provided by the pHel3 vector in the wild-type strain. Similar results were obtained when the experiment was performed in the absence of kanamycin, using the plasmid-free wild-type strain (data not shown). To compare the mutants and the complemented strains, the assays were performed in the presence of kanamycin and chloramphenicol. In this case, mutants harboring the pHel2 vector were used as references. (A) ▴, wild type plus pHel3; ▾, flaA1 mutant; ⧫, wbpB mutant. (B) •, flaA1 mutant plus pHel2; ▴, flaA1 mutant plus pHel2-flaA1; ▪, flaA1 mutant plus pHel2-wbpB. (C) ○, wbpB mutant plus pHel2; □, wbpB mutant plus pHel2-wbpB; ▵, wbpB mutant plus pHel2-flaA1.
Sensitivity of H. pylori NCTC 11637 to serum killing.
Assays were performed as described by Bacon et al. (6), using 0, 0.1, 0.5, 4, and 10% fresh or heat-inactivated (60 min, 56°C) rabbit serum in saline.
Motility assays.
Bacteria were harvested from a 2-day-old plate into 400 μl of BHI-YE or from a 1-ml overnight liquid culture, diluted 1:10 in BHI-YE saturated with 10% CO2-85% N2-5% O2, and allowed to grow for 7.5 h. The cultures were then diluted to an OD600 of 0.3 or 0.9 and were used to inoculate motility plates (0.4% agar in BHI-YE) by stabbing. Each dilution was spotted in triplicate, and dilutions were made in triplicate for each strain. The growth of the swimming halo was monitored after 4 days of incubation at 37°C. The remaining diluted culture was used to inoculate regular BHI-YE plates to estimate bacterial viability by colony counting and microscopic observation.
Suspension-clearing assays.
Liquid cultures (1 ml) in BHI-YE saturated with 10% CO2-85% N2-5% O2 were prepared by inoculation from 2-day-old plates. After overnight growth, the cultures were diluted 1:10 in 2 ml of BHI and grown again for 18 to 24 h. The cultures were then diluted to an OD600 of 1 (total volume, 2 ml) in BHI and left to sit at room temperature. The rate of suspension clearance was measured over 8 h by carefully removing 70 μl from the top of the suspension and measuring the OD600 using a microcell (path length, 1 cm). Two independent experiments using two dilutions of each strain were performed.
Microscopic observations.
Cells harvested from plates or liquid cultures were examined as wet mounts under a phase-contrast microscope (Zeiss; oil immersion, ×400 magnification). For electron microscopy, cells were harvested from 1- to 2-day-old plates and resuspended in 1% glutaraldehyde in 20 mM HEPES buffer (pH 7). They were analyzed in the negative staining mode using 1% uranyl acetate.
Urease activity assay.
A 10-μl volume of a bacterial suspension at an OD600 of 0.4 in saline was mixed with 100 μl of 0.33 M urea-0.001% phenol red in 0.005 M sodium phosphate-0.15 M NaCl (pH 6.7) (11). Urease activity was assayed by monitoring the OD565 over time in a 96-well plate. All experiments were done in triplicate. The data were normalized to the total amount of proteins present in the sample (Bio-Rad assay).
Growth curves.
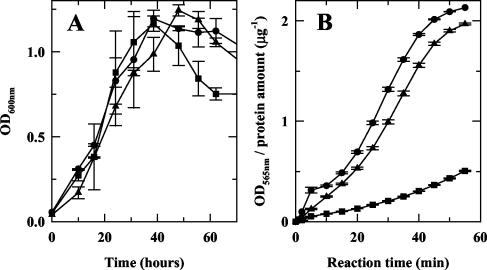
Bacterial suspensions (1 ml at an OD600 of 0.055) in BHI-YE were grown in 24-well plates with agitation under microaerophilic conditions, and the OD600 was monitored over 70 h. Data were recorded from four different plates with two repeats per strain on each plate. The data presented (see Fig. 2A) are the average of all the readings.
FIG. 2.
Effect of flaA1 or wbpB disruption on growth rate (A) and urease activity (B). Circles, wild type; squares, flaA1::kan; triangles, wbpB::kan.
Acid resistance assays.
For acid shock resistance assays, 10 μl of bacterial suspension at an OD600 of 9 in BHI-YE was diluted in 90 μl of PBS buffer adjusted to different pHs and containing 0 to 20 mM of urea in a 96-well plate. After 1 h of incubation at 37°C under microaerophilic conditions, 15 μl of the bacterial suspension was transferred to 195 μl of BHI-YE broth and the bacteria were incubated for 3 to 5 days before the OD600 was recorded. The data presented (see Fig. 3, left panels) are the average of two experiments, with duplicates for each condition tested in each experiment.
FIG. 3.
Effect of gene disruption on the ability of H. pylori to resist acid shock (A to C) and long-term acid exposure (D to F) in the presence of variable amounts of urea (a, b, c, d, and e correspond to 0, 5, 10, 15, and 20 mM urea, respectively). (A and D) wild type; (B and E) flaA1::kan; (C and F) wbpB::kan.
For long-term acid exposure, 75 μl of bacterial suspension at an OD600 of 1 in BHI-YE was added to 1 ml of BHI-YE adjusted to pH 5, 6, or 7 with 0 or 10 mM urea in 24-well plates. Bacterial growth was measured at 20 and 39 h by monitoring the OD600. The assay was performed twice with independent cultures, with two plates per experiment and duplicates for each condition tested on each plate. The data presented (see Fig. 3, right panels) are the average of the results obtained at 39 h.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the genes in this study are as follows: flaA1 (NCTC 11637), AY319294; flaA1 (SS1), AY319295; wbpB (NCTC 11637), AY319297; wbpB (SS), AY319296; flaA (NCTC 11637), AY319298; and flaB (NCTC 11637), AY319299.
RESULTS
The flaA1 and wbpB genes are highly conserved and expressed in several strains of H. pylori.
Southern blotting experiments performed using the flaA1 and wbpB genes from strain 26695 as probes indicated that strains SS1 and NCTC 11637 contained a single homologue of each gene (data not shown). After PCR amplification and DNA sequencing, the derived protein sequences were compared with those of strains 26695 and J99 (2, 65) (Table 1). Overall, the four strains harbored nearly identical FlaA1 and WbpB proteins. The only noticeable difference between strains was that WbpB from strain 26695 was slightly shorter than all its homologues.
TABLE 1.
Conservation of the FlaA1 and WbpB proteins among several strains of H. pylori
| H. pylori strain | FlaA1 protein
|
WbpB protein
|
||
|---|---|---|---|---|
| Size (aa)a | Identity (%)b | Size (aa) | Identity (%) | |
| 26695 | 333 | 100 | 289 | 100 |
| J99 | 333 | 98 | 315 | 96 |
| NCTC 11637 | 333 | 98 | 315 | 95 |
| SS1 | 333 | 99 | 315 | 97 |
aa, amino acids.
