Abstract
The bromodomain and extra terminal (BET) protein family member BRD4 is a transcriptional regulator, critical for cell cycle progression and cellular viability. Here, we show that BRD4 plays an important role in embryonic stem cell (ESC) regulation. During differentiation of ESCs, BRD4 expression is upregulated and its gene promoter becomes demethylated. Disruption of BRD4 expression in ESCs did not induce spontaneous differentiation but severely diminished hematoendothelial potential. Although BRD4 regulates c-Myc expression, our data show that the role of BRD4 in hematopoietic commitment is not exclusively mediated by c-Myc. Our results indicate that BRD4 is epigenetically regulated during hematopoietic differentiation ESCs in the context of a still unknown signaling pathway.
Keywords: bromodomain, BRD4, hematopoiesis, stem cells, differentiation
Introduction
The human BRD4 protein is a member of the bromodomain and extra terminal (BET) family characterized by the presence of 2 tandem bromodomains capable of binding to acetylated lysine in histone H3 and H4.1,2 It is a transcriptional regulator of growth inducing genes that interact with acetylated histones to enable the recruitment of P-TEFb elongation factor to targeted loci, which stimulates RNAP II-dependent transcription.3–5 The role of BRD4 in cell cycle progression is well described, and both overexpression and downregulation of endogenous BRD4 levels cause G1/S arrest in cultured human cells.6–8 Also, BRD4 is a proto-oncogene which is activated by fusion with the nuclear protein in testis (NUT), causing rare but extremely aggressive carcinomas.9,10 More recently, BRD4 has been identified as a major c-MYC regulator and pharmacological suppression of BRD4 has shown a strong anti-tumorigenic effect in acute myeloid leukemia.3,11,12 Nonetheless, overexpression of BRD4 represses tumor growth and metastasis in mice models,13,14 and ectopic expression of BRD4-NUT reduces in vitro proliferation in mouse cells.15 These controversial studies indicate that the role of BRD4 in cancer could be more complex than previously anticipated, and not only restricted to cell cycle progression. In fact, recent studies have demonstrated that BRD4-NUT protein interferes with the normal epithelial differentiation in NUT midline carcinomas.16,17
Besides the role of BRD4 in carcinogenesis, BRD4 deficiency has also been linked to developmental defects. While BRD4 knockout mice are not viable and die shortly after implantation, mice carrying a defective copy of BRD4 show post-natal anatomical abnormalities such as head malformations, absence of subcutaneous fat, low weight, and cataracts, suggesting a possible developmental role for BRD4.7 Here, we analyzed the contribution of BRD4 to hematopoietic specification from embryonic stem cells (ESCs) and report that epigenetic modulation of BRD4 is involved in human hematopoiesis.
Results
Human BRD4 is demethylated and upregulated during ESCs differentiation
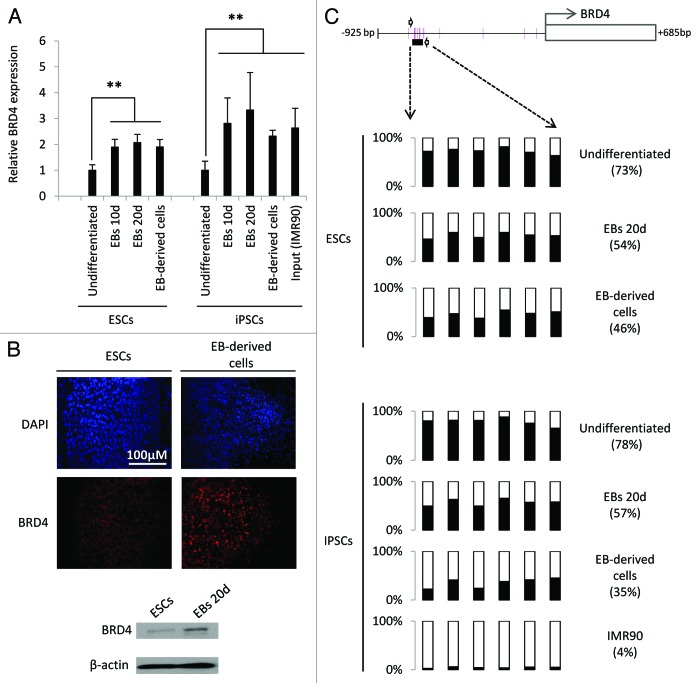
To study the regulation of BRD4 during ESC differentiation we first analyzed its RNA expression during spontaneous EB differentiation. We observed that RNA expression significantly increased upon differentiation of both ESCs and induced pluripotency stem cells (iPSCs) (Fig. 1A). BRD4 RNA levels were also higher in EB-derived cells obtained after plating of EBs on an adherent surface (Fig. 1A). Additionally, BRD4 was less expressed in iPSCs than in the IMR90 fibroblasts from which they were initially derived (Fig. 1A). Immunofluorescence staining and western blotting confirmed at the protein level that BRD4 is also upregulated in EBs and EB-derived cells as compared with undifferentiated ESCs (Fig. 1B).
Figure 1. Human BRD4 is demethylated and its expression upregulated during spontaneous differentiation of ESCs. (A) BRD4 expression in undifferentiated ES cells and embryoid bodies (EBs) at 10 and 20 d of differentiation and, EB-derived cells obtained by attachment of EBs on gelatin-coated plates. The iPSC line MSUH-002 and the somatic cell line originally reprogrammed (IRM90) were also included. BRD4 mRNA levels were analyzed by qRT-PCR and are represented as the mean value ± SD of three independent experiments (**P ≤ 0.01). (B) Immunofluorescence staining for BRD4 (red) in ES cells and EB-derived cells. Nuclei were labeled with DAPI (blue). Lower panel shows western blot analysis of BRD4 expression in undifferentiated ES cells and 20-d-old embryoid bodies. (C) Pyrosequencing analysis of DNA methylation in the BRD4 promoter region. Six CpG sites located approximately 600 bp upstream of the transcription initiation site were analyzed.
Developmental genes differentially expressed during cell differentiation are often subjected to epigenetic regulation.18,19 Since we have previously reported that BRD4 is partially regulated by DNA methylation in cancer cells,14 we hypothesized that upregulation of BRD4 during differentiation may be due to the loss of DNA methylation at its gene promoter. To test this hypothesis, we analyzed the promoter DNA methylation status of BRD4 in ESCs, iPSCs, EBs and EB-derived cells using bisulfite pyrosequencing. Results showed that BRD4 promoter underwent demethylation upon differentiation in all cell lines tested (Fig. 1C). This result was confirmed by bisulfite sequencing of multiple clones (Fig. S1A). Interestingly, BRD4 promoter in MSUH-002 cells is highly methylated (78%) as compared with that in IMR90 fibroblasts (4%) (Fig. 1C), suggesting that cellular reprogramming involves BRD4 DNA re-methylation. Additionally, ESCs treated with a demethylating agent (5-aza-2′-deoxycytidine) showed increased BRD4 expression, which provides further evidence of the role of DNA methylation in BRD4 regulation in ESCs (Fig. S1B).
