Abstract
A complex interplay between multiple chromatin modifiers is critical for cells to regulate chromatin structure and accessibility during essential DNA-templated processes such as transcription. However, the coordinated activities of these chromatin modifiers in the regulation of gene expression are not fully understood. We previously determined that the budding yeast histone H4 methyltransferase Set5 functions together with Set1, the H3K4 methyltransferase, in specific cellular contexts. Here, we sought to understand the relationship between these evolutionarily conserved enzymes in the regulation of gene expression. We generated a comprehensive genetic interaction map of the functionally uncharacterized Set5 methyltransferase and expanded the existing genetic interactome of the global chromatin modifier Set1, revealing functional overlap of the two enzymes in chromatin-related networks, such as transcription. Furthermore, gene expression profiling via RNA-Seq revealed an unexpected synergistic role of Set1 and Set5 in repressing transcription of Ty transposable elements and genes located in subtelomeric regions. This study uncovers novel pathways in which the methyltransferase Set5 participates and, more importantly, reveals a partnership between Set1 and Set5 in transcriptional repression near repetitive DNA elements in budding yeast. Together, our results define a new functional relationship between histone H3 and H4 methyltransferases, whose combined activity may be implicated in preserving genomic integrity.
Keywords: transcription, genetic interaction, Set5, Set1, histone methylation, telomeres, retrotransposons
Introduction
Eukaryotic genomes are packaged into discrete regions of transcriptionally active genes (euchromatin) and transcriptionally silenced genes (heterochromatin). The establishment and maintenance of these chromatin regions is essential for virtually all nuclear processes and requires the dynamic posttranslational modification of histones.1 In particular, distinct patterns of histone lysine methylation are associated with the function and structure of specific chromatin domains. For example, methylation of histone H3 at lysines 4 and 36 is abundant in euchromatic regions, whereas these marks are reduced in heterochromatin.2
The most well-studied lysine methyltransferases (KMTs) in budding yeast are Set1, Set2, and Dot1, which methylate lysines 4, 36, and 79 on histone H3, respectively. Despite the global presence of these marks in euchromatin, removal of any one of these enzymes results in limited changes in gene expression.3 However, it has been observed that combined depletion of more than one chromatin-regulatory protein often leads to more severe phenotypes,4,5 suggesting that chromatin regulators act in concert with one another to exert their functions. For example, maintenance of euchromatin-heterochromatin boundaries at telomeres requires the cooperative activities of Dot1, Set1, Set2, histone acetylation, and the histone variant Htz1.6,7
We recently identified Set5 as the first histone H4 methyltransferase in budding yeast.8 Set5 is evolutionarily conserved and targets the functionally important lysines 5, 8, and 12 of the histone H4 tail, which have demonstrated roles in several aspects of chromatin function, including transcription and DNA damage responses. However, the role of Set5-mediated methylation in regulating the genome remains unknown.8,9 Our initial genetic studies demonstrated a functional link between Set5 and Set1, as cells lacking both methyltransferases show decreased fitness when subjected to genotoxic and cellular stress. These results indicate that Set5 and Set1 might function in similar pathways.
Set1 is a conserved lysine methyltransferase largely known to contribute to the regulation of gene expression, participating in both transcriptional activation and repression.10 Interestingly, Set1 has specifically been shown to influence gene expression at specialized chromatin environments, including telomeres and Ty retrotransposons. Deletion of SET1 results in defects in telomeric silencing and derepression of subtelomeric genes.10-15 Although not entirely clear, it is postulated that the derepression of telomere-proximal genes in set1∆ cells is caused by the titration of the Sir silencing factors away from the telomere when the euchromatin-heterochromatin boundary is disrupted,15 or that Set1 may have a more direct effect on gene repression through the regulation of antisense transcription.10,14 In addition to its role at telomeres and other silenced chromatin domains, Set1 has been reported to be required for silencing of Ty element transcription,16 although the mechanism for this remains largely undefined.17
Given the well-established and diverse roles for Set1 in regulating transcriptional activity, we hypothesized that Set5 may function with Set1 in the control of gene expression. Here, we provide an analysis of the functional interplay between Set1 and Set5 in S. cerevisiae. We expand our genetic interaction studies and present an extensive synthetic genetic interaction profile of SET1 and SET5, revealing overlapping functions of these enzymes in critical cellular processes such as transcription regulation and telomere maintenance. RNA-Seq analysis in cells lacking both SET1 and SET5 uncovers a synergistic role for these enzymes in transcriptional repression near telomeres and at transposable elements. Thus, our findings define a cooperative relationship between Set5 and Set1 in transcriptional control through the repression of genes associated with repetitive DNA elements.
