Abstract
Klinefelter syndrome (KS) is the most common sex-chromosome aneuploidy in humans. Most affected individuals carry one extra X-chromosome (47,XXY karyotype) and the condition presents with a heterogeneous mix of reproductive, physical and psychiatric phenotypes. Although the mechanism(s) by which the supernumerary X-chromosome determines these features of KS are poorly understood, skewed X-chromosome inactivation (XCI), gene-dosage dysregulation, and the parental origin of the extra X-chromosome have all been implicated, suggesting an important role for epigenetic processes. We assessed genomic, methylomic and transcriptomic variation in matched prefrontal cortex and cerebellum samples identifying an individual with a 47,XXY karyotype who was comorbid for schizophrenia and had a notably reduced cerebellum mass compared with other individuals in the study (n = 49). We examined methylomic and transcriptomic differences in this individual relative to female and male samples with 46,XX or 46,XY karyotypes, respectively, and identified numerous locus-specific differences in DNA methylation and gene expression, with many differences being autosomal and tissue-specific. Furthermore, global DNA methylation, assessed via the interrogation of LINE-1 and Alu repetitive elements, was significantly altered in the 47,XXY patient in a tissue-specific manner with extreme hypomethylation detected in the prefrontal cortex and extreme hypermethylation in the cerebellum. This study provides the first detailed molecular characterization of the prefrontal cortex and cerebellum from an individual with a 47,XXY karyotype, identifying widespread tissue-specific epigenomic and transcriptomic alterations in the brain.
Keywords: Klinefelter syndrome; DNA methylation; gene expression; 47,XXY; prefrontal cortex; cerebellum
Introduction
Klinefelter syndrome (KS) is the most common sex-chromosome aneuploidy in humans, affecting approximately 1 in every 600 newborn males.1 Most individuals with KS carry one extra X-chromosome (karyotype 47,XXY), although other reported variants include 48,XXXY, 48,XXYY and 49,XXXXY.2 The condition presents with a broad range of phenotypes that often vary in severity.2 In addition to the well-characterized physical and physiological features—tall stature, gynecomastia, hypogonadism, and absent spermatogenesis3—KS is also often associated with psychiatric and neurodevelopmental phenotypes including language-based learning disabilities, decreased verbal intelligence and difficulties with task planning and inhibitory control.4 Of note, individuals with KS frequently exhibit symptoms related to schizophrenia including schizotypal traits,5 auditory hallucinations6 and verbal cognition impairment.7 Furthermore, several structural brain abnormalities are associated with the disorder including abnormal cerebral asymmetry8,9 and reductions in the size of specific brain regions,6,10,11 total brain volume,10,12,13 and white matter volume.9,10
The mechanism(s) by which the supernumerary X-chromosome determines the phenotypes evident in KS are poorly understood, although skewed X-chromosome inactivation (XCI), gene-dosage dysregulation, and the parental origin of the extra X-chromosome have all been implicated,14-16 suggesting that epigenetic processes play an important role. However, little is known about the specific epigenetic changes associated with KS, especially in tissues relevant to the KS phenotype. A study comparing global long interspersed nucleotide element-1 (LINE-1) DNA methylation in whole blood from individuals with Turner’s syndrome (45,XO), healthy males (46,XY), healthy females (46,XX) and KS patients (47,XXY) reported that increased chromosomal number was associated with hypomethylation across the genome.17 Studies of genome-wide patterns of DNA methylation in patients with trisomy 2118 and trisomy 819 also reveal large changes not limited to the supernumerary chromosome, indicating that widespread epigenetic changes may be a common feature of chromosomal aneuploidy. Moreover, differences in brain morphology have been associated with imprinting of the X-chromosome in Turner Syndrome, indicating that the parental origin of the X-chromosome may be important in mediating the psychiatric symptoms present in sex abnormalities.20
This study is the first to examine genome-wide patterns of DNA methylation and gene expression in two regions of the brain obtained post-mortem from a patient with a 47,XXY karyotype. We identify widespread tissue-specific epigenomic and transcriptomic alterations, providing potential clues about the molecular causes and consequences of KS.