Identity with respect to 26695 (set at 100%).
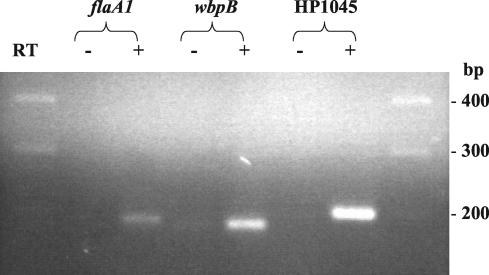
RT-PCR indicated that the flaA1 and wbpB genes were expressed in laboratory strains SS1, 26695, and NCTC 11637 (Fig. 1), since RT-PCR products of the expected size were obtained only in the presence of reverse transcriptase. Preliminary data obtained using formalin-fixed and paraffin-embedded human gastric biopsy specimens indicated that both genes were also expressed in the context of infection in the natural host (data not shown).
FIG. 1.
RT-PCR analysis of expression of flaA1 and wbpB in strain NCTC 11637. HP1045, encoding acetyl coenzyme A synthetase, was used as a positive control for constitutive expression. RT, reverse transcriptase. Signals obtained only in the samples where reverse transcriptase was added indicate expression of the genes of interest. Similar results were obtained with strains 26695 and SS1 (data not shown).
General characteristics of flaA1 and wbpB knockout mutants.
On plates, the flaA1 mutant grew less rapidly than the wbpB mutant, which, in turn, grew slightly less rapidly than the wild-type strain. This was not dependent on the presence of kanamycin in the growth media. In broth, all strains exhibited similar growth patterns, and the same maximal culture densities could be obtained after 40 h of growth (Fig. 2A). Interestingly, a significant decrease in OD600 was observed for the flaA1 mutant after 40 to 50 h of growth, suggesting that, in contrast to the wild type, this mutant did not survive in the stationary phase. This could explain the lower growth observed on plates for this mutant. Both mutants still produced urease as judged by the urea-phenol red assay (11). However, the levels of urease produced by the flaA1 mutant were much lower than those produced by the wbpB mutant or by wild-type bacteria (Fig. 2B).
Like the wild type, both mutants were able to resist acid shock in the absence of urea at pH ≥ 3 (Fig. 3A to C, bars a). The addition of urea allowed resistance to acid shock of all strains at pH 2 (bars b to e). This indicated that the limited urease activity of the flaA1 mutant was sufficient to support viability at pH 2. At pH ≥ 7, addition of urea became deleterious. In the flaA1 mutant, this toxic effect inherent to the chaotropic nature of urea appeared at lower urea concentrations (15 mM) (Fig. 3B, bars d and e) than for the wild type and for all pHs of >2. This was consistent with the lower levels of urease activity shown by this mutant (Fig. 2B).
Like wild-type bacteria, both mutants resisted long-term acid exposure in the absence of urea (Fig. 3D to F, bars a). The slight decrease in viability observed for all strains at pH 5 could be eliminated by the addition of 10 mM urea (bars c), indicating again that urease activity supports acid resistance under these conditions too.
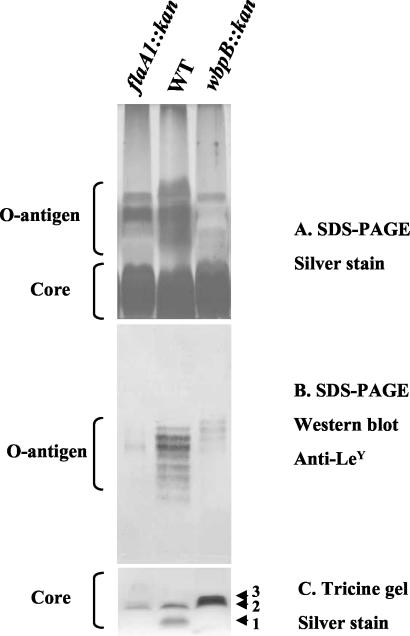
The flaA1 and wbpB knockout mutants exhibit altered LPS profiles.
SDS-PAGE analysis of wild-type and mutant LPS revealed that the mutants had lost most of their O antigen (Fig. 4A). The wbpB mutant produced slightly more Lewis Y-containing O antigen than did the flaA1 mutant. Also, the O chains present in the wbpB mutant appeared slightly shifted compared with the wild-type bands (Fig. 4B), suggesting that the core to which the O antigen was attached might have a slightly different structure. When the core LPS was analyzed on higher-resolution 10 to 20% gradient Tricine gels, the wild-type strain exhibited two well resolved bands, bands 1 and 2 (Fig. 4C). In contrast, in both mutants, the fast-migrating band (band 1) was missing and the slower-migrating band (band 2) was not affected. In addition, a third band (band 3) migrating slightly slower than band 2 appeared in the wbpB mutant only. The presence of band 3 was not dependent on the amount of sample loaded on the gel.
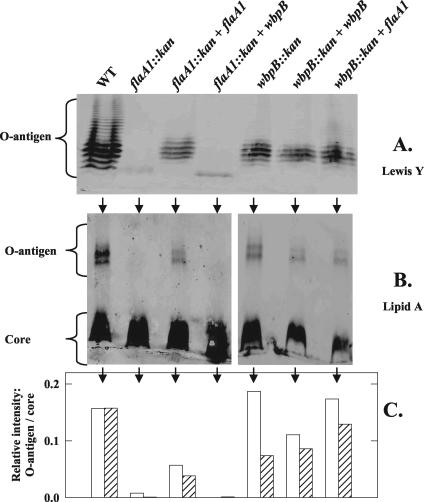
FIG. 4.
Gel electrophoresis analysis of the LPS of the flaA1 and wbpB mutants of H. pylori strain NCTC 11637. The LPS were analyzed on SDS-PAGE (12% polyacrylamide) gels (A and B) or 10 to 20% Tricine gradient gels (C) and detected by silver staining (A and C) or Western immunoblotting (B) with anti-Lewis Y antibody (Calbiochem). Only 1/10 of the amount of sample used for the SDS-PAGE gels was loaded on the Tricine gel to allow for good resolution of core bands. WT, wild type.
Quantitative analysis performed by anti-Lewis Y and anti-lipid A Western immunoblots confirmed that the flaA1 mutant produced very little, if any, Lewis Y O antigen and that the wbpB mutant produced more Lewis Y O antigen (hatched bar) than the flaA1 mutant but less than the wild type (Fig. 5A and C). It also revealed that the wbpB mutant produced high-molecular-weight O antigen that reacted with anti-lipid A antibody (Fig. 5B and C, white bars) but not with anti-Lewis Y antibody (hatched bars). This material could correspond to other Lewis antigens, which differ from Lewis Y by the number and position of fucose residues (40, 68).
FIG. 5.