Results
Set5 and Set1 cooperate to negatively regulate gene expression
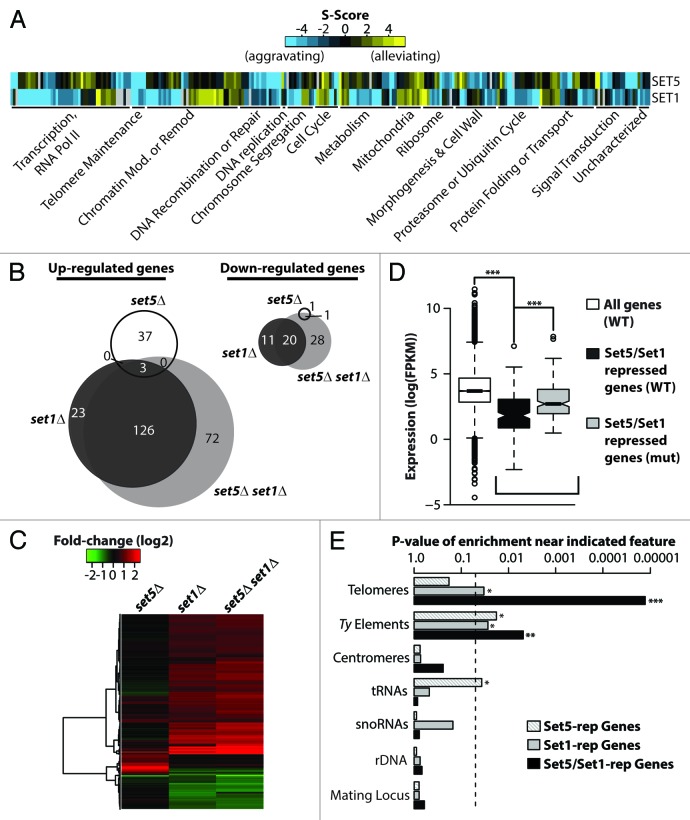
The majority of biological functions are accomplished by the coordinated action of multiple proteins. Synthetic genetic interaction maps provide an unbiased way to dissect the role of newly identified proteins by revealing their functional connections with other complexes.18 The E-MAP (Epistatic Miniarray Profile) approach has been particularly illuminating in defining functional pathways for a number of chromatin-associated factors in DNA-templated processes, especially transcription.18,19 We previously demonstrated a negative (aggravating) synthetic genetic interaction between Set1 and the recently characterized methyltransferase Set5, illustrating a functional relationship between the two proteins.8 Interestingly, it has been demonstrated that transcription factors with negative genetic interactions often regulate common gene sets.19 To further understand how the function of Set5 and Set1 intersect in transcription and other cellular processes, we generated a genetic interaction map of SET1 and SET5 using the E-MAP approach.20 We and others previously observed a limited genetic network using a set1∆ query strain,18,21 characteristic of a ‘noisy’ strain producing a high standard deviation of fitness among mutants that precludes quantitative analyses.20 As such, we generated a query strain containing a hypomorphic allele of SET1 using the Decrease Abundance by mRNA Perturbation (DAmP) technique, which removes the 3′ terminator region of a gene resulting in reduced mRNA stability and consequent protein abundance.22 The SET1-DAmP and set5∆ query strains were crossed to a previously defined subset of 1536 deletion or DAmP strains covering components of all major biological processes.23 The genetic interactome of our SET1-DAmP strain recapitulates, and expands upon, the interactions previously described for other members of the Set1-containing COMPASS complex.18 In agreement with its role in both transcriptional activation and repression,10,18 we find that SET1 interacts with factors involved in transcription regulation and chromatin modification or remodeling (Fig. 1A). Analysis of the genetic interactome for Set5 indicates a number of functional interactions with both nuclear and cytoplasmic factors, likely owing to its varied subcellular localization.8 Among the nuclear factors, Set5 shows functional links to the transcriptional machinery (Fig. 1A). Interestingly, although Set1 and Set5 display largely distinct sets of genetic interactions, they show overlapping functions with factors that influence telomere maintenance, DNA recombination or repair, and a subset of transcription-related genes (Fig. 1A).
Figure 1. Set5 and Set1 synergistically repress transcription of genes enriched near repetitive elements. (A) Heatmap representation of E-MAP genetic interaction scores (S-scores) of genes that show significant interactions with SET1 and SET5. Genes were grouped into previously described manually-curated categories.18 (B) Venn diagram of differentially regulated genes between wild type and indicated gene knockout strains with false discovery rate (FDR) < 0.05 and fold change > 1.7 in RNA-Seq analysis. (C) Expression heatmap and hierarchical clustering of strains described in (B). Color intensity represents fold-change (log2) in gene expression relative to wild type (WT). (D) Set5 and Set1 repress a subset of lowly expressed genes in WT cells. Boxplots depict the gene expression distributions of all genes in WT [white; “All Genes”], Set5/Set1-repressed genes in WT [black, “Set5/Set1-repressed genes (WT)”], and Set5/Set1-repressed genes in set5∆ set1∆ [gray; “Set5/Set1-repressed genes (mut)”]. “Repressed” genes represent significantly differentially upregulated genes in the double mutant compared with WT. Expression is shown as FPKM (Fragments Per Kilobase per Million mapped reads). (E) Set5/Set1-repressed genes are enriched near transposable elements and telomeres. Indicated chromosomal features are shown according to p-value of enrichment, described in Methods, for genes whose repression is dependent on Set5, Set1, or Set5/Set1. Dashed vertical line denotes P = 0.05. P values represent the two-sided probability value from the Wilcoxon rank sum test, *P < 0.05, ** P < 0.01, *** P < 0.001.
Due to the established link between Set1 and transcription regulation, and the functional overlap uncovered by our quantitative genetic analyses, we hypothesized that Set1 and Set5 might cooperate to regulate gene expression programs. We previously observed only minor changes in gene expression in cells lacking Set5,8 whereas investigations of the role of Set1 in gene expression have revealed that it contributes to transcriptional activation and repression.10 Recent genome-wide studies report that greater than 75% of Set1-dependent genes are upregulated in the set1∆ mutant compared with wild type, and these Set1-repressed genes are significantly enriched near telomere proximal regions.3,14,15,24
To investigate the combined role for Set1 and Set5 in gene expression, we performed whole-transcriptome sequencing (RNA-Seq) and analyzed the differential expression profiles of mutants lacking Set1, Set5, or both. Total mRNA from two independent biological replicates of set1∆, set5∆ or set1∆ set5∆ cells were used for single-end sequencing. Differential expression analysis of set1∆ mutants showed significant overlap with previous set1∆ microarray gene expression studies (data not shown).3,14
Our results illustrate that Set5 and Set1 cooperate in the repression of transcription. Specifically, of the 183 genes displaying at least a 1.7-fold change in expression in the set1∆ cells, the vast majority (83%) are upregulated in the mutant compared with wild type (Fig. 1B). Deletion of SET5 alone resulted in changes in the expression of a small subset of genes (42 genes total). With the exception of two genes (one of which is SET5), all of the Set5-regulated genes showed higher transcript levels in the mutant compared with the wild type strain. A similar differential expression profile was observed in a strain harboring an integrated point mutation of a conserved catalytic residue (Y402A) in the SET5 gene that disrupts its methyltransferase activity.8 Specifically, the 37 genes upregulated in set5∆ are also upregulated in the SET5Y402A mutant (Fig. S1), indicating that Set5′s catalytic activity is required for its function in gene repression.