Results and Discussion
As part of an integrated “-omics” study of schizophrenia (Pidsley et al., submitted), we examined genome-wide patterns of DNA methylation, gene expression, and genetic variation in post-mortem cerebellum and prefrontal cortex brain tissue samples from schizophrenia patients and controls. During the standard quality control steps of these data we identified a discrepancy between reported and measured sex for one schizophrenia patient who displayed the genomic characteristics expected of both male and female samples simultaneously. As expected, across the entire set of samples, males and females showed distinct levels of DNA methylation across probes on the X-chromosome, with the exception of one sample recorded as male, who clustered with the female samples (Fig. S1A). This individual also clustered with females for XIST gene expression (Fig. S2A) but with males when DNA methylation and gene expression across the Y-chromosome were assessed (Figs. S1B and S2B). A 47,XXY karyotype was confirmed via the high-resolution SNP genotyping array data (Figs. S3 and S4) and the presence of the Y-chromosome was confirmed via a PCR-based sex-typing assay (Fig. S5). A number of other genomic alterations were identified in this individual (Table S1), although there was no obvious excess burden of autosomal copy number variations (CNVs), except for one region with four copies of a region spanning the NKAIN2 locus. This is notable since copy number changes in this gene have previously been reported in neuropsychiatric phenotypes including neurodevelopmental disorders and schizophrenia.21-24 The patient’s autopsy report did not record KS, suggesting that the 47,XXY karyotype was undetected during the patient’s lifetime, although we did not have access to detailed pre-mortem medical records. In addition to schizophrenia, the autopsy report highlighted that the 47,XXY patient had a poor nutritional state, hepatomegaly, vascular spiders, nystagmus, dysdyadocokinesia, some degree of ataxia at the time of death, a known history of alcohol abuse and were prescribed the medications parentrocite and sulpiride.
Identifying structural brain abnormalities associated with a 47,XXY karyotype is of relevance given the established link between KS and several neuropsychiatric disorders including schizophrenia.5,25 Detailed records taken by the neuropathologist at autopsy show that although the 47,XXY patient had a similar total brain mass to other patients (47,XXY = 1417 g, all other samples = 1410 ± 182 g, other males = 1454 ± 201 g, females = 1325 ± 92 g) they had a markedly lower cerebellum mass (47,XXY = 111 g, all other samples = 170 ± 24 g, other males = 175 ± 27 g, females = 160 ± 15 g), equating to more than two standard deviations below the mean of the other samples (Fig. 1; Fig. S6). The reduced cerebellum mass is consistent with the patient’s autopsy report of movement disorders, and previous studies demonstrate an association between cerebellar ataxia and reduced cerebellar size.26 Reductions in cerebral volume and decreased cortical thickness in the left inferior frontal, temporal, and superior motor regions have been previously reported in KS,13 and a recent imaging study showed significant reductions in the volume of several brain regions, including the cerebellum, in KS patients compared with controls.10 Other studies have also described brain abnormalities in KS patients, including reductions in total brain volume,12 specific brain regions6,11 and white matter volume,9 as well as abnormal cerebral asymmetry.8,9 These latter findings are consistent with the theory that brain asymmetry and cerebral dominance in humans is determined by a XY homologous gene pair.27 Structural brain abnormalities have also been identified in patients with other types of X-chromosome aneuploidy28; a recent study reported increased cerebellum volume in Turner syndrome (45,XO) patients, contrasting with the decreased cerebellum volume in our patient (47,XXY).20 However, because reduced cerebellum mass has been associated with alcoholism29 it is possible that the reduction observed in the 47,XXY patient is related to the their alcohol abuse history.

Figure 1. Reduced cerebellar mass in a 47,XXY patient comorbid for schizophrenia. Shown is the average cerebellar mass (in grams) across all samples compared with the cerebellar mass of the 47,XXY patient. Shown are the minimum, maximum, median and interquartile range of the cerebellar mass.
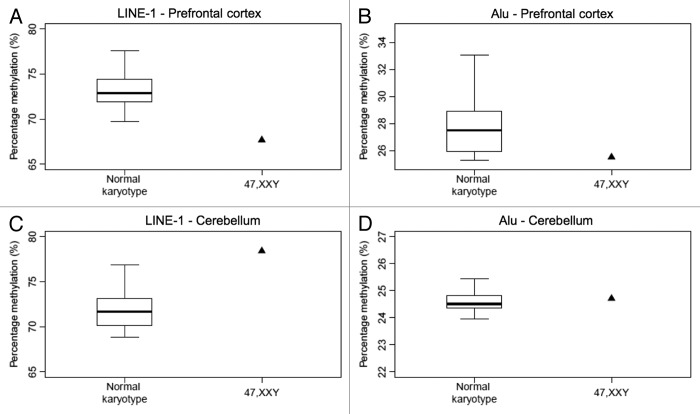
Global DNA methylation levels were estimated in both brain regions across all individuals using bisulfite-pyrosequencing assays targeting LINE-1 and Alu repeat elements across the genome, as described previously.30,31 Relative to other samples, the 47,XXY patient was a striking hypomethylated outlier across both LINE-1 (47,XXY = 67.7%, other samples = 73.0 ± 2.3%) and Alu (47,XXY = 25.5%, other samples = 28.1 ± 2.6%) repetitive elements in the prefrontal cortex (Fig. 2A and B; Fig. S7A and B). Conversely in the cerebellum, the 47,XXY individual showed notable hypermethylation compared with the other samples at LINE-1 repetitive elements (47,XXY = 78.4%, other samples = 71.9 ± 2.1%), although no significant difference was observed at Alu repetitive elements (47,XXY = 24.7%, other samples = 24.8 ± 0.8%) (Fig. 2C and D; Fig. S7C and D).