(A and B) SDS-PAGE analysis of the LPS of knockout mutants and complemented strains detected by Western immunoblotting using anti-Lewis Y (A) or anti-lipid A (B) antibodies. Detection was performed using goat anti-mouse immunoglobulin G conjugated with Alexa Fluor 680 and scanning with a Li-Cor Odyssey infrared imaging system. The anti-Lewis Y antibody detected the O antigen exclusively whereas the anti-lipid A antibody detected the lipid A associated both with the O antigen and with the core LPS. (C) Quantitative data were obtained by normalizing the amount of O antigen detected with either antibody (open bars, anti-Lipid A; hatched bars, anti-LeY) by the amount of core LPS detected with the anti-Lipid A antibody. WT, wild type.
Taken together, these data showed that the inactivation of either flaA1 or wbpB affected LPS synthesis in a similar, although not exactly identical, fashion. It did not affect the synthesis of LPS precursors per se since low levels of O antigen could still be detected in each mutant. However, it might have affected the transfer of the O units to the lipid A acceptor, as suggested by analysis of the core LPS, and, to a lesser extent, the activity of the transferases responsible for the synthesis of the O units, as shown by the presence of non-Lewis Y O antigens in the wbpB mutant.
Reintroducing the functional gene in trans partly restored the production of the O antigen in each mutant (Fig. 5), indicating that the loss of O-antigen production observed in each mutant was gene specific and not due to polarity effects. The copy number and poor stability of the complementation plasmid in strain NCTC 11637 might explain why complementation was not complete. In the flaA1-complemented flaA1 mutant, the O antigen produced was composed of Lewis Y-containing species, as in the wild type (white and hatched bars of similar size). Interestingly, in the wbpB-complemented wbpB mutant, the amount of Lewis Y-containing O antigen did not increase (hatched bar) but its proportion of the total O antigen (detected with anti-lipid A [white bar]) did increase, so that all O antigen produced was of the Lewis Y type, as in the wild type. Hence, qualitatively, reintroducing the functional wbpB gene into the wbpB mutant also restored a wild-type like O-antigen production made exclusively of Lewis Y O antigens. The identity of the Lewis Y-reacting band that appears below the O antigen in the flaA1::kan + wbpB and flaA1::kan lanes (Fig. 5A) is unknown, but it does not appear to be LPS related since it did not react with anti-lipid A antibody.
Disruption of flaA1 or wbpB affects the barrier properties of the outer membrane.
Defects in LPS synthesis often correlate with higher sensitivity of bacteria to killing by serum, detergents (SDS or bile salts) (29), or hydrophobic antibiotics (novobiocin) (67) and with decreased virulence properties in animal models (56, 70). Hence, the sensitivity of the flaA1 and wbpB mutants to each of these compounds was investigated.
The extreme sensitivity to serum of the NCTC 11637 strain used for these studies (7) prevented us from detecting any significant differences between the wild type and mutants. However, a significantly higher sensitivity of the mutants than of the wild type to SDS (Fig. 6A) and a slight increase in sensitivity to novobiocin and bile salt were observed (data not shown). These results indicate the higher accessibility of the outer membrane to antibiotic or detergents and its reduced stability, which is consistent with decreased O-antigen production.
Providing the functional gene in trans restored wild-type-like SDS sensitivity in each mutant (Fig. 6B and C), demonstrating the gene specificity of the increased SDS sensitivity phenotype.
flaA1 can cross-complement the wbpB inactivation.
We investigated whether the bifunctional dehydratase/reductase FlaA1 could complement the disruption of the putative reductase activity of WbpB by monitoring both the LPS production by and SDS sensitivity of cross-complemented mutants. Providing a functional copy of flaA1 in trans in the wbpB mutant did restore wild-type-like SDS sensitivity to the wbpB mutant (Fig. 6C). This correlated with an increased production of Lewis Y-containing O antigen in this complemented mutant (Fig. 5, hatched bars). This indicated that FlaA1 and WbpB have a redundant reductase activity and might be involved in the same biochemical pathway. However, providing wbpB in the flaA1 mutant did not restore wild-type levels of SDS sensitivity (Fig. 6B) or O-antigen production (Fig. 5) to the flaA1 mutant, indicating that the complementation effect observed previously was gene specific. This is consistent with the fact that WbpB is predicted to be monofunctional (reductase only) and cannot complement the lack of dehydratase function of the flaA1 mutant.
The flaA1 and wbpB disruptions abrogate flagellum-mediated motility.
On examination by phase-contrast microscopy, the wild-type bacteria were motile and exhibited corkscrew movements. In contrast, the flaA1 and wbpB mutants only oscillated and showed no corkscrew movement.
Motility assays performed with bacteria in soft agar showed a clear reduction of motility on disruption of the flaA1 and wbpB genes (Table 2). The results were not dependent on the length of incubation or on the density or nature (from broth or plate) of the inoculum.
TABLE 2.
Effect of the inactivation of flaA1 and wbpB on the motility of H. pylori NCTC 11637 as measured by the soft agar plate motility assaya
| OD600 | Diamb (mm) of motility halo of strain:
|
||
|---|---|---|---|
| WT | fla1::kan | wbpB::kan | |
| 0.3 | 9 ± 1 | 2 ± 1 | 3 ± 0 |
| 0.9 | 10 ± 2 | 3 ± 1 | 4 ± 1 |
The motility plates were inoculated with two different dilutions of bacterial culture and were typically read after 4 days of growth at 37°C. Longer incubation times did not result in larger motility halos. All cultures exhibited similar viability when plated on BHI-YE. WT, wild type.
Results are expressed as means ± standard deviations.
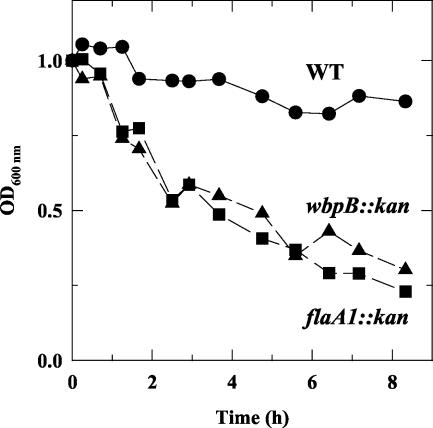
To further demonstrate the effects of the mutations on bacterial motility, suspension-clearing assays were performed. The wild-type strain exhibited good stability in the suspension, since the OD600 only decreased slightly over time (Fig. 7). In contrast, the mutants settled rapidly. These data show that disruption of flaA1 or wbpB resulted in a similar phenotype whereby the ability of the bacteria to maintain themselves in suspension by active movement was impaired. Taken together, the data presented above indicate that both mutants have impaired motility.
FIG. 7.