Analysis of the transcriptome of the set1∆ set5∆ mutants revealed a 40% increase in the number of genes significantly differentially expressed in the double mutant compared with the set1∆ mutant alone (250 Set5/Set1-dependent genes compared with 183 Set1-dependent genes) (Fig. 1B). Moreover, greater than 80% of the genes significantly differentially expressed in the double mutant are upregulated compared with wild type. Furthermore, of the genes that are upregulated both in set1∆ and set1∆ set5∆ cells, we observe an additional 22% increase, on average, in transcript levels in the double mutant (Fig. 1C). Unexpectedly, of the 40 genes that are upregulated in the set5∆ mutant, 18 genes are either downregulated (3 genes, 7.5%), or unchanged (15 genes, 37.5%) in the double mutant compared with wild type. The small sample size of this gene set precludes significant analysis, but suggests that Set5′s unique role in gene repression requires functional Set1.
In summary, our results demonstrate that combined deletion of SET1 and SET5 results in an exacerbation of the transcriptional derepression observed upon deletion of SET1 alone, and therefore prompted further analysis of the genes dependent on both Set5 and Set1 for repression (this gene set is hereafter referred to as “Set5/Set1-repressed genes”). Based on GO term analysis, there is no significant enrichment for specific functional categories or cellular processes within the Set5/Set1-repressed genes (data not shown). To further explore the characteristics of this subset of genes, we examined the relative transcript levels of the Set5/Set1-repressed genes by comparing their median expression distribution to global gene expression. Set5/Set1-repressed genes show significantly lower transcript levels in wild type cells compared with the genome-wide average (Fig. 1D), suggesting that Set1 and Set5 might selectively control genes located in regions of inherently low transcriptional activity. These observations prompted us to analyze the specific chromosomal locations of the Set5/Set1 co-regulated genes. We examined the distribution of the genes upregulated in set1∆, set5∆, and set1∆ set5∆ cells with respect to defined chromosomal features. Analysis of the single mutants revealed that genes regulated by Set5 alone are enriched near Ty elements and tRNAs, whereas genes regulated by Set1 alone are enriched near telomeres, as previously reported,10-15 and Ty elements. Interestingly, the Set5/Set1-repressed genes showed a distinct enrichment proximal to both telomeres and Ty elements (Fig. 1E), but were not enriched near other tested genomic features, such as centromeres and the mating-type locus. Importantly, the Set5 and Set1 repressed genes near Ty elements are dispersed genome-wide and not enriched near telomeric Ty elements, indicating that these are distinct, non-overlapping genomic enrichment patterns. Identical results were observed when analyzing the subset of 72 genes uniquely repressed in set1∆ set5∆ mutants—excluding genes also repressed in either of the single mutants (referred to as “Set5/Set1 unique”). Specifically, the 72 “Set5/Set1 unique” genes show lower transcription in wild type cells than global transcription levels (Fig. S2A), and significant enrichment near telomeres and Ty elements (Fig. S2B). Overall, these results indicate that Set5 and Set1 are acting together to repress gene expression near repetitive regions of the genome, and warrant a more detailed exploration of the regulation of gene expression near these elements.
Telomere proximal genes are co-regulated by Set1 and Set5
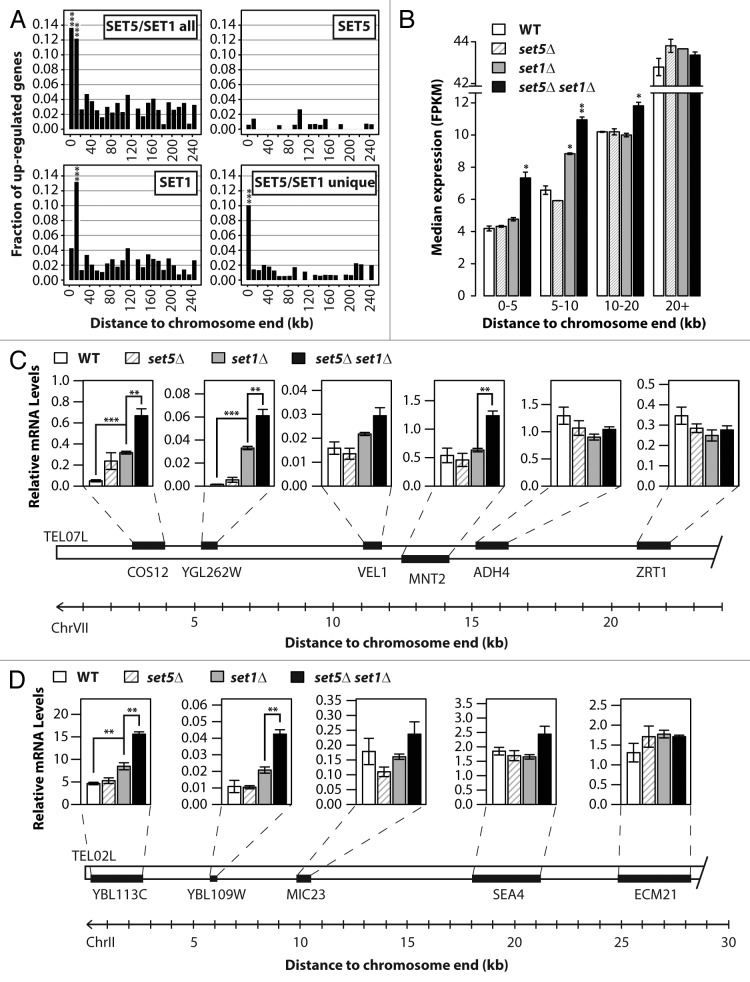
To gain insight into the cooperative role of Set1 and Set5 in regulating gene expression near telomeric regions, we analyzed the distribution of genes upregulated in the set1∆ set5∆ strain relative to the distance from chromosome ends. For the Set5/Set1-repressed genes, the largest fraction of genes is located within the first 20 kb from the chromosome end (Fig. 2A, upper left panel). The small number of genes repressed by Set5 alone shows a random distribution throughout the chromosomes (Fig. 2A, upper right panel), while Set1-repressed genes are found significantly enriched near telomeric boundaries (Fig. 2A, lower left panel), consistent with previous reports.14,15 The distribution of the Set5/Set1 unique genes showed a striking pattern of enrichment within 10 kb of the chromosome end (Fig. 2A, lower right panel). Furthermore, median transcript levels of genes positioned closer than 20 kb to the telomere are significantly higher in the double mutant compared with single mutants or wild type (Fig. 2B). Notably, the observed difference in transcription gradually diminishes in genes located beyond 10 kb from the end of the chromosome (Fig. 2B). Together, these results show that Set5 synergizes with Set1 to repress transcription of subtelomeric genes.