Figure 2. Tissue-specific differences in global DNA methylation in a 47,XXY patient comorbid for schizophrenia. The 47,XXY patient is significantly hypomethylated in the prefrontal cortex across both LINE-1 (A) and Alu repetitive elements (B) but hypermethylated in the cerebellum at LINE-1 elements (C), compared with the other samples. No significant difference was observed for DNA methylation across Alu repeat elements in the cerebellum (D). Shown are the minimum, maximum, median and interquartile range of the DNA methylation values.
Using the Infinium HumanMethylation450 BeadChip (Illumina Inc.) we identified numerous autosomal regions showing consistent differential DNA methylation in the 47,XXY patient compared with other samples (Tables 1 and 2). Of note, a region within the sperm-associated antigen 1 (SPAG1) gene is significantly hypermethylated in both the prefrontal cortex and the cerebellum, an interesting observation in the context of the infertility associated with KS given the role of this gene in spermatogenesis, fertilization, and infertility.32 Other differentially methylated regions (DMRs) were found to be tissue-specific, and/or only detectable relative to either female or male controls. Of potential relevance to KS, for example, are cerebellar DMRs in the vicinity of Piwi-like protein 1 (PIWIL1), which plays a role in the self-renewal of germline stem cells.33 Also of interest, given the cerebellar abnormalities seen in this individual, is evidence for cerebellum-specific hypermethylation of the LIM/homeobox 4 (LHX4) gene; mutations in this gene have been associated with altered brain development and cerebellar structure.34 Given the comorbid diagnosis of schizophrenia in this patient, it is interesting that several of the prefrontal cortex DMRs are located in close proximity to other neurobiologically-relevant genes, including NOTCH4, EPHB3 and KCNN1, that have been previously implicated in schizophrenia,35 brain development36-38 and regulation of microglia and neurons during neuroinflammation.38 The large differences in DNA methylation reported here are specific to the XXY individual; none of these regions are significantly differentially methylated in our analysis of schizophrenia and matched controls (Pidlsey et al., submitted).
Table 1. 47,XXY-associated differently methylated regions in the prefrontal cortex.
| Gene | DMR position (hg19) | Probes in DMR | 47,XXY vs. all other | 47,XXY vs. other males | 47,XXY vs. females | |||
|---|---|---|---|---|---|---|---|---|
| β difference | P value | β difference | P value | β difference | P value | |||
| VWA1 | chr1:1374601–1374669 | 2 | -0.05 | 6.28E-05 | ||||
| chr1:182669244–182669315 | 2 | 0.13 | 3.14E-06 | |||||
| chr1:247681102–247681931 | 8 | 0.16 | 1.35E-06 | |||||
| OR2L13 | chr1:248100585–248100614 | 4 | 0.3 | 7.88E-06 | ||||
| chr2:4931004–4931074 | 2 | -0.23 | 2.07E-08 | -0.23 | 3.66E-08 | -0.23 | 3.93E-09 | |
| MAP4K3 | chr2:39665105–39665186 | 2 | -0.01 | 4.03E-05 | ||||