Suspension-clearing assays comparing the abilities of the wild-type (WT) and mutant strains to maintain themselves dispersed in suspension by active swimming. Aliquots of a bacterial suspension were withdrawn at regular intervals from the top of the suspension, which was maintained without agitation under microaerophilic conditions, and the OD600 was recorded as an estimation of the cell density. This is a representative example of three independent experiments performed with different cultures.
Flagellum production is abrogated in the flaA1 mutant but not in the wbpB mutant.
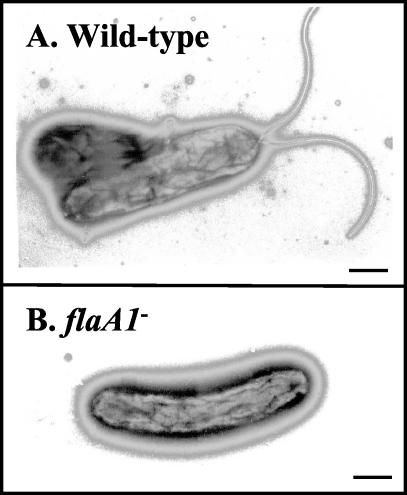
The wild-type and mutant strains were examined by electron microscopy to investigate if the gene disruptions had any effect on the production of flagella. The typical unipolar sheathed flagella of the wild-type strain could be readily detected by negative staining (Fig. 8A). In contrast, the flaA1 mutant was devoid of flagella, indicating that the production and/or assembly of flagellins was impaired in this mutant (Fig. 8B). The wbpB mutant still produced flagella (data not shown).
FIG. 8.
Electron micrographs of wild-type (A) and flaA1 mutant (B) strains obtained with 1% uranyl acetate staining. The sheathed unipolar flagella were easily detected in the wild-type strain as well as in the wbpB mutant (data not shown), but the flaA1 mutants were devoid of flagella. Bar, 0.5 μm.
The flaA1 mutant still produces flagellins.
To examine whether the lack of flagellum production observed in the flaA1 mutant was due to the lack of flagellin production or to a defect in their export and assembly, soluble cell extracts of the flaA1 mutant were compared with cell extracts from the wild type and the wbpB mutant in an anti-flagellin Western blotting assay.
First, we demonstrated that strain NCTC 11637 produces two flagellins, FlaA and FlaB, which were >99.5% identical to flagellins found in strain 26695 and 57.2% identical to one another. NCTC 11637 FlaA was 53.3 kDa in size, and FlaB was only slightly larger (54.0 kDa).
Second, we demonstrated that both flagellins could be readily detected by Western immunoblotting using a polyclonal antiserum raised in rabbits against overexpressed and purified NCTC 11637 FlaA flagellin. This was consistent with their high levels of similarity at the protein level. Note that in the gel system used, the two flagellins were not resolved from one another.
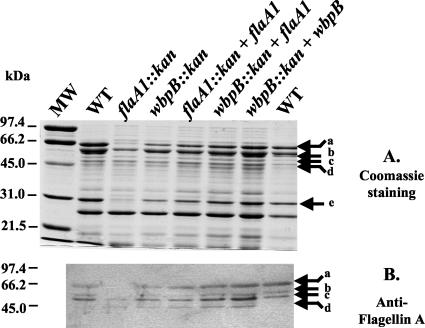
Third, the anti-FlaA antibody was used to investigate flagellin production in wild-type and mutant H. pylori strains. Figure 9 shows that even after adsorption against an E. coli lysate, the anti-FlaA antibody detected several proteins in wild-type H. pylori soluble extracts (bands a to d). Band c (53 kDa) had the strongest reactivity with the anti-FlaA antibody and migrated to the same position as pure overexpressed flagellins (data not shown), indicating that it corresponds to the flagellin(s) FlaA and/or FlaB. Band c was not affected by the flaA1 or wbpB disruption, indicating that both mutants still produced flagellin(s), including the aflagelatted flaA1 mutant. Hence, the data suggest that the export and/or assembly of the flagellins is impaired in the flaA1 mutant.
FIG. 9.
SDS-PAGE analysis of soluble cell extracts of wild-type (WT), mutant, and complemented strains by Coomassie staining (A) and anti-FlaA Western immunoblotting (B). The gels contained 8% acrylamide. The identities of the bands labeled a to e are described in Results. MW, molecular mass standards.
Band a (66 kDa) was shown to correspond to the large subunit of urease, UreB, by MS analysis. The cross-reactivity of the anti-FlaA antibody with UreB could be explained by the existence of a 113-amino-acid stretch where the two proteins exhibit 54% homology. Interestingly, a strong reduction in the amount of band a was observed in the flaA1 mutant only. This was consistent with the reduction in urease activity described above for this mutant (Fig. 2B). MS analysis also indicated that the reduction in UreB production was accompanied by a reduction in the production of the small subunit of urease, UreA, as observed on Coomassie-stained gels (band e, 31 kDa). The production of bands a and e was restored in the complemented mutant, indicating that the effect was gene specific.
MS analysis indicated that band d contained multiple proteins unrelated to flagellins. Since this band was not affected by the flaA1 or wbpB disruptions, its analysis was not pursued further.
Finally, the production of band b (58 kDa) was abrogated in the flaA1 mutant only, and the effect was gene specific since band b reappeared in the flaA1-complemented flaA1 mutant (Fig. 9). Considering that several strains of H. pylori harbour glycosylated flagellins (34, 60), the possibility that band b might correspond to posttranslationally modified flagellins was investigated by MS analysis. However, contamination of band b with catalase (58.6 kDa) that was present both in the wild type and in the flaA1 mutant (a negative control that does not contain band b) prevented us from demonstrating the presence of flagellins within band b. The experiments below aim at clarifying this question by refining the purification and detection of the flagellins and by investigating the glycosylation status of the flagellins in the band b-producing wild-type and wbpB mutant strains.
The flagellins of wild-type H. pylori strain NCTC 11637 are not glycosylated.
To examine the glycosylation status of the NCTC 11637 flagellins, flagellins obtained by glycine extraction were analyzed by Western immunoblotting as well as Coomassie and Ponceau staining (Fig. 10). Three major bands (bands A to C) in the size range of interest were detected by Coomassie (Fig. 10C) or Ponceau (Fig. 10A) staining. As described above (Fig. 9), band A is thought to be UreB. A significant reduction in reactivity of the serum with band A was obtained using serum that had been purified by affinity on a flagellin A column and adsorbed against a lysate of a urease-producing H. pylori flagellin A mutant (compare the intensity of band B in Fig. 10B and D).
FIG. 10.