Figure 2. Subtelomeric genes are transcriptionally upregulated in set5∆set1∆ double mutants. (A) Histograms show the fraction of genes in 10 kb intervals plotted as a function of their distance to the nearest chromosome end. “SET5/SET1 all” refers to all genes that are upregulated in set5∆ set1∆ mutants compared with wild type; “SET5” are genes upregulated in set5∆; “SET1” are genes upregulated in set1∆; “SET5/SET1 unique” are genes upregulated in the set5∆ set1∆ strain but not in the single mutants. (B) RNA-Seq median FPKM expression of all genes within 0–5 kb, 5–10 kb, 10–20 kb, or 20+ kb of the nearest chromosome end in the indicated wild type (WT) and mutant strains. (C andD) Quantitative RT-PCR measurements of mRNA levels of the indicated ORFs on the left arm of chromosome VII (C) or the left arm of chromosome II (D). Relative mRNA values were calculated according to a reference gene, described in Methods. Data are represented as mean ± SEM (standard error of the mean) for 3 biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001.
To further investigate the transcriptional derepression observed in the set1∆ set5∆ mutants, we directly analyzed the regional transcription profile of genes near the telomeres on the left arm of chromosomes II and VII (abbreviated TEL02L and TEL07L, respectively). At these particular telomeres, our transcriptome analyses identified clusters of Set5/Set1-repressed genes. Similar gene clusters were previously described in cells lacking Htz1, a histone variant that localizes to euchromatin-heterochromatin boundaries to prevent the spread of gene silencing factors.25 In line with earlier studies,15 reverse transcription coupled with quantitative real-time PCR (qRT-PCR) confirmed a modest increase in expression of TEL07L and TEL02L proximal genes in cells lacking SET1 (Fig. 2C and D). As predicted by our RNA-Seq analysis, this effect is exacerbated upon deletion of SET5 at the genes closest to the chromosome ends, whereas genes located further from the telomeres showed little transcriptional regulation by Set1 or Set5 (Fig. 2C and D).
To determine whether Set5 methyltransferase activity is required for gene expression, we generated a set1Δ strain harboring the integrated SET5Y402A catalytic mutant and analyzed the expression levels of the subtelomeric genes at TEL07L and TEL02L in these cells (Fig. S3). Our qRT-PCR analyses show similar or even enhanced transcriptional upregulation compared with that of set5Δ set1Δ cells, indicating that the cooperative role of Set1 and Set5 in transcription repression is dependent on the catalytic activity of Set5 (Fig. S3).
In agreement with these observations, analysis of the shared genetic interactions between SET1 and SET5 demonstrates a common role in telomere maintenance (Fig. S4). In total, our transcriptional and genetic results support the conclusion that Set1 and Set5 cooperatively maintain transcriptional repression at telomere-proximal regions.
Repression of Ty element transcription by Set1 and Set5
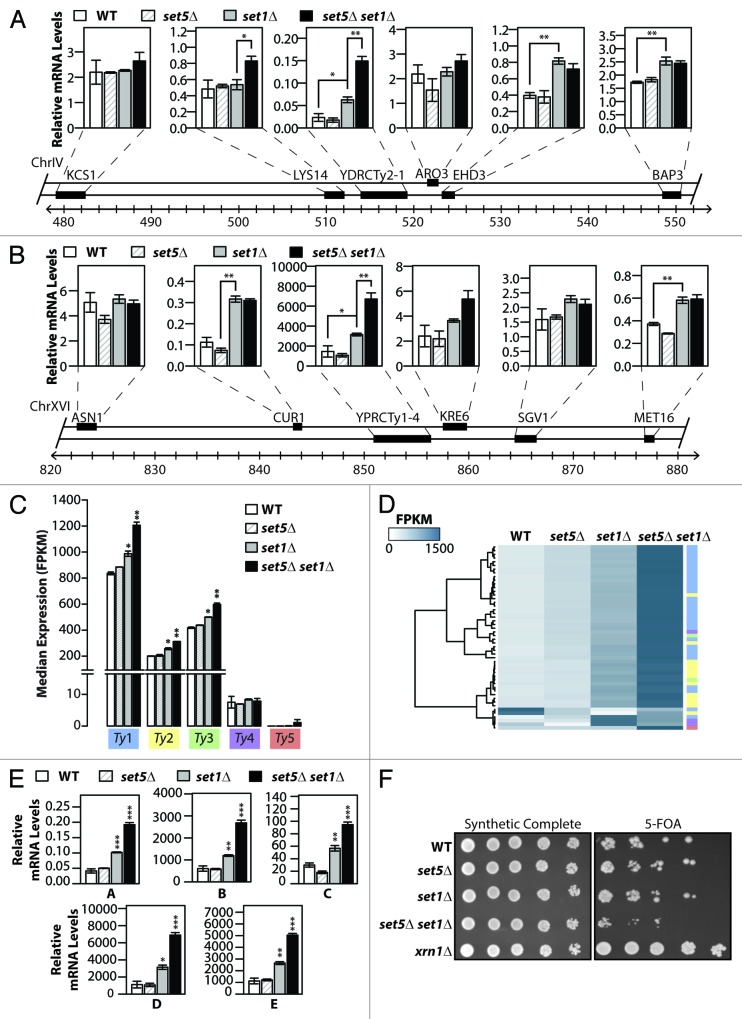
In addition to an enrichment of Set5/Set1-repressed genes in telomere proximal regions, analysis of our RNA-Seq results indicates that Set1 and Set5 are also influencing gene expression near Ty elements. We therefore used qRT-PCR to directly analyze the transcript levels of the genes surrounding two Ty elements as well as the Ty elements themselves. As shown in Figure 3A and B, we observe an increase in transcription of the genes proximal to Ty elements in set1∆ cells, with one of the adjacent genes (LYS14) showing a greater dependence on both Set1 and Set5 compared with Set1 alone. Interestingly, a stronger upregulation of the Ty elements themselves was observed in the double mutant compared with set1∆ cells (YDRCTy2–1 in Fig. 3A and YPRCTy1–4 in Fig. 3B). In addition, transcriptional upregulation at these Ty retrotransposons and neighboring genes is also dependent on Set5′s catalytic activity in set1∆ cells (Fig. S5).