| PCBP1 | chr2:70313772–70313833 | 2 | 0.01 | 2.57E-05 | ||||
| LOC388965 | chr2:84517321–84517950 | 9 | -0.03 | 3.14E-05 | ||||
| CXCR1 | chr2:219031640–219031719 | 2 | -0.05 | 2.61E-05 | ||||
| IHH | chr2:219922729–219922998 | 2 | 0.02 | 6.33E-07 | ||||
| chr2:240530497–240530569 | 2 | -0.16 | 2.46E-05 | |||||
| WNT5A | chr3:55515168–55515541 | 3 | 0.03 | 2.18E-05 | ||||
| NEK11 | chr3:130745442–130745959 | 13 | 0.04 | 7.82E-05 | 0.04 | < 2.00E-16 | ||
| DVL3 | chr3:183887905–183888477 | 4 | -0.05 | 4.49E-05 | ||||
| EPHB3 | chr3:184297380–184297522 | 3 | -0.07 | 1.33E-05 | ||||
| chr3:195578011–195578280 | 6 | 0.22 | 2.18E-05 | |||||
| chr4:53588360–53588850 | 6 | 0.03 | 1.68E-06 | |||||
| SNHG8 | chr4:119199621–119200372 | 11 | 0.04 | 2.30E-07 | 0.04 | 3.82E-11 | 0.04 | 2.16E-06 |
| chr5:784832–784915 | 3 | 0.5 | < 2.00E-16 | 0.5 | < 2.00E-16 | 0.5 | < 2.00E-16 | |
| RIPK1 | chr6:3077011–3077041 | 2 | -0.16 | 9.23E-05 | ||||
| NOTCH4 | chr6:32179862–32179971 | 2 | -0.02 | 1.43E-05 | ||||
| TAPBP | chr6:33269769–33269832 | 2 | -0.2 | 1.29E-08 | ||||
| chr6:168120556–168120635 | 2 | -0.06 | 3.07E-06 | -0.05 | 1.94E-06 | |||
| MAD1L1 | chr7:2144559–2144767 | 3 | -0.09 | 8.00E-07 | ||||
| chr7:157075207–157075303 | 2 | -0.1 | 2.74E-05 | |||||
| PTPRN2 | chr7:157744316–157744347 | 2 | -0.06 | < 2.00E-16 | -0.06 | < 2.00E-16 | -0.06 | < 2.00E-16 |
| chr8:58055876–58056026 | 2 | 0.39 | 1.20E-05 | 0.4 | 2.33E-06 | 0.38 | 3.12E-05 | |
| C8orf71 | chr8:58192753–58192883 | 2 | -0.22 | 9.11E-05 | ||||
| SPAG1 | chr8:101224915–101225361 | 5 | 0.13 | < 2.00E-16 | 0.13 | < 2.00E-16 | 0.14 | < 2.00E-16 |
| SPAG1 | chr8:101225800–101225902 | 2 | 0.22 | < 2.00E-16 | 0.22 | 6.54E-12 | 0.23 | < 2.00E-16 |
| chr10:2978126–2978687 | 5 | -0.1 | 7.97E-09 | |||||
| chr10:8090846–8090924 | 2 | 0.04 | 7.87E-05 | |||||
| STK32C | chr10:134062522–134062614 | 2 | -0.21 | 1.95E-05 | ||||
| TMBIM4 | chr12:66563381–66563928 | 10 | 0.03 | 3.12E-06 | 0.03 | 2.10E-07 | 0.03 | 4.13E-08 |
| GALNT9 | chr12:132904540–132904689 | 2 | -0.11 | 3.24E-06 | ||||
| NDRG2 | chr14:21493406–21493410 | 2 | 0.03 | 2.15E-06 | ||||
| chr14:95837801–95837929 | 3 | -0.05 | 1.77E-06 | -0.06 | 2.54E-06 | -0.05 | 3.52E-07 | |
| LBXCOR1 | chr15:68125566–68125599 | 2 | -0.2 | 7.84E-06 | ||||
| chr15:68126065–68126178 | 2 | -0.04 | 8.16E-05 | |||||
| chr19:17504632–17504972 | 3 | 0.15 | 2.61E-07 | |||||
| KCNN1 | chr19:18077727–18077834 | 3 | 0.02 | 4.85E-08 | ||||
| AXL | chr19:41731934–41732589 | 5 | 0.03 | 3.79E-06 | ||||
| KDELR1 | chr19:48894694–48895030 | 9 | 0.03 | 4.30E-05 | 0.03 | 3.79E-11 | ||
| chr21:44573854–44574022 | 3 | -0.09 | 1.02E-07 | -0.09 | 9.20E-08 | -0.08 | 9.79E-10 | |
Light gray boxes indicate non-significant results. Dark gray boxes indicate that no comparison was made (X-chromosome-linked genes were compared with females only whereas Y-chromosome-linked genes were compared with males only).
Table 2. 47,XXY-associated differently methylated regions in the cerebellum.