Analysis of the wild-type (WT) and wbpB mutant flagellins by SDS-PAGE and Western immunoblotting. (A and B) Optimization of the sample preparation method showing enrichment of the samples in the flagellin band (band C) in glycine extracts (G) compared to soluble extracts (S); Ponceau red staining (A) and Western immunoblotting with anti-FlaA serum (B) after affinity purification and adsorption against an H. pylori flagellin A (flaA−) knockout mutant were used. (C and D) Enzymatic deglycosylation of flagellum preparations obtained by glycine extraction. + and − indicate the presence and absence of enzymatic deglycosylation, respectively. Coomassie staining (C) and Western immunoblotting with anti-FlaA antibody adsorbed against an E. coli lysate (D) were used. C. jejuni (CJ, strain 81-176) flagellum preparations obtained under the same conditions and bovine fetuine (BF) were used as positive controls for deglycosylation. Note that the anti-FlaA antibody readily detects C. jejuni glycosylated flagellins. All gels contained 10% acrylamide, and overexpressed and purified flagellins (FlaA and FlaB) were used as positive controls independently or as a mixture (A+B). MW, molecular mass standards.
Band C migrated at the expected size for FlaA and FlaB, as seen using purified (nonglycosylated) overexpressed flagellins as controls, and reacted strongly with the anti-FlaA antiserum (Fig. 10B and D), confirming that it corresponds to FlaA and/or FlaB. Consistent with this assignment, band C was very faint in the flaA mutant (Fig. 10B), since this mutant produces only the minor flagellin FlaB. Note that although affinity purification and adsorption of the antibody against the H. pylori flaA mutant cell extract (Fig. 10B) significantly increased its specificity toward flagellins, it resulted in a significant titer loss as indicated by the lower reactivity of the serum toward purified flagellins (compare Fig. 10D and B) and by the very weak detection of the flagellins in soluble extracts (compare Fig. 10B with Fig. 9B).
Enzymatic deglycosylation using a combination of five different enzymes had no effect on the migration or amount of band C, indicating that it corresponds to nonglycosylated flagellins. C. jejuni glycosylated flagellines prepared and treated under the same conditions were included as a positive control and showed a significant shift in size after deglycosylation. Band B could be readily deglycosylated enzymatically (Fig. 10C). However, since it did not react with the anti-FlaA antibody and did not comigrate with band C after deglycosylation, it does not correspond to glycosylated flagellins. Note that no equivalent of the anti-FlaA-reacting band b from Fig. 9 was observed in glycine extracts, indicating that the glycine extraction method eliminated cross-reacting bands that were not flagellum related.
Overall, this analysis excludes the existence of any glycosylated flagellins in the wild-type strain and suggests that the wbpB mutant produces wild-type-like flagellins. However, these data did not determine whether the lack of motility of the wbpB mutant could be due to a lack of production of one of the two flagellins since FlaA and FlaB could not be resolved on these gels, and even after adsorption against a flaA knockout mutant, the anti-FlaA antibody still detected both flagellins without discrimination (Fig. 10B). Attempts at refining the purity of the flagellins by ultracentrifugation/acid dissociation and affinity chromatography to answer this question by direct MS analysis were unsuccessful due to the precipitation and therefore the loss of the flagellins.
DISCUSSION
The goal of this study was to determine the biological function of two novel genes encoding sugar-nucleotide-modifying enzymes, flaA1 and wbpB, in H. pylori. Whereas flaA1 exhibits homologues in numerous medically relevant bacteria, wbpB is a rather rare gene with only two significant homologues in bacterial genomes. The data provided in this study showed that both genes were conserved and expressed (Fig. 1; Table 1) in all H. pylori strains examined, suggesting an important and conserved function. Each gene could be inactivated in strain NCTC 11637 by insertion of a kanamycin resistance cassette, indicating that neither of them was essential in this strain. This is in contrast to a recent report indicating that disruption of HP0840 (FlaA1) might be lethal in other strains (60).
The two genes are distant from one another on the H. pylori chromosome (2, 65). The similarity of the phenotypes observed after disruption of either gene suggested that they belonged to the same functional pathway despite the absence of genetic linkage. Indeed, each gene affects LPS synthesis, outer membrane barrier function, and flagellum-mediated motility. However, subtle differences in the phenotypes of each mutant could be observed, and the phenotypes were proven to be gene specific by complementation experiments.
The functional link between the two genes is consistent with the fact that in the few bacteria that harbor a homologue for each gene, both genes were found within a single operon and participated in the same biological function (1, 9). This is also consistent with their biochemical activities, which might be partly complementary. Indeed, the unidirectional complementation of the disruption of wbpB by flaA1 supports the hypothesis that this functional complementation involves the common reductase activity associated with each enzyme. While this awaits further biochemical evidence, the reason for such redundancy of the reductase activity is currently not clear. The chromosomally encoded copy of flaA1 only partially complemented the wbpB mutation (Fig. 5 and 6C), so that the phenotype observed in the wbpB mutant was intermediate between those of the wild type and the flaA1 mutant (with the presence of flagella and higher levels of residual O antigens in the wbpB mutant). Full complementation of the wbpB mutation was observed only when multiple copies of flaA1 were provided using a complementation vector. This suggests that the reductase activity of FlaA1 is not very efficient, so that there is no actual functional redundancy in vivo. In addition, 4-keto sugar derivatives can be regarded as “pluripotent,” since they can undergo various enzymatic modifications (52, 62, 71). Thus, the 4-keto-6-deoxy-UDP-GlcNAc generated by FlaA1 might be involved in multiple biosynthetic pathways and its reduction by WbpB might serve as a channeling mechanism to prevent its diversion toward other pathways. Because the wbpB gene is fairly rare in the bacterial world but is conserved and expressed in all H. pylori strains examined, the pathway at stake might be related to specific features of the biology and/or pathogenesis of H. pylori.
LPS structure (4, 5, 46) and biochemical data (15, 30, 36, 44) indicated that FlaA1 and WbpB were not LPS biosynthetic enzymes per se in H. pylori. Disruption of their genes nevertheless significantly reduced O-antigen production and resulted in the formation of altered core LPS (Fig. 4). In the absence of structural information for these mutants, it is reasonable to assume that band 1 observed on Tricine gels could correspond to the core and that the slower-migrating band 2 could correspond to the “core plus one O-chain unit.” Band 3, which migrates only slightly slower than band 2, could represent a “core plus one” structure that carries an additional sugar residue on the terminal galactose of the O-chain unit. This additional sugar residue is likely to be l-fucose since the resulting difucosylated lactosamine motif is commonly found as a terminal structure in the H. pylori LPS (40, 46). This incorporation of an extra residue would explain the altered reactivity of the O chain produced by the wbpB mutant with the anti-Lewis Y antibody (Fig. 4). The activity of the transferase responsible for addition of this terminal sugar residue must be impaired or down-regulated in the flaA1 mutant so that no band 3 is present in this mutant and all O chains produced are of the Lewis Y type.