Figure 3. Loss of SET5 and SET1 results in increased transcription of transposable elements. (AandB) Quantitative RT-PCR measurements of mRNA levels of the Ty element YDRCTy2–1 (gene YDR034C-D) and YPRCTy1–4 (genes YPR158C-D and YPR158C-C) and the indicated ORFs located nearby on chromosome IV (A) and chromosome XVI (B) for wild type (WT) and mutant strains. Relative mRNA values were calculated relative to a reference gene, as described in Methods. (C) RNA-Seq median FPKM expression of full-length yeast transposable (Ty) elements grouped by family (color-coded) for wild type (WT) and indicated mutant cells. (D) Hierarchically clustered heatmap of full-length Ty element FPKM expression in indicated strains. Color code on right represents each of the families depicted in (C). (E) qRT-PCR was used to measure the mRNA levels of the indicated Ty elements, calculated relative to the TFC1 reference gene, in WT and mutant cells. Data are represented as mean ± SEM for 3 biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001. (F) set1∆ set5∆ cells show increased expression of a reporter gene fused to the TY1(ML2) promoter. Serial dilutions of yeast containing the Ty1(ML2)::URA3 reporter were spotted on plates with either synthetic complete media or synthetic media supplemented with 5-fluoroorotic acid (5-FOA). Increased URA3 expression results in decreased growth on 5-FOA containing media.
To further investigate the role of Set1 and Set5 in Ty element transcription itself, we performed a comprehensive transcriptional analysis of all the currently annotated Ty elements in budding yeast.26 Expression levels of RNA intermediates (co-transcribed GAG and POL genes) from individual retrotransposons were quantified and grouped by family (Ty1–5) (Fig. S6A). Median gene expression of Ty1, Ty2, and Ty3 families of retrotransposons shows an increase in set1∆ set5∆ double mutants compared with the single mutants or wild type (Fig. 3C). Furthermore, when individually examining each Ty element of the yeast genome, increased transcript levels were observed in cells lacking both Set1 and Set5 compared with Set1 or Set5 alone (Fig. 3D; Fig. S6B). Consistent with previous reports and our observations from Figure 3A and B, there is an increase in transcript levels of a majority of Ty retrotransposons in set1∆ mutants,16 and an even greater increase in set1∆ set5∆ double mutants (Fig. 3D; Fig. S6B).
To directly validate the results from our RNA-Seq analysis, we designed qRT-PCR probes to specifically quantify the transcript levels of several Ty1 and Ty2 retrotransposons, including the Ty elements also tested in Figure 3A and B. Due to the high homology between Ty coding sequences, each of the selected primer sets (labeled A – F) amplify more than one Ty1 or Ty2 element (see Table S1 for details on primer design). Deletion of SET1 alone resulted in a modest upregulation of Ty elements compared with wild type (Fig. 3E). In the absence of both Set1 and Set5, at least a 2-fold increase in GAG-POL transcript levels was observed in all regions tested (Fig. 3E).
A number of reporter assays have been previously developed to monitor Ty transposon expression levels in cells. A strain carrying the URA3 gene under the control of the highly expressed Ty1(ML2) native promoter was used to assess Ty1 transcription in our mutants.16 Cells expressing URA3 display slow growth in media containing 5-fluoroorotic acid (5-FOA). Deletion of the exoribonuclease Xrn1 results in reduced Ty1 mRNA levels and thus, an xrn1∆ strain is used as a control for low Ty1 transcription.16 We plated serial dilutions of wild type, set1∆, set5∆, set1∆ set5∆, and xrn1∆ cells on synthetic complete and 5-FOA plates and examined their growth. As expected, wild type cells show impaired growth on 5-FOA, due to normal levels of the Ty1(ML2)::URA3 transcript, while the xrn1∆ strain grew robustly on 5-FOA, due to minimal URA3 expression (Fig. 3F). Furthermore, set1∆ and set5∆ single mutant strains show similar growth to that observed for wild type. Importantly, and consistent with our gene expression results, set1∆ set5∆ double mutant cells display further growth impairment on 5-FOA compared with the single mutants alone, indicating an increase in URA3 expression levels and suggesting coordinated regulation of the Ty1 promoter by Set1 and Set5. In addition, analysis of the genetic interactomes indicates that Set5 and Set1 have common genetic interactions with factors involved in the regulation of Ty elements (Fig. S7). Taken together, these data demonstrate that Set1 and Set5 act in concert to locally repress transcription at Ty retrotransposons.