| Gene | Position (hg19) | Probes in DMR | 47,XXY vs. all other | 47,XXY vs. males | 47,XXY vs. females | |||
|---|---|---|---|---|---|---|---|---|
| β difference | P value | β difference | P value | β difference | P value | |||
| LHX4 | chr1:180201891–180201893 | 2 | 0.26 | 5.42E-05 | ||||
| LHX4 | chr1:180202256–180202784 | 2 | 0.09 | 2.92E-09 | ||||
| LHX4 | chr1:180203135–180203143 | 8 | 0.05 | 1.49E-06 | ||||
| LAMB3 | chr1:209799191–209799353 | 4 | -0.14 | 7.94E-05 | -0.14 | 3.66E-07 | ||
| TTC15 | chr2:3469154–3469529 | 2 | -0.19 | 2.47E-07 | ||||
| TET3 | chr2:74328923–74329082 | 2 | -0.07 | 5.17E-05 | ||||
| chr3:87137933–87138700 | 2 | 0.11 | < 2.00E-16 | 0.11 | < 2.00E-16 | 0.1 | 2.66E-12 | |
| CLDN18 | chr3:137728810–137729296 | 9 | -0.13 | 2.25E-05 | ||||
| NHEDC1 | chr4:103940711–103941300 | 2 | 0.08 | 1.96E-07 | ||||
| chr5:784832–784915 | 2 | 0.43 | < 2.00E-16 | 0.43 | < 2.00E-16 | 0.43 | < 2.00E-16 | |
| chr5:68628240–68628738 | 2 | 0.1 | 3.90E-06 | |||||
| RIPK1 | chr6:3077011–3077041 | 3 | -0.17 | 6.98E-05 | ||||
| HIST1H3C | chr6:26045532–26045663 | 13 | 0.1 | 3.02E-12 | 0.1 | 4.68E-09 | 0.1 | < 2.00E-16 |
| PTPRN2 | chr7:157744316–157744347 | 4 | -0.05 | 7.10E-10 | ||||
| chr8:58055876–58056175 | 3 | 0.43 | 8.50E-05 | 0.44 | 1.55E-06 | |||
| C8orf71 | chr8:58191386–58192065 | 6 | -0.25 | 4.89E-08 | -0.26 | 3.82E-09 | -0.25 | 1.38E-07 |
| SPAG1 | chr8:101224915–101225361 | 6 | 0.13 | < 2.00E-16 | 0.13 | < 2.00E-16 | 0.13 | < 2.00E-16 |
| SPAG1 | chr8:101225800–101225902 | 11 | 0.21 | < 2.00E-16 | 0.21 | < 2.00E-16 | 0.21 | < 2.00E-16 |
| C9orf64 | chr9:86571409–86572014 | 3 | 0.1 | 6.98E-05 | 0.1 | 6.25E-06 | ||
| KIAA1274 | chr10:72254314–72254335 | 2 | -0.11 | 1.70E-06 | ||||
| CALHM1 | chr10:105218160–105218286 | 2 | -0.1 | 6.26E-05 | ||||
| CALCB | chr11:15093613–15093769 | 2 | -0.16 | 2.67E-06 | -0.16 | 1.66E-06 | -0.18 | 3.66E-07 |
| TP53AIP1 | chr11:128812804–128813442 | 2 | -0.19 | 1.24E-05 | ||||
| TP53AIP1 | chr11:128812846–128813008 | 3 | -0.19 | 7.61E-05 | ||||
| PIWIL1 | chr12:130822286–130822818 | 2 | -0.25 | 4.26E-05 | ||||
| NBEA | chr13:36044860–36045352 | 2 | 0.05 | 3.26E-05 | 0.05 | 2.21E-06 | 0.05 | 8.17E-05 |
| chr15:26489846–26490045 | 2 | -0.05 | 8.82E-05 | |||||
| KRTAP17–1 | chr17:39472114–39472340 | 2 | -0.15 | 9.52E-05 | ||||
| chr17:55213563–55213600 | 5 | -0.15 | 9.68E-05 | |||||
| chr17:77680078–77680232* | 2 | -0.19 | 6.73E-05 | -0.19 | 3.31E-07 | |||
| LRP5L* | chr22:25758621–25758749 | 5 | -0.1 | 8.18E-05 | ||||
| MXRA5 | chrX:3264517–3265089 | 2 | 0.04 | 1.92E-09 | ||||
| TTTY14 | chrY:21238886–21239607 | 2 | 0.09 | 1.18E-11 | ||||
| * CNV with copy number of 3 overlapping with the region (see Table S1) | ||||||||
Light gray boxes indicate non-significant results. Dark gray boxes indicate that no comparison was made (X-chromosome-linked genes were compared with females only whereas Y-chromosome-linked genes were compared with males only).
Numerous genes were found to be differentially expressed (DE) in the 47,XXY prefrontal cortex and cerebellum compared with other samples (Tables 3 and 4) with a Bonferroni-corrected z-score P value < 0.05. In both brain regions the vast majority of DE genes were characterized by increased expression in the 47,XXY patient (prefrontal cortex: 12 loci significantly upregulated, 1 locus significantly downregulated; cerebellum: 18 loci significantly upregulated, 0 loci significantly downregulated), suggesting that the supernumerary X-chromosome may be upregulating transcription at multiple autosomal loci across the genome. Although some DE genes were observed only in comparisons with both 46,XY males and 46,XX females, most were sex-comparison-specific. Strikingly, across both brain regions, many more 47,XXY DE genes were observed in comparison with females than males in both tissues (prefrontal cortex: 47,XXY vs females = 59, 47,XXY vs males = 18; cerebellum: 47,XXY vs females = 50, 47,XXY vs males = 21). Furthermore, although some changes were consistently observed in both the prefrontal cortex and cerebellum (e.g., CAMP, EPCAM and LOC441208), the majority were tissue-specific to a single brain region. The large differences in gene expression reported here are specific to the 47,XXY individual; none of these transcripts are differentially expressed in our analysis of schizophrenia and matched controls (Pidlsey et al., submitted).