Both mutants were nonmotile (Fig. 7; Table 2), and the lack of motility correlated directly with the lack of flagellum production in the flaA1 mutant whereas the wbpB mutant still produced wild-type-like sheathed flagella. Western immunoblotting showed that flagellin(s) was produced by the flaA1 mutant, suggesting that it was the export or assembly of the flagellins into a flagellum that was impaired in this mutant. Flagellin O glycosylation has been found in other strains of H. pylori (34, 60) and has been proposed to be concomitant with its export to the bacterial surface (34). Considering our observation that the O-glycosylation sites described in the flagellin sequence of strain 1061 were conserved in the flagellins of strain NCTC 11637 and considering that glycosylation of surface appendage within one bacterial species is often strain dependent (3, 20, 59), we investigated if flagellins were also glycosylated in strain NCTC 11637. Our results obtained using enzymatic deglycosylation treatment and Western immunoblotting indicated that the flagellins of wild-type strain NCTC 11637 were not glycosylated. Thus, a defect in the posttranslational modification of the flagellins cannot be responsible for the lack of production of flagella in the flaA1 mutant or for the lack of functionality of the wbpB mutant flagella.
It has been reported previously that H. pylori mutants lacking the major flagellin, FlaA, do not produce flagella whereas mutants lacking the minor flagellin, FlaB, produce nonfunctional flagella (63). The possibility that FlaA would be missing in the flaA1 mutant and would prevent flagellum assembly cannot be excluded since the gel system and antibody used for these studies did not allow for discrimination between both flagellins. Similarly, the possibility that FlaB would be missing from the wbpB mutant could explain the presence of nonfunctional flagella. Alternatively, the lack of motility of the wbpB mutant might be due to a defect within a structural component of the basal body or motor of the flagella that has yet to be identified.
The defects in LPS production, membrane barrier properties, and flagellum-mediated motility associated with inactivation of flaA1 or wbpB suggested that both genes might be important for the virulence of H. pylori. In addition, the down-regulation of urease production observed in the flaA1 mutant (Fig. 2B and 9, bands a and e) might further compromise its survival in vivo, although the residual levels of urease produced were sufficient to sustain acid resistance in vitro (Fig. 3). Hence, FlaA1 and WbpB could represent valuable targets for the development of novel therapeutic agents against H. pylori. Since inactivating these enzymes abolishes the production of several virulence factors, it is unlikely that resistance to FlaA1- or WbpB-specific inhibitors could arise easily, in contrast to what is observed with currently available antibiotics (18, 31, 41). Mouse colonization assays are under way to investigate whether inactivation of flaA1 or wbpB results in defects in host colonization or survival in vivo.
The effect of a single gene on the synthesis of several virulence factors has been demonstrated previously (10, 20, 39) and involves direct incorporation of specific sugars within the structure of different virulence factors. In contrast, the sugars produced by FlaA1 (and WbpB) are not integral part of the virulence factors affected by disruption of either gene. We propose that these sugars are instead part of glycosylation motifs that target proteins involved in LPS or flagellum assembly and function and regulate their activity. Such regulation of protein function by their glycosylation has been observed previously in other bacterial species (17, 53, 55).
Several of the C. jejuni N-glycosylated proteins carry bacillosamine, a 2,4-diacetamido-2,4,6-trideoxyglucopyranose (72). Based on the biochemical function of FlaA1 (15) and considering the close relationship between H. pylori and C. jejuni, we propose that bacillosamine-dependent protein glycosylation also occurs in H. pylori and that FlaA1 is the first enzyme responsible for bacillosamine biosynthesis. While this work was in progress, it was shown that FlaA1 could complement the function of CJ1293, a homologue found in C. jejuni, and that CJ1293 was involved in the production of flagella (25). We also demonstrated that CJ1293 has the same biochemical activity as FlaA1 and performs a UDP-GlcNAc C-6 dehydration consistent with the first step of bacillosamine biosynthesis (12). Hence, these new data corroborate our hypothesis that FlaA1, like CJ1293, is involved in bacillosamine biosynthesis. Potentially glycosylated enzymes involved in LPS and flagellum synthesis are currently being analyzed in the context of the flaA1 mutation to confirm this hypothesis.
In summary, we demonstrate herein that FlaA1 and WbpB are functionally linked in the dual control of LPS biosynthesis and flagellum production and/or function in H. pylori and propose that this dual control involves protein glycosylation. This work is the first report to establish the existence of a regulatory network connecting the LPS and flagellum biosynthesis machineries and suggests that FlaA1- and WbpB-dependent protein glycosylation might be the underlying mechanism.
Acknowledgments
This work was supported by operating grant MOP-62775 from the Canadian Institutes of Health Research to C.C., as well as by a joint grant from the Canadian Association of Gastroenterology/Abbott Laboratories and CIHR to C.C. C.C. is the recipient of a University Faculty Award from the Natural Sciences and Engineering Research Council of Canada and of a Premier's Research Excellence Award (Ontario).
We thank D. E. Taylor (University of Alberta) and S. Logan (NRC, Ottawa, Canada) for providing strains 26695, NCTC 11637, and SS1 and R. Haas (Max von Pettenkofer Institute for Hygiene and Medicinal Microbiology, Munich, Germany) for providing plasmids pHel2 and pHel3. We thank J. S. Lam (University of Guelph) for support at the onset of this project and for providing anti-lipid A antibody, and we thank M. Smith and V. Somalinga for technical assistance. We thank D. Moyles (University of Guelph) for the electron microscopy analyses and A. Doherty-Kerby (University of Western Ontario) for MS analyses. Also, we also thank M. Valvano (University of Western Ontario) for the use of his Li-Cor Scanning system and S. Koval (University of Western Ontario) for the use of her microscope.