BRD4 deregulation does not induce spontaneous differentiation of ES cells
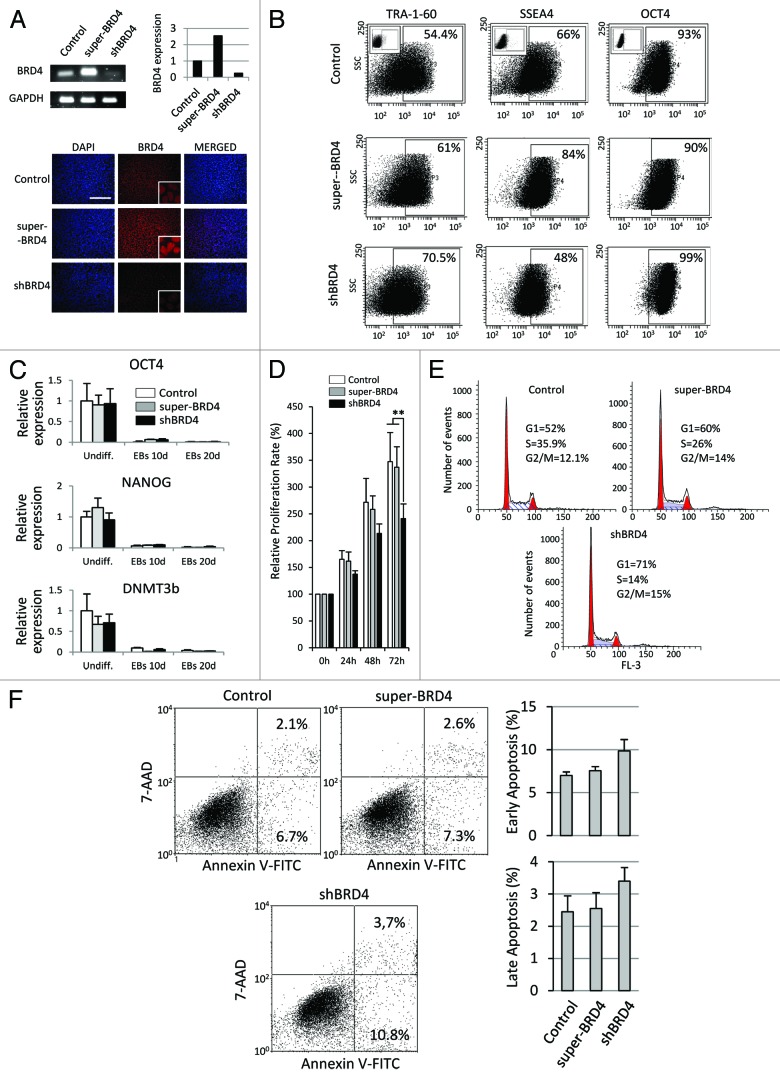
To further explore the role of BRD4 in ESCs differentiation, we stably upregulated and downregulated BRD4 levels in ESCs verifying BRD4 expression by real-time PCR, immunofluorescence (Fig. 2A) and western blot (Fig. S2). Flow cytometry analysis showed that neither overexpression nor downregulation of BRD4 induced a clear loss of expression of the pluripotency markers OCT4 and TRA-1–60 (Fig. 2B). Because SSEA4 levels were apparently reduced in shBRD4 cells (Fig. 2B), we analyzed expression of the ST3 β-galactoside α-2, 3-sialyltransferase 2 (ST3GAL2), which is an indicator of SSEA4 synthesis,20 by quantitative PCR, although we did not observe gene expression changes (Fig. S3). Thus, the current data do not clearly support the notion that BRD4 is regulating SSEA4 expression in ESCs. In line with this, mRNA levels of the pluripotency markers OCT4, NANOG, and DNMT3b were not affected either (Fig. 2C). As BRD4 activity has previously been associated with proliferation and cell cycle progression in cancer cells, we examined whether BRD4 is also regulating cell cycle in ESCs. Super-BRD4 cells proliferated at a similar rate to control cells. However, shBRD4 ESCs showed a proliferation rate slightly slower than control cells, albeit the difference was statistically significant (Fig. 2D). FACS analysis of DNA content showed the greatest accumulation of cells at G1 phase in shBRD4 ESCs (71%) as compared with super-BRD4 cells (60%) and to control cells (52%) (Fig. 2E). Inhibition and downregulation of BRD4 has previously been shown to exert a pro-apoptotic effect on cancer cells. Using annexin V staining we observed a slightly higher incidence of both early and late apoptotic cells in shBRD4 ESC cultures but it was not was statistically significant (Fig. 2F).
Figure 2. BRD4 alteration affected cell cycle progression but it did not induce spontaneous differentiation. (A) BRD4 mRNA levels analyzed by qRT-PCR and semi-quantitative PCR in shBRD4 and super-BRD4 ESCs. Lower panels show immunofluorescence staining for BRD4 (red). Nuclei were labeled with DAPI (blue). (B) Flow-cytometric analysis for OCT4, TRA-1–60, and SSEA4 in shBRD4 and super-BRD4 cells. (C) qRT-PCR analysis of OCT4, NANOG, and DNMT3b in undifferentiated ES cells, EBs at 10 and 20 d (control, shBRD4, and super-BRD4 cells). (D) Proliferation of shBRD4 and super-BRD4 cells compared with control cells, analyzed by MTT assay and represented as the mean value ± SD of three independent experiments (**P ≤ 0.01). (E) FACS analysis of DNA content in undifferentiated ESCs. (F) Apoptosis analysis by FITC-Annexin V staining. Left panels show representative dot plots. Graphs show mean values ± SD of three independent experiments.
BRD4 expression affects mesodermal differentiation of ESCs
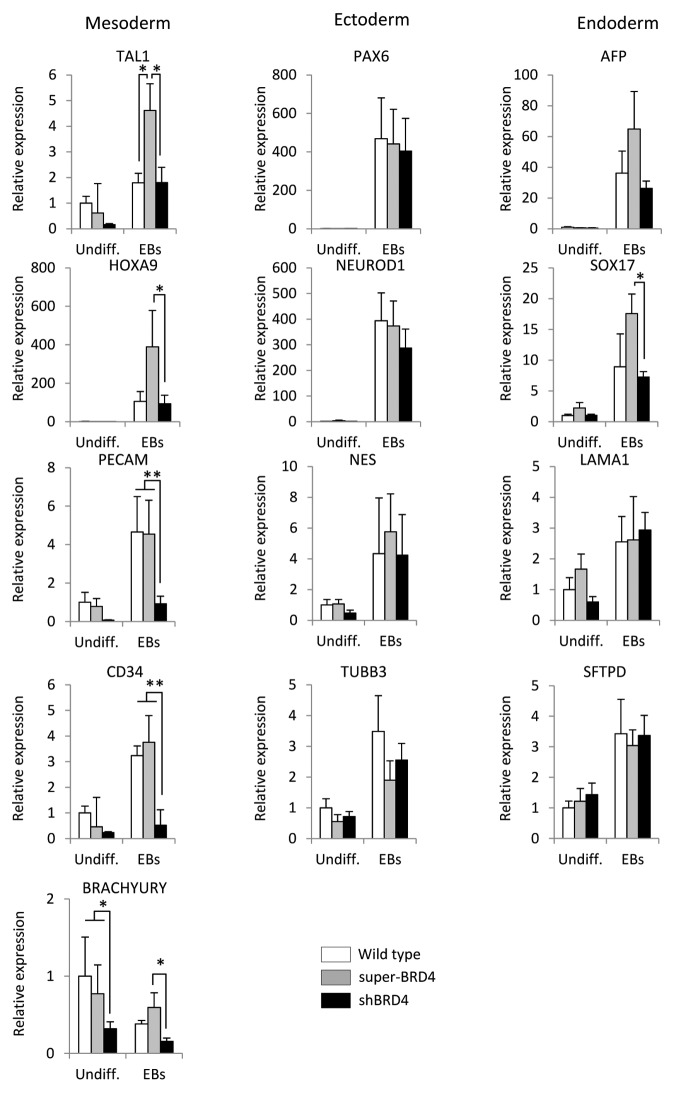
Because pluripotency markers were not affected by BRD4 regulation, we wanted to study in greater detail the functional role of BRD4 during differentiation. We induced EB formation and analyzed the emergence of developmental markers during spontaneous differentiation (Fig. 3). Alteration of BRD4 levels showed no effect on neuroectodermal markers (PAX6, NEUROD1, NESTIN, and β-III-TUBULIN), and only one endodermal marker (SOX17) was significantly affected. In contrast, all the mesodermal markers analyzed were affected by BRD4 expression: TAL-1, HOXA9, and BRACHYURY were upregulated in super-BRD4 cells, while BRACHYURY, CD31/PECAM, and CD34 were downregulated in shBRD4. These results indicate that BRD4 might play an important role during mesodermal specification.