Table 3. 47,XXY-associated differentially expressed genes in the prefrontal cortex.
| nuID | Gene | Probe position (hg19) | Strand | 47,XXY vs. all other | 47,XXY vs. males | 47,XXY vs. females | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| fold change | P value | fold change | P value | fold change | P value | |||||
| fVedl_tOxJHgKDgkUk | ANGPTL7 | chr1:11255703–11255752 | + | 0.17 | 1.11E-16 | 0.17 | 5.11E-15 | 0.17 | < 2.00E-16 | |
| Zl4IEsHlNCOIgeii9c | VCAM1 | chr1:101204019–101204068 | + | 0.12 | 1.78E-08 | |||||
| cqSF81At6xB7XyHjiQ | PRG4 | chr1:186282872–186282921 | + | 0.2 | < 2.00E-16 | |||||
| lojns5Hg3o6bW_X07 g | CFH | chr1:196648872–196648921 | + | 0.23 | 1.43E-07 | |||||
| Qo0T0_REI6OtlJLFOU | CFH | chr1:196658692–196658741 | + | 0.38 | 1.11E-16 | |||||
| NIDSs_kv.U.0.u7g.8 | H3F3A | chr1:226259333–226259382 | + | 0.33 | 7.90E-07 | |||||
| NfqgAIRfuk8Lfn_vAk | EXOC8 | chr1:231468728–231468777 | - | 0.21 | 1.68E-07 | |||||
| ZoXhLA6oEHo3o160B8 | EPCAM | chr2:47606943–47606992 | + | 0.69 | 1.11E-16 | |||||
| BnrvPnvPuvuuMkvvno | EPCAM | chr2:47607046–47607095 | + | 1.04 | 2.26E-08 | |||||
| cpAs4gp4jAojq4jkyo | EPCAM | chr2:47612328–47612349: 47613711–47613738 |
+ | 0.2 | 1.65E-09 | 0.2 | 2.98E-07 | 0.21 | < 2.00E-16 | |
| xd6Z60LRKEWCAU0bEY | EFEMP1 | chr2:56149495–56149544 | - | 0.39 | 6.29E-10 | |||||
| 9l31SOkN7XN3uegjnI | AOX1 | chr2:201535434–201535483 | + | 0.08 | < 2.00E-16 | |||||
| Zl7s26IBCIUH.09XlU | COL6A3 | chr2:238232942–238232991 | - | 0.39 | 6.62E-14 | |||||
| QXNDUD1d6L9O85e_Sk | COL6A3 | chr2:238233095–238233144 | - | 0.13 | 6.77E-10 | |||||
| rItcu7lcV6dKfep3iA | CAMP | chr3:48266905–48266954 | + | 0.2 | < 2.00E-16 | 0.2 | < 2.00E-16 | 0.2 | < 2.00E-16 | |
| 69KffSiAkLEesX6HuQ | SLC25A20 | chr3:48900042–48900091 | - | 0.36 | 5.69E-11 | |||||
| lnxCeHgiilejtJhR.U | DNASE1L3 | chr3:58179070–58179093: 58178505–58178530 |
- | 0.13 | < 2.00E-16 | |||||
| 6h.tJeHqxD3.vD_S7o | PHLDB2 | chr3:111694722–111694770 | + | 0.27 | 3.50E-09 | |||||
| ommprt5mtriFZmriGc | FAIM | chr3:138327937–138327986 | + | 0.11 | 3.54E-07 | |||||
| ln0vn7fesf3vOp.6ok | TP63 | chr3:189614936–189614985 | + | 0.21 | < 2.00E-16 | |||||
| KpJwqU0.HokiEUlMrU | NULL | chr4:144496759–144496808 | - | 0.09 | 5.31E-07 | |||||
| 35QRHIUd.o9CJHuzh4 | GPX8 | chr5:54460446–54460495 | + | 0.15 | < 2.00E-16 | |||||
| riWe5en0.0i4hP7Huk | BTNL9 | chr5:180488062–180488111 | + | 0.2 | 3.57E-12 | |||||
| QfyhEl_TCJbggfqkvk | LOC401233 | chr6:3019504–3019553 | - | 0.15 | 7.79E-07 | |||||
| fajeo6Sh6PMkUgoxnk | BMP5 | chr6:55625255–55625272: 55623882–55623913 |
- | 0.19 | 2.30E-14 | 0.18 | 7.69E-12 | 0.19 | < 2.00E-16 | |
| K6oCu8IOLU3SUMufrs | TBX18 | chr6:85444495–85444544 | - | 0.12 | 6.56E-07 | |||||