REFERENCES
- 1.Allen, A., and D. Maskell. 1996. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol. Microbiol. 19:3752. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aspinall, G. O., and M. A. Monteiro. 1996. Lipopolysaccharides of Helicobacter pylori strains P466 and MO19: structures of the O antigen and core oligosaccharide regions. Biochemistry 35:2498-2504. [DOI] [PubMed] [Google Scholar]
- 5.Aspinall, G. O., M. A. Monteiro, H. Pang, E. J. Walsh, and A. P. Moran. 1996. Lipopolysaccharide of the Helicobacter pylori type strain NCTC 11637 (ATCC 43504): structure of the O antigen chain and core oligosaccharide regions. Biochemistry 35:2489-2497. [DOI] [PubMed] [Google Scholar]
- 6.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 7.Berstad, A. E., K. Hogasen, G. Bukholm, A. P. Moran, and P. Brandtzaeg. 2001. Complement activation directly induced by Helicobacter pylori. Gastroenterology 120:1108-1116. [DOI] [PubMed] [Google Scholar]
- 8.Bhasin, N., A. Albus, F. Michon, P. J. Livolsi, J. S. Park, and J. C. Lee. 1998. Identification of a gene essential for O-acetylation of the Staphylococcus aureus type 5 capsular polysaccharide. Mol. Microbiol. 27:9-21. [DOI] [PubMed] [Google Scholar]
- 9.Burrows, L. L., D. F. Charter, and J. S. Lam. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481-495. [DOI] [PubMed] [Google Scholar]
- 10.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479-26485. [DOI] [PubMed] [Google Scholar]
- 11.Clyne, M., A. Labigne, and B. Drumm. 1995. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect. Immun. 63:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creuzenet, C. S. 2004. Characterization of CJ1293, a new UDP-GlcNAc C6 dehydratase from Campylobacter jejuni. FEBS Lett. 559:136-140. [DOI] [PubMed]
- 13.Creuzenet, C., M. Belanger, W. W. Wakarchuk, and J. S. Lam. 2000. Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 275:19060-19067. [DOI] [PubMed] [Google Scholar]
- 14.Creuzenet, C., and J. S. Lam. 2001. Topological and functional characterization of WbpM, an inner membrane UDP-GlcNAc C6 dehydratase essential for lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 41:1295-1310. [DOI] [PubMed] [Google Scholar]
- 15.Creuzenet, C., M. J. Schur, J. Li, W. W. Wakarchuk, and J. S. Lam. 2000. FlaA1, a new bifunctional UDP-GlcNAc C6 Dehydratase/C4 reductase from Helicobacter pylori. J. Biol. Chem. 275:34873-34880. [DOI] [PubMed] [Google Scholar]
- 16.Creuzenet, C., R. V. Urbanic, and J. S. Lam. 2002. Structure-function studies of two novel UDP-GlcNAc C6 dehydratases/C4 reductases: variation from the SYK dogma. J. Biol. Chem. 277:26769-26778. [DOI] [PubMed] [Google Scholar]
- 17.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dailidiene, D., M. T. Bertoli, J. Miciuleviciene, A. K. Mukhopadhyay, G. Dailide, M. A. Pascasio, L. Kupcinskas, and D. E. Berg. 2002. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob. Agents Chemother. 46:3940-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Kievit, T. R., and J. S. Lam. 1994. Monoclonal antibodies that distinguish inner core, outer core, and lipid A regions of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 176:7129-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiGiandomenico, A., M. J. Matewish, A. Bisaillon, J. R. Stehle, J. S. Lam, and P. Castric. 2002. Glycosylation of Pseudomonas aeruginosa 1244 pilin: glycan substrate specificity. Mol. Microbiol. 46:519-530. [DOI] [PubMed] [Google Scholar]
- 21.Dunn, B. E., G. P. Campbell, G. I. Perez-Perez, and M. J. Blaser. 1990. Purification and characterization of urease from Helicobacter pylori. J. Biol. Chem. 265:9464-9469. [PubMed] [Google Scholar]
- 22.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 23.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fomsgaard, A., M. A. Freudenberg, and C. Galanos. 1990. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J. Clin. Microbiol. 28:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50:659-671. [DOI] [PubMed] [Google Scholar]
- 26.Graham, D. Y. 1991. Helicobacter pylori: its epidemiology and its role in duodenal ulcer disease. J. Gastroenterol. Hepatol 6:105-113. [DOI] [PubMed] [Google Scholar]
- 27.Heuermann, D., and R. Haas. 1998. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. Mol. Gen. Genet. 257:519-528. [DOI] [PubMed] [Google Scholar]
- 28.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh, M., K. Wada, S. Tan, Y. Kitano, J. Kai, and I. Makino. 1999. Antibacterial action of bile acids against Helicobacter pylori and changes in its ultrastructural morphology: effect of unconjugated dihydroxy bile acid. J. Gastroenterol. 34:571-576. [DOI] [PubMed] [Google Scholar]
- 30.Jarvinen, N., M. Maki, J. Rabina, C. Roos, P. Mattila, and R. Renkonen. 2001. Cloning and expression of Helicobacter pylori GDP-l-fucose synthesizing enzymes (GMD and GMER) in Saccharomyces cerevisiae. Eur. J. Biochem. 268:6458-6464. [DOI] [PubMed] [Google Scholar]
- 31.Jenks, P. J., R. L. Ferrero, and A. Labigne. 1999. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J. Antimicrob. Chemother. 43:753-758. [DOI] [PubMed] [Google Scholar]
- 32.Josenhans, C., K. A. Eaton, T. Thevenot, and S. Suerbaum. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect. Immun. 68:4598-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177:3010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Josenhans, C., L. Vossebein, S. Friedrich, and S. Suerbaum. 2002. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett. 210:165-172. [DOI] [PubMed] [Google Scholar]
- 35.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 1999. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol. 181:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon, D. H., J. S. Woo, C. L. Perng, M. F. Go, D. Y. Graham, and F. A. El-Zaatari. 1998. The effect of galE gene inactivation on lipopolysaccharide profile of Helicobacter pylori. Curr. Microbiol. 37:144-148. [DOI] [PubMed] [Google Scholar]
- 37.Labigne, A., and H. de Reuse. 1996. Determinants of Helicobacter pylori pathogenicity. Infect. Agents Dis. 5:191-202. [PubMed] [Google Scholar]
- 38.Leclerc, G., S. P. Wang, and B. Ely. 1998. A new class of Caulobacter crescentus flagellar genes. J. Bacteriol. 180:5010-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linton, D., M. Gilbert, P. G. Hitchen, A. Dell, H. R. Morris, W. W. Wakarchuk, N. A. Gregson, and B. W. Wren. 2000. Phase variation of a beta-1,3 galactosyltransferase involved in generation of the ganglioside GM1-like lipo-oligosaccharide of Campylobacter jejuni. Mol. Microbiol. 37:501-514. [DOI] [PubMed] [Google Scholar]