Figure 3. BRD4 expression affected mesodermal markers during spontaneous differentiation of ESCs. qRT-PCR analysis of differentiation markers associated with the three embryonic germ layers. Results are represented as the mean value ± SD of three independent experiments. Asterisks are employed to denote statistical differences between groups (* P ≤ 0.05) (**P ≤ 0.01)
Downregulation of BRD4 reduces early mesodermal specification and hemogenic potential
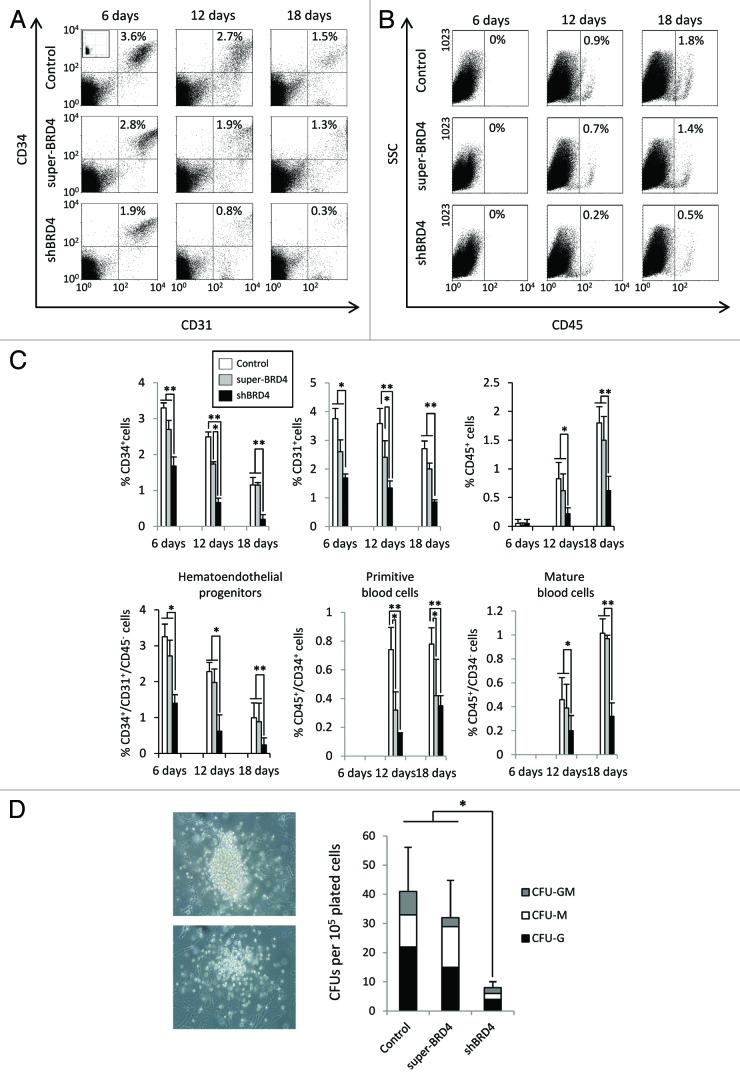
To study the potential role of BRD4 in mesodermal specification in more detail, we co-cultured shBRD4 and super-BRD4 ESCs on OP9, which has been shown to induce mesodermal differentiation.21 We then analyzed the levels of early embryonic hematoendothelial progenitors expressing CD31 (PECAM) and CD34 but negative for CD45 (Fig. 4A and B), which are responsible for endothelial and hematopoietic development during ESC differentiation.21–25 After 6 d of differentiation, CD34+/CD31+/CD45- population was significantly underrepresented in shBRD4 cells (1.4%) compared with control cells (3.25%) or super-BRD4 cells (2.72%) (P < 0.05) (Fig. 4A and C). This hemogenic population progressively decreased over time and after 18 d of differentiation, it was almost completely absent in shBRD4 cells (>0.3%) although still notably present (>1%) (P < 0.01) in controls and super-BRD4 ESCs. Primitive blood cells CD45+/CD34+did not emerge prior to day 6. After 12 d, the percentages of CD45+/CD34+ cells in control cells was around 0.74%, notably higher than in super-BRD4 (0.32%) and shBRD4 cells (0.15%) (P < 0.05). At day 12 of OP9 differentiation, mature blood cells, represented by the CD45+/CD34- population, were found at similar levels in control (0.46%) and super-BRD4 cells (0.39%) but were considerably lower in shBRD4 (0.19%) (P < 0.01). The percentage of CD45+/CD34- cells increased at 18 d but differences between control (1.01%) and shBRD4 (0.32%) cells were maintained (P < 0.01), and no significant difference were observed between control and super-BRD4 cells (0.97%) (Fig. 4B and C). To further confirm a role for BRD4 in hematopoietic differentiation, we analyzed the differentiation potential of hematopoietic progenitors derived from transfected ESCs by quantitative colony forming unit (CFU) assay (Fig. 4D). The number of CFUs derived from control ESC-hematopoietic derivatives was on average 4-fold higher than that generated from shBRD4 ESC-hematopoietic derivatives (P < 0.05). Nonetheless, the CFU potential of hematopoietic derivatives obtained from control ESC and super-BRD4 ESCs was very similar. In order to confirm the specificity of BRD4 in mesodermal differentiation, we next induced differentiation of ESCs into early neuroectodermal progenitors by EB formation and quantified neural colony emergence on MS-5 co-culture (Fig. S4). We did not observe either differential expression of neural markers after neural induction or significant changes in the neural potential on MS-5 cell co-culture, suggesting that neuroectodermal differentiation is not affected by BRD4 disruption.
Figure 4. Downregulation of BRD4 impairs hematoendothelial specification. (A and B) Representative flow cytometry dot plots of hematopoietic markers during ESC differentiation on OP9 co-culture. The percentage of CD34+/CD31+ and CD45+ cells corresponding to each dot plot are shown in the upper right corner. (C) Percentage of CD34+, CD31+, and CD45+ cells during hematopoietic differentiation. Primitive hematoendothelial progenitors were identified as CD34+/CD31+/CD45-, immature blood cells as CD34+/CD45+ and mature blood cells as CD45+/CD34-. Percentages are represented as the mean value ± SD of three independent experiments. (D) Colony forming unit (CFU) potential of control, shBRD4, and super-BRD4 ESCs. Results are represented as the mean value ± SD of three independent experiments (*P ≤ 0.05) (**P ≤ 0.01).