Discussion
Our data show that Set5 and Set1 cooperate to repress transcription near telomeres and Ty elements. Loss of Set1 was previously associated with abnormal spreading of the silencing factors beyond heterochromatic boundaries, resulting in derepression of subtelomeric genes.11,15 Although the mechanisms are unclear, several chromatin modifying complexes and Set1 are also involved in regulating chromatin structure at Ty1 elements.16,27,28 Both at telomeres and Ty elements, transcriptional silencing requires the target of Set1’s enzymatic activity, H3K4.15,16 The present study suggests that Set5 (and its catalytic activity on histone H4 lysines 5, 8, and 12) and Set1 cooperate to regulate transcription, perhaps by establishing a specialized chromatin structure in subtelomeric regions and Ty elements. A comprehensive characterization of the chromatin marks that decorate subtelomeric regions and Ty elements and proximal genes will be necessary to investigate this model. Furthermore, generation of ChIP-grade antibodies will be instrumental to directly test the presence of methylated H4 at these genomic locations in the future.
The establishment of chromatin architecture at telomeres and Ty elements is dependent on multiple histone modifiers, chromatin remodeling complexes, and non-coding transcription.5,17,29 There are a number of possible mechanisms by which Set1 and Set5 might cooperatively regulate gene expression at these regions. For example, hypoacetylation of histone H4 lysines 5, 8, and 12 is required for silencing at telomeres,30 and although the mechanisms are still unclear, some evidence indicates that histone acetylation plays a role in Ty1 retrotransposon silencing.16 Since acetylation and methylation at the same lysines are mutually exclusive, perhaps H4 methylation by Set5 is dynamically regulating the levels of H4 acetylation at lysines 5, 8, and 12. Set5 may then work together with the Set1-dependent H3K4 methyl mark at or near repetitive elements to maintain a repressive chromatin environment.
Recent research suggests that noncoding RNA molecules play a fundamental role in mediating heterochromatic gene silencing at telomeres and can prevent the mobilization of transposable elements (for a review see ref. 31). Interestingly, Set1 and H3K4 methylation contribute to non-coding transcription at Ty elements and telomeres.14,16,32 It is therefore possible that Set5 may cooperate with Set1 in the control of non-coding transcription near repetitive DNA sequences.
Interestingly, deletion of SET1 results in defects in telomere length.10,33,34 It is, however, unclear whether telomere shortening causes transcriptional de-repression of subtelomeric genes, or is a consequence of it.10,35 Regardless, the cooperative role of Set5 and Set1 in repressing transcription at telomeres suggests the possibility that telomere shortening may be exacerbated in set5∆ set1∆ mutants. Future examination of the telomere length of set5∆ set1∆ mutants will be needed to investigate the relationship between transcription and telomere length in these cells.
Whether the mechanisms are direct or indirect, the coordinated activity of Set1 and Set5 brings a new level of regulation to gene expression at telomeres and Ty elements. Our combined genetic and gene expression data provide novel and important insight into Set5′s cooperative role with Set1 in the regulation of transcription near repetitive elements across the genome. Future studies, including analysis of non-coding transcripts and genome-wide mapping of histone modifications, will be needed to characterize the cooperative effect of both Set5 and Set1 on the chromatin landscape.
Materials and Methods
Yeast strains and media
S. cerevisiae strains used in this study are listed in Table 1. Deletion strains were obtained from the Yeast Knockout Collection (Open Biosystems) or generated by standard PCR-mediated gene disruption. The SET5Y402A and corresponding SET5WT strains and derivatives are described in ref. 8. Strains YAM698 and YAM700 were kindly provided by A. Morillon (Institut Curie Paris).16 Standard YPD and SC growth media and plates were used. For the Ty1(ML2)::URA3 assay, plates were supplemented with 1mg/mL 5-FOA (Sigma; cat. no. F5013).
Table 1. Yeast strains used in this study.
| Strain | Genotype | Background | Reference |
|---|---|---|---|
| Wild type | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | BY4741 | YKO |
| YGM6 | MATa set5Δ::KANMX | BY4741 | YKO |
| YGM76 | MATa set5Δ::NATMX | BY4741 | 8 |
| YGM2 | MATa set1Δ::KANMX | BY4741 | YKO |
| YGM77 | MATa set5Δ::NATMX set1Δ::KANMX | BY4741 | 8 |
| YGM167 | MATa SET5::SET5wt::NATMX | BY4741 | 8 |
| YGM168 | MATa SET5::SET5Y402A::NATMX | BY4741 | 8 |
| YGM169 | MATa SET5::SET5wt::NATMX set1Δ::KANMX | BY4741 | 8 |
| YGM170 | MATa SET5::SET5Y402A::NATMX set1Δ::KANMX | BY4741 | 8 |
| YEG113 | MATa set5Δ::HIS3MX6 | BY4741 | 8 |
| YAM698 | MATa ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 can1–100 TY1(ML2)::URA3 | W303 | 16 |
| YAM700 | MATa Ty1(ML2)::URA3 xrn1Δ::KANMX | W303 | 16 |
| YEG180 | MATa Ty1(ML2)::URA3 set5Δ::KANMX | W303 | This study |
| YEG195 | MATa Ty1(ML2)::URA3 set1Δ::NATMX | W303 | This study |
| YEG197 | MATa Ty1(ML2)::URA3 set5Δ::KANMX set1Δ::NATMX | W303 | This study |
| E-MAP wild type |
MATalpha his3Δ1 leu2Δ0 LYS2+ met15Δ0 ura3Δ0 can1Δ::MATaPr-HIS3 lyp1Δ::MATαPr-LEU2 | YMS196H | 22 |
| YS550 | MATalpha set5::NAT clone#1 | YMS196H | This study |
| YS551 | MATalpha set5::NAT clone#2 | YMS196H | This study |
| YS553 | MATalpha set5::NAT clone#3 | YMS196H | This study |
| YS455 | MATalpha SET1 3′UTR::NAT | YMS196H | This study |
E-MAP experiments