- 40.Logan, S. M., J. W. Conlan, M. A. Monteiro, W. W. Wakarchuk, and E. Altman. 2000. Functional genomics of Helicobacter pylori: identification of a beta-1,4 galactosyltransferase and generation of mutants with altered lipopolysaccharide. Mol. Microbiol. 35:1156-1167. [DOI] [PubMed] [Google Scholar]
- 41.Marais, A., C. Bilardi, F. Cantet, G. L. Mendz, and F. Megraud. 2003. Characterization of the genes rdxA and frxA involved in metronidazole resistance in Helicobacter pylori. Res. Microbiol. 154:137-144. [DOI] [PubMed] [Google Scholar]
- 42.Marshall, B. J., and J. R. Warren. 1984. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet i:1311-1315. [DOI] [PubMed]
- 43.McGee, D. J., C. A. May, R. M. Garner, J. M. Himpsl, and H. L. Mobley. 1999. Isolation of Helicobacter pylori genes that modulate urease activity. J. Bacteriol. 181:2477-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGowan, C. C., A. Necheva, S. A. Thompson, T. L. Cover, and M. J. Blaser. 1998. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol. Microbiol. 30:19-31. [DOI] [PubMed] [Google Scholar]
- 45.Mobley, H. L., M. J. Cortesia, L. E. Rosenthal, and B. D. Jones. 1988. Characterization of urease from Campylobacter pylori. J. Clin. Microbiol. 26:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monteiro, M. A., B. J. Appelmelk, D. A. Rasko, A. P. Moran, S. O. Hynes, L. L. MacLean, K. H. Chan, F. S. Michael, S. M. Logan, J. O'Rourke, A. Lee, D. E. Taylor, and M. B. Perry. 2000. Lipopolysaccharide structures of Helicobacter pylori genomic strains 26695 and J99, mouse model H. pylori Sydney strain, H. pylori P466 carrying sialyl Lewis X, and H. pylori UA915 expressing Lewis B classification of H. pylori lipopolysaccharides into glycotype families. Eur. J. Biochem. 267:305-320. [DOI] [PubMed] [Google Scholar]
- 47.Monteiro, M. A., K. H. Chan, D. A. Rasko, D. E. Taylor, P. Y. Zheng, B. J. Appelmelk, H. P. Wirth, M. Yang, M. J. Blaser, S. O. Hynes, A. P. Moran, and M. B. Perry. 1998. Simultaneous expression of type 1 and type 2 Lewis blood group antigens by Helicobacter pylori lipopolysaccharides. Molecular mimicry between H. pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. J. Biol. Chem. 273:11533-11543. [DOI] [PubMed] [Google Scholar]
- 48.Monteiro, M. A., P. Y. Zheng, B. J. Appelmelk, and M. B. Perry. 1997. The lipopolysaccharide of Helicobacter mustelae type strain ATCC 43772 expresses the monofucosyl A type 1 histo-blood group epitope. FEMS Microbiol. Lett. 154:103-109. [DOI] [PubMed] [Google Scholar]
- 49.Newton, D. T., and D. Mangroo. 1999. Mapping the active site of the Haemophilus influenzae methionyl-tRNA formyltransferase: residues important for catalysis and tRNA binding. Biochem. J. 339:63-69. [PMC free article] [PubMed] [Google Scholar]
- 50.Nilsson, I., M. Utt, H. Nilsson, A. Ljungh, and T. Wadstrom. 2000. Two-dimensional electrophoretic and immunoblot analysis of cell surface proteins of spiral-shaped and coccoid forms of Helicobacter pylori. Electrophoresis 21:2670-2677. [DOI] [PubMed] [Google Scholar]
- 51.Peterson, W. L. 1991. Helicobacter pylori and peptic ulcer disease. N. Engl. J. Med. 324:1043-1048. [DOI] [PubMed] [Google Scholar]
- 52.Pfoestl, A., A. Hofinger, P. Kosma, and P. Messner. 2003. Biosynthesis of dTDP-3-acetamido-3,6-dideoxy-alpha-d-galactose (dTDP-d-Fucp3NAc) in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 278:26410-26417. [DOI] [PubMed] [Google Scholar]
- 53.Qa'Dan, M., L. M. Spyres, and J. D. Ballard. 2001. pH-enhanced cytopathic effects of Clostridium sordellii lethal toxin. Infect. Immun. 69:5487-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasko, D. A., M. Keelan, T. J. Wilson, and D. E. Taylor. 2001. Lewis antigen expression by Helicobacter pylori. J. Infect. Dis. 184:315-321. [DOI] [PubMed] [Google Scholar]
- 55.Richard, J. F., L. Petit, M. Gibert, J. C. Marvaud, C. Bouchaud, and M. R. Popoff. 1999. Bacterial toxins modifying the actin cytoskeleton. Int. Microbiol. 2:185-194. [PubMed] [Google Scholar]
- 56.Rioux, S., C. Begin, J. D. Dubreuil, and M. Jacques. 1997. Isolation and characterization of LPS mutants of Actinobacillus pleuropneumoniae serotype 1. Curr. Microbiol. 35:139-144. [DOI] [PubMed] [Google Scholar]
- 57.Sau, S., and C. Y. Lee. 1996. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J. Bacteriol. 178:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaffer, C., T. Wugeditsch, H. Kahlig, A. Scheberl, S. Zayni, and P. Messner. 2002. The surface layer (S-layer) glycoprotein of Geobacillus stearothermophilus NRS 2004/3a. Analysis of its glycosylation. J. Biol. Chem. 277:6230-6239. [DOI] [PubMed] [Google Scholar]
- 60.Schirm, M., E. C. Soo, A. J. Aubry, J. Austin, P. Thibault, and S. M. Logan. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 61.Sherburne, R., and D. E. Taylor. 1995. Helicobacter pylori expresses a complex surface carbohydrate, Lewis X. Infect. Immun. 63:4564-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stern, R. J., T. Y. Lee, T. J. Lee, W. Yan, M. S. Scherman, V. D. Vissa, S. K. Kim, B. L. Wanner, and M. R. McNeil. 1999. Conversion of dTDP-4-keto-6-deoxyglucose to free dTDP-4-keto-rhamnose by the rmIC gene products of Escherichia coli and Mycobacterium tuberculosis. Microbiology 145:663-671. [DOI] [PubMed] [Google Scholar]
- 63.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 65.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 66.Uemura, N., S. Okamoto, S. Yamamoto, N. Matsumura, S. Yamaguchi, M. Yamakido, K. Taniyama, N. Sasaki, and R. J. Schlemper. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784-789. [DOI] [PubMed] [Google Scholar]
- 67.Walsh, A. G., M. J. Matewish, L. L. Burrows, M. A. Monteiro, M. B. Perry, and J. S. Lam. 2000. Lipopolysaccharide core phosphates are required for viability and intrinsic drug resistance in Pseudomonas aeruginosa. Mol. Microbiol. 35:718-727. [DOI] [PubMed] [Google Scholar]
- 68.Wang, G., Z. Ge, D. A. Rasko, and D. E. Taylor. 2000. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol. Microbiol. 36:1187-1196. [DOI] [PubMed] [Google Scholar]
- 69.Warren, J., and B. Marshall. 1983. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet i:1273-1275. [PubMed]
- 70.Yethon, J. A., J. S. Gunn, R. K. Ernst, S. I. Miller, L. Laroche, D. Malo, and C. Whitfield. 2000. Salmonella enterica serovar typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect. Immun. 68:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida, Y., Y. Nakano, T. Nezu, Y. Yamashita, and T. Koga. 1999. A novel NDP-6-deoxyhexosyl-4-ulose reductase in the pathway for the synthesis of thymidine diphosphate-d-fucose. J. Biol. Chem. 274:16933-16939. [DOI] [PubMed] [Google Scholar]
- 72.Young, N. M., J. R. Brisson, J. Kelly, D. C. Watson, L. Tessier, P. H. Lanthier, H. C. Jarrell, N. Cadotte, F. St Michael, E. Aberg, and C. M. Szymanski. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]