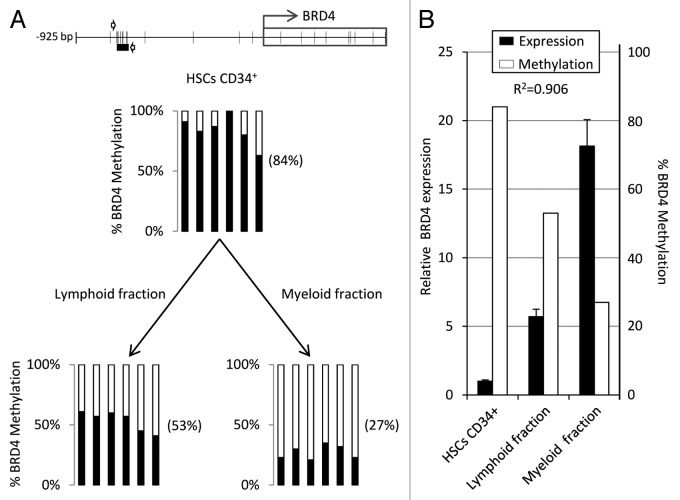
To further investigate the role of BRD4 in hematopoiesis, we analyzed BRD4 methylation and expression in human CD34+ hematopoietic stem and progenitor cells (HSPCs) derived from cord blood and in both myeloid and lymphoid fractions (Fig. 5A). BRD4 promoter was more highly methylated in CD34+ HSPCs (84%) than in terminally differentiated myeloid (53%) and lymphoid cells (27%). Finally, to determine the relationship between DNA methylation and gene expression we used qRT-PCR to analyze the BRD4 levels in these same samples and found very low BRD4 expression in CD34+ HSPCs and high expression in the both myeloid and lymphoid fractions, showing a good correlation with methylation status (R2 = 0.906) (Fig. 5B) which points to DNA methylation indeed playing a role in BRD4 regulation in somatic hematopoiesis.
Figure 5. Human BRD4 is differentially methylated and expressed in somatic CD34+ HSPCs and myeloid/lymphoid cells. (A) Pyrosequencing analysis of DNA methylation in the BRD4 promoter region of CB-derived CD34+ HSPCs and myeloid and lymphoid cell fractions. (B) Correlation between BRD4 expression and DNA methylation in CD34+ HSPCs, myeloid, and lymphoid cells (R2 = 0.906). DNA methylation is represented as the average percentage of methylation of the 6 CpG sites analyzed by pyrosequencing.
Disruption of BRD4 in ESCs is associated with c-MYC expression
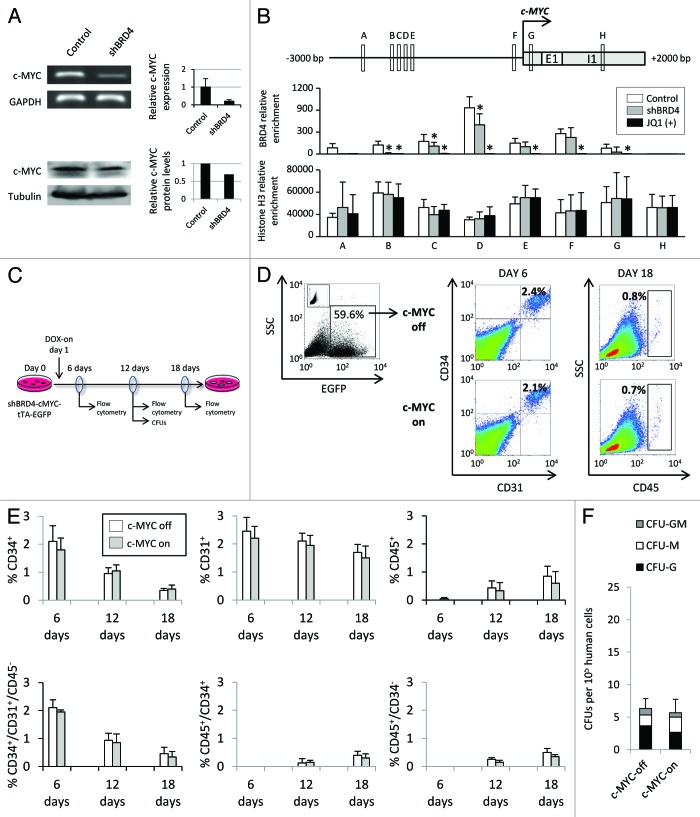
To investigate the molecular mechanism by which epigenetic regulation of BRD4 modulates hematopoiesis, we focused our attention on c-MYC, a known BRD4 target that has previously been shown to play an important role in hematoendothelial specification.26–28 The BRD4 mRNA and protein levels in shBRD4 cells were 21% and 69% that of normal cells, respectively (Fig. 6A). To establish a functional relationship between BRD4 and c-MYC in the ESCs, we used chromatin immunoprecipitation (ChIP) to assess c-MYC promoter occupancy by BRD4. We analyzed 8 DNA regions of around 200 bp (Fig. 6B) distributed between 2700 bp upstream (6) and 1100 bp downstream (2) of the c-MYC TSS. We found BRD4 enrichment in control vs. shBRD4 cells in the 5′ region of the TSS and also at 200 bp of the TSS in the 3′region (Fig. 6B). These results suggest that BRD4 directly binds c-MYC promoter region in ESCs and that the role of BRD4 in hematopoiesis could be mediated, at least in part, by c-MYC. In order to explore this possibility, we used a doxycycline-inducible c-MYC expression system packed in a lentiviral vector. Lentivirus particles generated with this vector efficiently infected ESCs (up to 60% of transduction efficiency) and overexpressed c-MYC in response to doxycycline (Fig. S5A and B). Additionally, upon doxycycline treatment, shBRD4-cMYC ESCs showed an improved proliferation rate, similar to that found in wild type cells (Fig. S5C), indicating that proliferation impairment was mostly mediated by c-MYC downregulation. To test the role of c-MYC in hematopoietic specification, ESCs were infected 3 times with 12 h intervals to achieve an infection efficiency of around 60%. Human c-MYC overexpression was induced 12 h after plating on OP9 stromal cells and was maintained for 18 d (Fig. 6C). In wild type cells, c-MYC overexpression did not impair or even slightly promoted the appearance of early CD34+/CD31+ hemoendothelial progenitors at different time points but impaired robustly the generation of CD45+ blood cells (Fig. S6). Furthermore, overexpression of c-MYC in shBRD4 cells neither improved the emergence of hematopoietic cells (Fig. 6D and E) nor CFU generation (Fig. 6F). Thus, c-MYC overexpression in shBRD4 ESCs did not rescue on its own the hemogenic/hematopoietic potential, indicating that BRD4 repression-mediated hematopoietic impairment from ESCs is not exclusively owing to c-MYC downregulation.
Figure 6. Disruption of BRD4 in ESCs reduced c-MYC expression. (A) RNA expression and western blot analysis of c-MYC in control and shBRD4 ESCs. mRNA levels were quantified by qRT-PCR and are represented as the mean value ± SD of three independent experiments in control and shBRD4 transfected cells. Relative c-MYC protein levels were quantified by densitometric analysis of western blot images. (B) Quantitative ChIP determination of BRD4 occupancy at the 5′ promoter region of c-MYC. ESCs treated with JQ1, a potent inhibitor of bromodomain binding to acetylated histones, were used as negative control. An intragenic region located at 1100 bp from the TSS was also analyzed. Relative enrichment is represented as the mean value ± SD of four independent experiments. (C) Schematic representation of the experimental design for the rescue experiment in shBRD4 ESCs infected with an inducible c-MYC lentiviral vector and end point analyses. (D) Representative dot plots of hematopoietic differentiation after c-MYC overexpression. (E) Percentage of CD34+, CD31+, and CD45+ cells at different time points during hematopoietic differentiation. (F) Colony forming unit (CFU) potential of shBRD4 ESCs transduced with an inducible c-MYC lentiviral vector, with or without doxycycline. Results are represented as the mean value ± SD of three independent experiments.