Strain construction for the Epistatic Miniarray Profile experiment was performed as previously described using a Singer RoToR replica pinning robot (Singer Instruments).36,37 After photographing plates, colony sizes were measured using HT Colony Grid Analyzer software, and genetic interaction scores (S-scores) were quantified using the E-MAP toolbox for MATLAB.36 Significant genetic interactions were determined using previously established thresholds, with S ≤ –2.5 demarcating significant negative (synthetic sick or lethal) interactions and S ≥ 2.0 indicating significant positive (alleviating or epistatic) interactions. Tables S1–3 include the complete list of E-MAP processed scores and categories for Set5/Set1-significantly interacting genes. Genes were manually curated into functional categories as previously described.18
RNA-Seq library preparation
Total RNA was extracted and purified with the MasterPureTM Yeast RNA Purification Kit following the manufacturer’s instructions (Epicenter; cat. no. QER09015). RNA quantity and integrity were determined on an Agilent Technologies 2100 Bioanalyzer (RNA Integrity Number ≥ 6.2 for each sample). Poly-A based mRNA enrichment was performed via the Illumina TruSeqTM RNA Sample Preparation v2 Low-Throughput (LT) protocol (Illumina; cat. no. RS-122-2001). Briefly, poly-A containing mRNA molecules were purified from 8 µg of total RNA using poly-T oligo-attached magnetic beads and two rounds of enrichment were performed before thermal mRNA fragmentation. Fragmented mRNA was subjected to cDNA synthesis per instructions in the LT protocol. Specifically, SuperScript II reverse transcriptase and random primers were used for first strand cDNA synthesis before converting to double stranded DNA via supplied reagents. Ends were repaired and adenylated (3′) before subjecting the ds cDNA to multiple indexing adaptor ligation. DNA fragments with ligated adapters were enriched via PCR and the resulting library quality was verified on an Agilent Technologies 2100 Bioanalyzer.
RNA-Seq and analysis
Indexed libraries were sequenced on an Illumina HiSeq2000 platform according to the manufacturer’s protocols (Illumina). Reads were processed using the Tuxedo software suite (Bowtie v2.1.0, TopHat v2.0.5, and Cufflinks v2.0.2) following the “Quantification of reference annotation only” protocol as described in ref. 38. Briefly, single 101 bp reads were mapped to an S. cerevisiae reference genome (Ensembl EF4) with TopHat, specifying ‘–no-novel-juncs’. Expression and differential transcription were determined using Cufflinks and results were accessed in the R statistical computing environment using CummeRbund (v2.0.0). All RNA-Seq data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE52086 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE52086).
Statistical analysis
FPKM expression values represent Fragments Per Kilobase of transcript per Million mapped reads.39 Significantly differentially expressed (SDE) genes were identified at a false discovery rate (FDR)-adjusted P value of 0.05 and restricted to genes with fold-change relative to WT greater than 1.7, to ensure only robust differences were considered in the downstream analyses. Expression values for SDE genes were hierarchically clustered and plotted in R (v3.0.1) (Fig. 1C). The complete list of genes differentially expressed in each strain and their corresponding FPKM values is listed in Tables S4–6. To assess gene set enrichment near certain genomic regions, locations of chromosomal features were downloaded from the Saccharomyces Genome Database (SGD) using YeastMine query builder, and distances from gene transcription start sites (TSS) to each chromosomal feature were calculated. Significance of enrichment near specific features was assessed using the Wilcoxon rank sum (WRS) test in R. The WRS test is a non-parametric analog to the t test, and enables comparisons of distribution location shift in FPKM expression values from two populations of genes. The distribution of gene distances to nearest feature of the indicated geneset was compared with the genome-wide distribution of distances. Reported P values are from the two-sided test, with the alternative hypothesis that the true location shift is not equal to zero. For all reported significant P values in Figure 1E, the geneset distributions were shifted closer to the indicated feature than would be expected by chance given the genome-wide distribution.
RNA extraction for qRT-PCR analyses
The standard hot phenol extraction procedure was used to extract total RNA from the indicated yeast strains.40 Genomic DNA was eliminated using the Ambion DNase I kit per manufacturer’s instructions (Ambion-Invitrogen; cat. no. AM1906). cDNA was obtained using Superscript III first-strand synthesis (Invitrogen; cat. no. 18080-051) with random primers starting with 5 µg of total RNA. The Roche Universal Probe library (UPL) system was used for the quantitative transcript analysis. Primer pairs and their corresponding probe were designed using the assay Design Center. qRT-PCR information including all primer sequences used in this study is listed in Tables S7–9. Real-time amplification was performed on the LightCycler 480 instrument II (Roche) using the Roche probes master mix. The comparative threshold method was used for relative mRNA level quantification with TFC1 as a normalizing control.
Supplementary Material
Disclosure of Potential Conflicts of Interest
OPG is a co-founder of EpiCypher, Inc. The other authors declare no financial or non-financial competing interests.
Acknowledgments
The authors wish to thank the members of the Morrison, Gozani, and Chua labs for helpful discussions and critical reading of the manuscript. We thank A. Morillon (Institut Curie Paris) for generously providing yeast strains. We are grateful for the guidance and RNA-Seq reagents supplied by the Lab of Hunter Fraser at Stanford University, and the services provided by the Protein and Nucleic Acid facility at Stanford University. Martín GM was supported by the Generalitat de Catalunya Beatriu de Pinós award BP-A 2010. Morrison AJ is supported by funding from NIH (R00 GM085212). This work was supported in part by a grant from the NIH to Gozani O (R01 CA172560). Gozani O is a recipient of an Ellison Senior Scholar in Aging Award.