| 07q5ey9ScdKKeKSiDg | IKBIP | chr6:168224491–168224540 | - | -0.69 | 2.87E-07 | |||||
| 0uHF8y.yziXFfoICwo | TMEM196 | chr7:19759195–19759244 | - | 0.21 | 4.12E-09 | |||||
| Zn50ZUiIgAvCeeFeCk | INMT | chr7:30797037–30797086 | + | 0.12 | 3.31E-12 | |||||
| cyxSgtKKIa_cDsvc3o | LOC441208 | chr7:32768986–32769035 | + | 0.32 | < 2.00E-16 | 0.33 | < 2.00E-16 | 0.32 | 7.88E-15 | |
| iX3qB7qexxL0p66.jk | FGL2 | chr7:76825768–76825817 | - | 0.43 | 2.49E-11 | |||||
| KmAXs1qVBeAE1SUIHo | COL1A2 | chr7:94057677–94057726 | + | 0.35 | < 2.00E-16 | |||||
| 9jUT7yueFyEiOB4rX4 | COL1A2 | chr7:94060111–94060160 | + | 0.45 | < 2.00E-16 | |||||
| QyCF6hIoee3ld3rRXk | DEFA1 | chr8:6856660–6856709 | - | 0.12 | 1.38E-13 | 0.12 | 1.57E-12 | 0.12 | 1.11E-15 | |
| xKBJBHXp4QcJQzpXK0 | SCARA5 | chr8:27727946–27727995 | - | 0.26 | 9.82E-07 | |||||
| 0LiHngASd5JSA633Ro | SDCBP | chr8:59492276–59492306 | + | 0.15 | 6.15E-07 | |||||
| K7con83.RdfSKOc6rU | BNC2 | chr9:16416697–16416746 | - | 0.17 | 1.83E-13 | |||||
| 6oXSUI4JlkkvBc4W5I | CCL19 | chr9:34689927–34689940: 34689801–34689836 |
- | 0.32 | < 2.00E-16 | |||||
| WrlIsC.cAwT13R_xVE | OGN | chr9:95146774–95146823 | - | 0.77 | 4.11E-15 | |||||
| BDhFLxNahZPgiMZeio | OGN | chr9:95147978–95148027 | - | 0.88 | < 2.00E-16 | |||||
| ZjdFPOE1NVcnRRMkTI | OMD | chr9:95176881–95176930 | - | 0.45 | 3.73E-07 | |||||
| Kc4qiig6iKqdKpQwHc | SLC27A4 | chr9:131123437–131123486 | + | 0.09 | 3.26E-08 | |||||
| oKAoV4glIpSToLgqkg | ITIH2 | chr10:7788602–7788651 | + | 0.2 | 1.24E-09 | |||||
| lqMskrfH4OAt4z_eOk | OTUD1 | chr10:23730535–23730584 | + | 0.16 | 9.87E-11 | |||||
| xqKT0lF6D5OJ2dL_o8 | PDE6H | chr12:15134355–15134404 | + | 0.16 | 2.54E-08 | |||||
| ZfKOiSBOiKCP0.wnXU | SLCO1C1 | chr12:20905935–20905984 | + | 0.13 | 2.83E-10 | |||||
| fX9eh..pR5XocuAg6E | PKP2 | chr12:32944450–32944499 | - | 0.21 | 2.92E-07 | |||||
| cjif7cfQC.v58VfSXU | GJB2 | chr13:20761910–20761959 | - | 1 | < 2.00E-16 | |||||
| ciAGgiIoDiuq_igFTo | EDNRB | chr13:78470881–78470930 | - | 0.18 | 1.49E-07 | 0.18 | 1.11E-16 | |||
| xuigGSeQeCaIdwFSfk | PTGDR | chr14:52742862–52742911 | + | 0.15 | < 2.00E-16 | |||||
| rl55P5uN0IXUlILV9Q | PTGDR | chr14:52743020–52743069 | + | 0.32 | < 2.00E-16 | |||||
| 0knRyVFXXc.6sIg.HE | TMEM30B | chr14:61744888–61744937 | - | 0.45 | < 2.00E-16 | |||||
| TP6d2kdUp6iejF5XpI | LOC388152 | chr15:84871644–84871693 | - | 0.12 | 5.26E-09 | |||||
| 3eh0.Qkv5.70DbjiAU | NUDT21 | chr16:56463489–56463538 | - | 0.15 | 1.04E-07 | 0.15 | 6.69E-08 | 0.15 | 5.92E-07 | |
| KsuX1EryiLsDi3rL_0 | TMEM220 | chr17:10617182–10617231 | - | 0.09 | 2.07E-08 | 0.09 | 2.60E-09 | 0.08 | 3.79E-08 | |
| x.Sd_F7Vd6eXeLeDdU | TOP2A | chr17:38545067–38545116 | - | 0.11 | 1.49E-07 | 0.12 | < 2.00E-16 | |||