Discussion
In this work, our aim was to study the possible role of the BET family member BRD4 in ESC differentiation. Although BET proteins are essential for many biological processes and tissue homeostasis their contribution to early embryonic development remains elusive. Recent work has identified a diverse range of in vivo target genes and specific cell functions that suggest that BET proteins may act as key regulators of cell cycle and differentiation. For instance, it has recently been reported that BRD3 associates with acetylated GATA1 to promote erythroid differentiation.29 Additionally, BRDT, BRD2, and BRD4 knockout mice have been generated and a possible developmental role for these BET proteins has been suggested in each case. Indeed, BRDT null mice exhibit defects in male germ cell differentiation30,31 while BRD2 knockout mice show neural tube abnormalities linked to neuroepithelial differentiation.32 Furthermore, complete elimination of BRD4 in mice leads to early death after implantation. Nonetheless, mice carrying one single functional copy of BRD4 show developmental abnormalities which are initially associated with a lower proliferative potential in vivo,7 although defects associated with deregulation of developmental genes cannot be ruled out. The role of BRD4 on cell differentiation is further supported by the role of BRD4-NUT fusion protein in NUT-midline carcinomas, which inhibits the association of wild type BRD4 to the c-fos promoter blocking epithelial differentiation.17 Our results extend these findings by showing that BRD4 is involved in the differentiation of ESCs. First, we observed that BRD4 undergoes demethylation and its expression is upregulated upon differentiation, a type of epigenetic control often observed in key developmental genes.18,19,33 Additionally, this BRD4 downregulation in ESCs interferes with cell cycle progression and mildly reduces the proliferation rate, in line with the previously described role of BRD4 in cell cycle progression.6–8 In these studies, BRD4 disruption strongly impaired cell proliferation. Nonetheless, it has been observed that treatment with JQ1, a potent inhibitor of BRD4 binding to acetylated histones, has a strong antiproliferative effect on leukemia cells but only mildly affects non-cancerous cells.12 This result suggests that some cell types are more vulnerable to BRD4 disruption than others and may explain why BRD4 reduction in ES cells is not so critical for cell proliferation. In any case, this was not enough to induce spontaneous in vitro ESC differentiation, suggesting that BRD4 is not an essential determinant of pluripotency. Thus, shBRD4 ESCs, which display BRD4 expression levels around 20% of that of wild type ESCs retain the undifferentiated state during in vitro culture, consistent with the reported viability of mice with only one defective copy of the BRD4 gene.7 We also demonstrated that mesodermal differentiation is affected by BRD4 alteration during differentiation of ESCs. The emergence of hematoendothelial progenitor cells (CD34+/CD31+/CD45-), primitive blood cells (CD45+/CD34+), mature blood cells (CD45+/CD34-) and CFU potential were greatly diminished by BRD4 reduction during co-culture with OP9 cells. Interestingly, super-BRD4 cells showed a similar hemogenic potential to control cells. Additionally, we did not observe a detrimental effect on neural differentiation, although a possible role in this process cannot be completely excluded. A putative role for BRD4 in hematopoiesis is further suggested by the differential expression and methylation status in the somatic hematopoietic cells. In CD34+ HSPCs we found low BRD4 expression and high levels of methylation in the promoter region compared with myeloid and lymphoid cell fractions in which BRD4 expression was upregulated and its promoter severely demethylated. Gene demethylation is often observed during lineage specification of HSPCs34–36 indicating that BRD4 promoter demethylation and subsequent gene overexpression could be a hallmark in blood cell specification from ESCs.
We also explored the molecular mechanism underlying the hematopoietic impairment after BRD4 disruption in ESCs. Recent studies have demonstrated that BRD4 is a key regulator of c-MYC expression through its direct association to the promoter region.11,12,26 c-MYC deregulation provides a suitable mechanism to explain hematopoietic impairment in shBRD4 cells since the implication of c-MYC in hematopoietic development and leukemogenesis has been clearly established.37 c-MYC is expressed in almost all proliferating cells and represses cell differentiation in many cell types. Interestingly, c-Myc null mice die in the second week of embryonic development and exhibit vascular defects and impaired hematopoiesis.38 Also, a conditional knockout in which c-Myc has been targeted in hematopoietic precursors, exhibits severe cytopenia due to impaired HSPC differentiation.39 Our ChIP assays indicate that BRD4 might regulate c-Myc expression in ESCs, suggesting therefore that BRD4-dependent c-MYC induction may be required for hematoendothelial specification. In wild type cells, c-MYC overexpression did not impair the emergence of early hematoendothelial specification (CD34+/CD31+/CD45- progenitors) but did reduce terminal CD45+ blood differentiation. On the other hand, shBRD4 ESCs showed no enhanced hemogenic/hematopoietic differentiation, suggesting that BRD4 may also target alternative critical genes, other than c-MYC, involved in hematopoiesis. Additionally, these results highlight that the anti-tumorigenic effect of BET inhibitors in myeloid leukemia11,12 may not be entirely mediated by c-MYC downregulation, and that BRD4 impairment could be also influencing hematopoietic differentiation in cancer cells. In summary, our results show that epigenetic regulation of BRD4 modulates hematopoietic specification of ESCs, extending previous evidence for the developmental role of the BET protein family members, and will help to understand the putative role of BRD4 in cell differentiation and hematological malignancies.
Materials and Methods
Cell culture
The human embryonic cell line H9 was obtained from WiCell following approval from the Spanish National Embryo Ethical Committee. DNA and RNA samples from the cell line MSUH-002 were kindly donated by Professor JB Cibelli at the Michigan State University. IMR90, MS-5, and OP9 stromal cells were obtained from the American Type Culture Collection (ATCC). IMR90 and MS-5 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco) supplemented with 10% fetal calf serum (FCS) (HyClone). OP9 cells were cultured in α-MEM (Gibco) supplemented with 20% FCS. Human ES cells were maintained in matrigel using mesenchymal stromal cell-conditioned medium supplemented with 8 ng/ml basic fibroblast growth factor (bFGF) (Sigma) as previously described.40 Media were changed daily.
Stem cell differentiation
For embryoid body formation, cells were treated with collagenase IV (Gibco) and scraped off of the Matrigel-coated plates (BD Biosciences). For spontaneous differentiation, cell aggregates were cultured in low-attachment plates (Sterilin) in differentiation medium consisting in KO-Dulbecco’s modified Eagle’s medium supplemented with 20% KO serum replacement (Gibco), nonessential amino acids (Gibco), glutamine (Sigma), and 0.1 mM β-mercaptoethanol (Sigma), the medium being changed every 2 d. After 20 d, EBs were plated on gelatin-coated plates and cultured in DMEM supplemented with 20% FCS for an additional 10 d, passaging with TrypLE express (Gibco).