Glossary
Abbreviations:
- RNA-Seq
RNA sequencing
- KMT
lysine methyltransferase
- E-MAP
Epistatic Miniarray Profile
- DAmP
Decrease Abundance by mRNA Perturbation
- GO
gene ontology
- qRT-PCR
quantitative real-time PCR
- 5-FOA
5-fluoroorotic acid
- ChIP
Chromatin Immunoprecipitation
- YKO
Yeast Knock Out
- FPKM
fragments per kilobase per million mapped reads
- SDE
significantly differentially expressed
- FDR
false discovery rate
- SGD
Saccharomyces Genome Database
- TSS
transcription start site
- WRS
Wilcoxon rank sum
- UPL
Universal Probe Library
- SEM
standard error of the mean
- ORF
open reading frame
- MIPS
Munich Information Center for Protein Sequences
- LTR
long terminal repeat
- PR
protease
- IN
integrase
- RT
reverse transcriptase
- RH
RNAse H
- NA
RNA binding coding domain
References
- 1.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell. 2011;42:536–49. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Y, Rodriguez AM, Stanton JD, Kitazono AA, Wyrick JJ. Simultaneous mutation of methylated lysine residues in histone H3 causes enhanced gene silencing, cell cycle defects, and cell lethality in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:6832–41. doi: 10.1128/MCB.00745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verzijlbergen KF, Faber AW, Stulemeijer IJ, van Leeuwen F. Multiple histone modifications in euchromatin promote heterochromatin formation by redundant mechanisms in Saccharomyces cerevisiae. BMC Mol Biol. 2009;10:76. doi: 10.1186/1471-2199-10-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 7.Schoeftner S, Blasco MAA. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28:2323–36. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green EM, Mas G, Young NL, Garcia BA, Gozani O. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nat Struct Mol Biol. 2012;19:361–3. doi: 10.1038/nsmb.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green EM, Morrison AJ, Gozani O. New marks on the block: Set5 methylates H4 lysines 5, 8 and 12. Nucleus. 2012;3:335–9. doi: 10.4161/nucl.20695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dehé PM, Géli V. The multiple faces of Set1. Biochem Cell Biol. 2006;84:536–48. doi: 10.1139/o06-081. [DOI] [PubMed] [Google Scholar]
- 11.Santos-Rosa H, Bannister AJ, Dehe PM, Géli V, Kouzarides T. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J Biol Chem. 2004;279:47506–12. doi: 10.1074/jbc.M407949200. [DOI] [PubMed] [Google Scholar]
- 12.Mueller JE, Canze M, Bryk M. The requirements for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006;173:557–67. doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fingerman IM, Wu CL, Wilson BD, Briggs SD. Global loss of Set1-mediated H3 Lys4 trimethylation is associated with silencing defects in Saccharomyces cerevisiae. J Biol Chem. 2005;280:28761–5. doi: 10.1074/jbc.C500097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margaritis T, Oreal V, Brabers N, Maestroni L, Vitaliano-Prunier A, Benschop JJ, van Hooff S, van Leenen D, Dargemont C, Géli V, et al. Two distinct repressive mechanisms for histone 3 lysine 4 methylation through promoting 3′-end antisense transcription. PLoS Genet. 2012;8:e1002952. doi: 10.1371/journal.pgen.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A. 2007;104:16609–14. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–26. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wery M, Kwapisz M, Morillon A. Noncoding RNAs in gene regulation. Wiley Interdiscip Rev Syst Biol Med. 2011;3:728–38. doi: 10.1002/wsbm.148. [DOI] [PubMed] [Google Scholar]
- 18.Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 19.Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. Genetic analysis of variation in transcription factor binding in yeast. Nature. 2010;464:1187–91. doi: 10.1038/nature08934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins SR, Roguev A, Krogan NJ. Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol. 2010;470:205–31. doi: 10.1016/S0076-6879(10)70009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandyopadhyay S, Mehta M, Kuo D, Sung MK, Chuang R, Jaehnig EJ, Bodenmiller B, Licon K, Copeland W, Shales M, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–9. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–52. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 23.Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A, Bonneil E, Thibault P, Verreault A, Festenstein RJ. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011;7:e1001354. doi: 10.1371/journal.pgen.1001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–36. doi: 10.1016/S0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 26.Carr M, Bensasson D, Bergman CM. Evolutionary genomics of transposable elements in Saccharomyces cerevisiae. PLoS One. 2012;7:e50978. doi: 10.1371/journal.pone.0050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morillon A, Bénard L, Springer M, Lesage P. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol Cell Biol. 2002;22:2078–88. doi: 10.1128/MCB.22.7.2078-2088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Servant G, Pennetier C, Lesage P. Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol Cell Biol. 2008;28:5543–54. doi: 10.1128/MCB.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 2008;178:197–214. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carmen AA, Milne L, Grunstein M. Acetylation of the yeast histone H4 N terminus regulates its binding to heterochromatin protein SIR3. J Biol Chem. 2002;277:4778–81. doi: 10.1074/jbc.M110532200. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 32.Caslini C, Connelly JA, Serna A, Broccoli D, Hess JL. MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol Cell Biol. 2009;29:4519–26. doi: 10.1128/MCB.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corda Y, Schramke V, Longhese MP, Smokvina T, Paciotti V, Brevet V, Gilson E, Géli V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–8. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 34.Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–36. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandell LL, Gottschling DE, Zakian VA. Transcription of a yeast telomere alleviates telomere position effect without affecting chromosome stability. Proc Natl Acad Sci U S A. 1994;91:12061–5. doi: 10.1073/pnas.91.25.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–52. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 38.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ares M. Isolation of total RNA from yeast cell cultures. Cold Spring Harb Protoc. 2012;2012:1082–6. doi: 10.1101/pdb.prot071456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.