Hematopoietic differentiation was performed as previously described.21,41–43 Briefly, undifferentiated hESCs were harvested with collagenase IV (Gibco) and plated at low density onto OP9 confluent plates in α-MEM (Gibco) supplemented with 10% FCS and 0.1 mM β-mercaptoethanol (Sigma). The hESC/OP9 co-cultures were maintained for 18 d with half the medium being changed every 2 d. Cells were harvested at 6, 12, and 18 d after 1 h incubation with collagenase IV (Gibco) followed by 30 min incubation with TrypLE express (Gibco). Each time, after gentle pipetting, cells were filtered through a 100-µm cell strainer to achieve a single cell suspension and used for flow-cytometric and CFU assays. Dissociated cells were stained with anti-CD34-phycoerythrin, anti-CD31-allophycocyanin and anti-CD45-fluorescein (all from Biolegend). Hematoendothelial progenitors were identified as CD34+/CD31+/CD45-, immature blood cells as CD34+/CD45+ and mature blood cells as CD45+/CD34-. Data acquisition and analysis was performed using a BD FACScaliburTM Cytometer/CellQuest Pro Software package (Becton Dickinson). The experiments were replicated three times and significant differences were assessed by a two-tailed independent samples t test.
For early neural differentiation, hEBs were grown for 4 d in KO-DMEM supplemented with 20% KO serum replacement (Gibco), nonessential amino acids (Gibco), glutamine (Sigma), 0.1 mM β-mercaptoethanol (Sigma), and 4 ng/ml bFGF (Sigma). They were then grown in suspension for an additional 10 d in DMEM/F12 (Gibco) supplemented with nonessential amino acids (Gibco), 2 µg/ml heparin (Sigma), 20 ng/ml bFGF (Sigma), and neural cell supplement N2 (Gibco). In parallel, neural differentiation was also induced by hESC/MS-5 co-culture as previously described.44 Briefly, undifferentiated hESCs were plated at low density on confluent MS-5 plates to avoid contact between colonies during the differentiation process. These hESC/MS-5 co-cultures were maintained in DMEM/F12 supplemented with 15% KO serum replacement (Gibco), nonessential amino acids (Gibco), glutamine (Sigma), and 0.1 mM β-mercaptoethanol. After 14 d the medium was changed to DMEM/F12 (Gibco) supplemented with nonessential amino acids (Gibco), 2 µg/ml heparin (Sigma), 20 ng/ml bFGF (Sigma), and neural cell supplement N2 (Gibco) for an additional week. The experiments were replicated five times and neural emergence was quantified by morphological criteria and mRNA expression. Significant differences were assessed by a two-tailed independent samples t test.
CB collection and CD34+ HSPC isolation
Fresh umbilical cord blood (CB) units from healthy newborns were obtained from The Andalusian Public Cord Blood Bank upon approval by our local Ethics and Biozahard Board Committee. CB samples were pooled to reduce variability among individual CB units. Mononuclear cells were isolated using Ficoll-Hypaque. After lysing the red cells (StemCell Technologies), CD34+ cells were purified by magnetic bead separation using the human CD34 MicroBead kit and the AutoMACS Pro separator (Miltenyi) following the manufacturer’s instructions. Purity of the CD34+ fraction was assessed by flow cytometry using anti-CD34-PE (Becton Dickinson -BD-) and only CD34+ fractions showing purity over 95% were used.45,46 Lymphoid and myeloid fractions were isolated using Histopaque-1077 (Sigma) following the manufacturer’s instructions, and cell purity was checked with the ADVIA 2120i Hematology System (Siemens) to ensure a minimal purity of 95% prior to analysis.
RNA purification and quantitative real-time RT-PCR analysis
RNA was isolated with TRIzol Reagent (Invitrogen) and reverse transcription was performed using Superscript II (Invitrogen) following the manufacturer’s protocols. Quantitative real-time RT PCR (qPCR) was performed by TaqMan Gene Expression Assays (Applied Biosystems) and SYBR green universal PCR master mix (Applied Biosystems). Primers are described in Table S1. Significant differences in gene expression were assessed by a two-tailed independent samples t test.
Cell proliferation assay
Cells (2 × 104) were seeded onto 96-well plates, allowed to attach, and 50 µg of 3-(4,5-dimethyl-2-yl)-2,5-ditetrazolium bromide (MTT) were added to medium (100 µl/well; 3 h, 37 °C, 5% CO2). Cell medium was removed and MTT-formazan crystals dissolved in DMSO (100 µl/well). Absorbance at 595 nm was determined with a Power Wave WS automated microtiter plate reader (BioTek). Optical density was directly proportional to cell number up to the density attained by the end of the assay. Results are expressed as the mean ± SD (n = 3). Significant differences were assessed by a two-tailed independent samples t test.
Cell treatment with 5-aza-2′-deoxycytidine
The human ES cell line H9 was treated with 2 μmol/L 5-aza-2′-deoxycytidine (AZA; A3656, Sigma) for 2 d to achieve demethylation and then mechanically harvested for RNA purification. Significant differences in gene expression were assessed by a two-tailed independent samples t test.
Genomic DNA extraction and DNA methylation assay
Genomic DNA was extracted with a Wizard Genomic DNA Purification Kit (Promega) following the manufacturer’s instructions. Sodium bisulfite modification of 500 ng DNA was performed with the EZ DNA methylation kit (D5002, Zymo Research) following the protocol recommended by the manufacturer.
Pyrosequencing was performed using the PyroMark kit (Qiagen). Primers used were: F1 (5′-GTGAAGGAGG ATTAAGGTTT TTAAG-3′), R1 biotinylated (5′-CACAAATAAA ATTACTTTTC CATCTAA-3′), and the sequencing primer S1 (5′-GTGAAGGAGG ATTAAGG-3′). These primers amplify a region 607 bp upstream of the BRD4 transcription initiation site, containing six CpG sites. The biotinylated PCR product was purified with the Pyrosequencing Vacuum Prep Tool (Qiagen). Methylation levels were quantified by using the PyroMark Q24 system (Biotage).
Bisulfite genomic sequencing of multiple clones was performed as previously described33 using a minimum of six independent clones per sample. Primers used were: Forward (5′-TGGGGGGTAG GGTGTATATA TA -3′) and Reverse (5′-ATCAAAAAAT CTATTCCCCT CC -3′), which amplify a 267 bp region in the BRD4 promoter containing 10 CpG sites and located 776 bp from the TSS.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde, permeabilized with 1% Triton X-100 in Dulbecco's phosphate-buffered saline (DPBS) (10 min, room temperature), and blocked with 10% donkey serum (2 h, room temperature). Then, the cells were incubated with anti-BRD4 antibody (Bethyl Laboratories) overnight at 4 °C. Cells were washed in DPBS containing 0.1% Triton X-100 (Sigma) and incubated with Alexa-594-conjugated secondary antibody (Life Technologies). DNA was stained with DAPI (4’, 6-diamidino-2-phenylidole) and immunofluorescence preparations imaged using a digital camera connected to a Leika DMRXA microscope.
Cell cycle analysis
Human ESCs were harvested after 10 min incubation with TrypLE express (Gibco) and filtered through a 100-µm cell strainer to achieve a single cell suspension. After washing with DPBS, cells were fixed in 70% ethanol. Fixed cells were stained with propidium iodide (40 µg/ml) and treated with RNase A (Roche Applied Science) for 1 h at room temperature. Analysis was performed with ModFit LT software.
Annexin v staining
Apoptosis was analyzed with the FITC-Annexin V staining kit (Biolegend) following the manufacturer’s instructions. Briefly, cells were washed with DPBS and transferred to the cell staining buffer containing FITC- annexin V and 7-Amino-actinomycin D (7-AAD). Stained cells were analyzed by flow cytometry. Significant differences were assessed by a two-tailed independent samples t test.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation was performed with control H9 and shBRD4 H9 cells. Control H9 cells were incubated with the BRD4 inhibitor JQ1 (500 nM) for 24 h and used as the negative control in the immunoprecipitation protocol. In brief, cells were fixed with 1% formaldehyde (Sigma-Aldrich) for 30 min at 4 °C, lysed in sodium dodecyl sulfate (SDS) lysis buffer [1% SDS, 10 mM EDTA (EDTA), and 50 mM Tris–HCl pH 8.1], and sonicated. Chromatin immunoprecipitation was performed overnight at 4 °C with anti-H3 (Millipore) and anti-BRD4 (Bethyl Laboratories) using 5x106 cells per reaction. Normal IgG (Abcam) was used as the negative control. Antibody–chromatin complexes were precipitated with Salmon Sperm DNA/Protein A-Agarose beads (Upstate Biotechnologies), then washed and eluted from beads using an elution buffer (1% SDS, 0.1 M NaHCO3). DNA was extracted with phenol–chloroform and was ethanol-precipitated. Immunoprecipitated DNA was analyzed in triplicate by real-time PCR from 1 μl eluted DNA. The primers used are described in Table S2. Results are presented as x-fold enrichment of precipitated DNA associated with BRD4, relative to a 1:200 dilution of unbound fraction. Significant differences were assessed by a two-tailed independent samples t test.
Colony Forming Unit (CFU) quantitative assay
CFU quantitative assays were performed by plating 50 000 cells from 12 d-old hESC/OP9 co-cultures into semisolid methylcellulose medium supplemented with recombinant human Stem Cell Factor (SCF), Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF), Colony-Stimulating Factor 3 (CSF3), IL-3 and IL-6 and Erythropoietin (Methocult H4435, StemCell Technologies, Inc.). Colonies were counted 10 d later using standard morphological criteria. Significant differences were assessed by two-tailed independent samples t test.
Western blot
Cell lysates were prepared by SDS lysis extraction in the presence of a complete protease inhibitor cocktail (Roche). After SDS-PAGE separation, proteins were detected by western blot analysis using antibodies against α-tubulin (1:3000, Sigma-Aldrich), β-actin (1:3000, Sigma-Aldrich), BRD4 (1:2000, Bethyl Laboratories), and c-MYC (1:2000, Santa Cruz). Secondary antibodies conjugated to horseradish peroxidase (HRP) were from GE Healthcare Life Sciences. Western blot images were quantified with Multi Gauge Software (Fujifilm). Background subtraction was applied individually to each lane and values were normalized with the loading control.
Plasmid construction and transfection
In order to overexpress BRD4 in ESCs, the BRD4 coding sequence was subcloned into pCEP4-puro vector from the plasmid pCDNA-BRD4-zeocin (addgene plasmid 14441) by NotI/HindII digestion. BRD4 knockdown was achieved by transfection with the plasmid pSUPER-shBRD4 (CT), donated by Dr Peter M Howley. This vector was successfully used in previous studies and demonstrated to have an efficient capacity to downregulate BRD4 in human cells.47 Nucleofection was performed with Amaxa Nucleofector (Lonza) using 4 µg of each plasmid following the manufacturer’s instructions. Selection of transfected clones was performed with 0.5 µg/ml of puromycin (Invitrogen) or 37.5 µg/ml hygromycin B (Invitrogen) for pCEP4-BRD4 and pSUPER-shBRD4, respectively. The inducible c-MYC lentiviral vector (LV-TRE-cMyc-Ubc-tTA-I2G) was kindly gifted by Dr Tomoyuki Yamaguchi and Dr Koji Eto. Lentiviral supernatants were obtained from transfected 293T/17 cells (ATCC) and concentrated with Amicon Ultracel 100K filter units (Millipore). H9 cells were infected 3 times at 12 h intervals and c-MYC expression was induced with 0.25, 0.5, or 1 µg/ml doxycycline (Sigma-Aldrich) added to the culture medium.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work has been financially supported by the FIS/FEDER (PI12/01080 to M.F.F., PI11/01728 to AF.F. and PI12/02587 to C.L.L.), the ISCIII (CP11/00131 to A.F.F.), the IUOPA (to R.M.R.), the FICYT (to E.G.T.), the Red de Investigación Renal (REDinREN) (to C.L.L.), and the Ramon Areces Foundation (to M.F.F.). The IUOPA is supported by the Obra Social Cajastur, Spain.
References
- 1.Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol Cell Biol. 2000;20:6537–49. doi: 10.1128/MCB.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang CM. Brd4 engagement from chromatin targeting to transcriptional regulation: selective contact with acetylated histone H3 and H4. F1000 Biol Rep. 2009;1:98. doi: 10.3410/B1-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–34. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 4.Dey A, Nishiyama A, Karpova T, McNally J, Ozato K. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol Biol Cell. 2009;20:4899–909. doi: 10.1091/mbc.E09-05-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–45. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maruyama T, Farina A, Dey A, Cheong J, Bermudez VP, Tamura T, Sciortino S, Shuman J, Hurwitz J, Ozato K. A Mammalian bromodomain protein, brd4, interacts with replication factor C and inhibits progression to S phase. Mol Cell Biol. 2002;22:6509–20. doi: 10.1128/MCB.22.18.6509-6520.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol Cell Biol. 2002;22:3794–802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You J, Li Q, Wu C, Kim J, Ottinger M, Howley PM. Regulation of aurora B expression by the bromodomain protein Brd4. Mol Cell Biol. 2009;29:5094–103. doi: 10.1128/MCB.00299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French CA, Miyoshi I, Kubonishi I, Grier HE, Perez-Atayde AR, Fletcher JA. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 2003;63:304–7. [PubMed] [Google Scholar]
- 10.French CA, Kutok JL, Faquin WC, Toretsky JA, Antonescu CR, Griffin CA, Nose V, Vargas SO, Moschovi M, Tzortzatou-Stathopoulou F, et al. Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol. 2004;22:4135–9. doi: 10.1200/JCO.2004.02.107. [DOI] [PubMed] [Google Scholar]
- 11.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA, Bergeron L, Sims RJ., 3rd Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci U S A. 2011;108:16669–74. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crawford NP, Alsarraj J, Lukes L, Walker RC, Officewala JS, Yang HH, Lee MP, Ozato K, Hunter KW. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008;105:6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez RM, Huidobro C, Urdinguio RG, Mangas C, Soldevilla B, Domínguez G, Bonilla F, Fernandez AF, Fraga MF. Aberrant epigenetic regulation of bromodomain BRD4 in human colon cancer. J Mol Med (Berl) 2012;90:587–95. doi: 10.1007/s00109-011-0837-0. [DOI] [PubMed] [Google Scholar]
- 15.Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Gonzalez A, Gazdar AF, Minna JD, Carbone DP, Dang TP. Cloned fusion product from a rare t(15;19)(q13.2;p13.1) inhibit S phase in vitro. J Med Genet. 2005;42:558–64. doi: 10.1136/jmg.2004.029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.French CA, Ramirez CL, Kolmakova J, Hickman TT, Cameron MJ, Thyne ME, Kutok JL, Toretsky JA, Tadavarthy AK, Kees UR, et al. BRD-NUT oncoproteins: a family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene. 2008;27:2237–42. doi: 10.1038/sj.onc.1210852. [DOI] [PubMed] [Google Scholar]
- 17.Yan J, Diaz J, Jiao J, Wang R, You J. Perturbation of BRD4 protein function by BRD4-NUT protein abrogates cellular differentiation in NUT midline carcinoma. J Biol Chem. 2011;286:27663–75. doi: 10.1074/jbc.M111.246975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borgel J, Guibert S, Li Y, Chiba H, Schübeler D, Sasaki H, Forné T, Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 20.Saito S, Aoki H, Ito A, Ueno S, Wada T, Mitsuzuka K, Satoh M, Arai Y, Miyagi T. Human alpha2,3-sialyltransferase (ST3Gal II) is a stage-specific embryonic antigen-4 synthase. J Biol Chem. 2003;278:26474–9. doi: 10.1074/jbc.M213223200. [DOI] [PubMed] [Google Scholar]
- 21.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–26. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Li L, Shojaei F, Levac K, Cerdan C, Menendez P, Martin T, Rouleau A, Bhatia M. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21:31–41. doi: 10.1016/j.immuni.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Ramos-Mejia V, Melen GJ, Sanchez L, Gutierrez-Aranda I, Ligero G, Cortes JL, Real PJ, Bueno C, Menendez P. Nodal/Activin signaling predicts human pluripotent stem cell lines prone to differentiate toward the hematopoietic lineage. Mol Ther. 2010;18:2173–81. doi: 10.1038/mt.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bueno C, Montes R, Melen GJ, Ramos-Mejia V, Real PJ, Ayllón V, Sanchez L, Ligero G, Gutierrez-Aranda I, Fernández AF, et al. A human ESC model for MLL-AF4 leukemic fusion gene reveals an impaired early hematopoietic-endothelial specification. Cell Res. 2012;22:986–1002. doi: 10.1038/cr.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bueno C, Ayllón V, Montes R, Navarro-Montero O, Ramos-Mejia V, Real PJ, Romero-Moya D, Araúzo-Bravo MJ, Menendez P. FLT3 activation cooperates with MLL-AF4 fusion protein to abrogate the hematopoietic specification of human ESCs. Blood. 2013; 121:3867-78, S1-3 [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–76. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois NC, Adolphe C, Ehninger A, Wang RA, Robertson EJ, Trumpp A. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development. 2008;135:2455–65. doi: 10.1242/dev.022707. [DOI] [PubMed] [Google Scholar]
- 28.Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, Frazier J, Chau BN, Loboda A, Linsley PS, et al. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci U S A. 2012;109:9545–50. doi: 10.1073/pnas.1121119109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamonica JM, Deng W, Kadauke S, Campbell AE, Gamsjaeger R, Wang H, Cheng Y, Billin AN, Hardison RC, Mackay JP, et al. Bromodomain protein Brd3 associates with acetylated GATA1 to promote its chromatin occupancy at erythroid target genes. Proc Natl Acad Sci U S A. 2011;108:E159–68. doi: 10.1073/pnas.1102140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang E, Nickerson HD, Wen D, Wang X, Wolgemuth DJ. The first bromodomain of Brdt, a testis-specific member of the BET sub-family of double-bromodomain-containing proteins, is essential for male germ cell differentiation. Development. 2007;134:3507–15. doi: 10.1242/dev.004481. [DOI] [PubMed] [Google Scholar]
- 31.Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, Hery P, Jounier S, Depaux A, Vitte AL, et al. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012;31:3809–20. doi: 10.1038/emboj.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsume M, Kimura-Yoshida C, Mochida K, Shibukawa Y, Amazaki S, Wada Y, Hiramatsu R, Shimokawa K, Matsuo I. Brd2 is required for cell cycle exit and neuronal differentiation through the E2F1 pathway in mouse neuroepithelial cells. Biochem Biophys Res Commun. 2012;425:762–8. doi: 10.1016/j.bbrc.2012.07.149. [DOI] [PubMed] [Google Scholar]
- 33.Calvanese V, Horrillo A, Hmadcha A, Suarez-Alvarez B, Fernandez AF, Lara E, Casado S, Menendez P, Bueno C, Garcia-Castro J, et al. Cancer genes hypermethylated in human embryonic stem cells. PLoS One. 2008;3:e3294. doi: 10.1371/journal.pone.0003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bocker MT, Hellwig I, Breiling A, Eckstein V, Ho AD, Lyko F. Genome-wide promoter DNA methylation dynamics of human hematopoietic progenitor cells during differentiation and aging. Blood. 2011;117:e182–9. doi: 10.1182/blood-2011-01-331926. [DOI] [PubMed] [Google Scholar]
- 36.Calvanese V, Fernández AF, Urdinguio RG, Suárez-Alvarez B, Mangas C, Pérez-García V, Bueno C, Montes R, Ramos-Mejía V, Martínez-Camblor P, et al. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic Acids Res. 2012;40:116–31. doi: 10.1093/nar/gkr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delgado MD, León J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–16. doi: 10.1177/1947601910377495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He C, Hu H, Braren R, Fong SY, Trumpp A, Carlson TR, Wang RA. c-myc in the hematopoietic lineage is crucial for its angiogenic function in the mouse embryo. Development. 2008;135:2467–77. doi: 10.1242/dev.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–63. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montes R, Ligero G, Sanchez L, Catalina P, de la Cueva T, Nieto A, Melen GJ, Rubio R, García-Castro J, Bueno C, et al. Feeder-free maintenance of hESCs in mesenchymal stem cell-conditioned media: distinct requirements for TGF-beta and IGF-II. Cell Res. 2009;19:698–709. doi: 10.1038/cr.2009.35. [DOI] [PubMed] [Google Scholar]
- 41.Choi KD, Vodyanik M, Slukvin II. Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nat Protoc. 2011;6:296–313. doi: 10.1038/nprot.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Real PJ, Ligero G, Ayllon V, Ramos-Mejia V, Bueno C, Gutierrez-Aranda I, Navarro-Montero O, Lako M, Menendez P. SCL/TAL1 regulates hematopoietic specification from human embryonic stem cells. Mol Ther. 2012;20:1443–53. doi: 10.1038/mt.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji J, Vijayaragavan K, Bosse M, Menendez P, Weisel K, Bhatia M. OP9 stroma augments survival of hematopoietic precursors and progenitors during hematopoietic differentiation from human embryonic stem cells. Stem Cells. 2008;26:2485–95. doi: 10.1634/stemcells.2008-0642. [DOI] [PubMed] [Google Scholar]
- 44.Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, Harrison NL, Studer L. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2004;101:12543–8. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bueno C, Montes R, Martín L, Prat I, Hernandez MC, Orfao A, Menendez P. NG2 antigen is expressed in CD34+ HPCs and plasmacytoid dendritic cell precursors: is NG2 expression in leukemia dependent on the target cell where leukemogenesis is triggered? Leukemia. 2008;22:1475–8. doi: 10.1038/leu.2008.134. [DOI] [PubMed] [Google Scholar]
- 46.Bueno C, Catalina P, Melen GJ, Montes R, Sánchez L, Ligero G, García-Pérez JL, Menendez P. Etoposide induces MLL rearrangements and other chromosomal abnormalities in human embryonic stem cells. Carcinogenesis. 2009;30:1628–37. doi: 10.1093/carcin/bgp169. [DOI] [PubMed] [Google Scholar]
- 47.Schweiger MR, You J, Howley PM. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J Virol. 2006;80:4276–85. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.