| EnpItS.SAMZeiUiS2E | FAM20A | chr17:66533566–66533615 | - | 0.12 | 2.70E-08 | 0.12 | 4.20E-09 | |||
| 9k3mzbqMPhOKn4iB1I | CD177 | chr19:43867372–43867421 | + | 0.14 | 6.18E-14 | 0.13 | 8.74E-12 | 0.14 | < 2.00E-16 | |
| 3CBVEhgxeipOOJilWo | MYL9 | chr20:35176437–35176486 | + | 0.11 | 1.34E-07 | |||||
| No174RVAVBCigl6guU | SLPI | chr20:43882216–43882241: 43881769–43881792 |
- | 0.27 | < 2.00E-16 | |||||
| ojLV_BETnlid6ABVEk | UBE2C | chr20:44444504–44444552: 44445348–44445348 |
+ | 0.1 | < 2.00E-16 | |||||
| r37yu690k8Sk.uedqI | LOC401397 | chr20:57523176–57523225 | - | 0.11 | 2.69E-08 | |||||
| TdSCif1KMn_KMJ5o4k | LOC96610 | chr22:22664198–22664247 | + | 0.09 | 6.12E-08 | |||||
| luuljtLxeOlOiCJCmE | ADRBK2 | chr22:25817204–25817253 | + | 0.1 | 1.11E-16 | |||||
| TADKWp66dGXsNUkf6Q | GGA1 | chr22:38013841–38013890 | + | -0.83 | 9.84E-07 | -0.86 | 2.61E-09 | |||
| Z5K0omXVEuL9VRHadc | RRP7A | chr22:42907975–42908024 | - | 0.08 | < 2.00E-16 | |||||
| EqZLp0Zez1SR.qKUKQ | RRP7B | chr22:42969512–42969561 | - | -0.55 | 6.55E-10 | |||||
| 0szegLje1eiuskL9Ro | NPM1 | chrX:123414788–123414837 | + | 0.2 | 8.35E-08 | |||||
| lLh40p.7RHpRI4TceU | GYG2P1 | chrY:14518992–14518993: 14518689–14518736 |
- | 0.11 | 1.21E-07 | |||||
Light gray boxes indicate non-significant results. Dark gray boxes indicate that no comparison was made (X-chromosome-linked genes were compared with females only whereas Y-chromosome-linked genes were compared with males only). Annotation data for each probe obtained using the Bioconductor package illuminaHumanv4.db.39
Table 4. 47,XXY-associated differentially expressed genes in the cerebellum.
| nuID | Gene | Probe position (hg19) | Strand | 47,XXY vs. all other | 47,XXY vs. other males | 47,XXY vs. other females | |||
|---|---|---|---|---|---|---|---|---|---|
| fold change | P value | fold change | P value | fold change | P value | ||||
| uO7tSXg9R5ohX55GF4 | TNFRSF18 | chr1:1140794–1140843 | - | 0.15 | 1.01E-14 | ||||
| TQ5MLOicLl.6KP36CI | FAM76A | chr1:28087875–28087924 | + | 0.07 | 2.70E-07 | ||||
| 3VHqE.9440_ek4J75o | PCNXL2 | chr1:233275460–233275463: 233270891–233270936 | - | 0.14 | 6.48E-12 | ||||
| ZoXhLA6oEHo3o160B8 | EPCAM | chr2:47606943–47606992 | + | 0.54 | 5.47E-12 | ||||
| BnrvPnvPuvuuMkvvno | EPCAM | chr2:47607046–47607095 | + | 0.9 | 3.44E-13 | ||||
| 6KDbq30yk7941631Kg | KCNH7 | chr2:163228344–163228393 | - | 0.12 | 7.42E-07 | ||||
| 6R96hdG6D63d4rLojk | LOC401052 | chr3:10048205–10048254 | - | 0.22 | 1.16E-10 | ||||
| TQeEon6idqJ_oip878 | ARPP21 | chr3:35722564–35722613 | + | 0.1 | 1.46E-12 | ||||
| rItcu7lcV6dKfep3iA | CAMP | chr3:48266905–48266954 | + | 0.09 | 3.46E-12 | 0.09 | 1.86E-09 | 0.1 | < 2.00E-16 |
| QEKvDrJSV50hXQ4Rno | ANKRD17 | chr4:74005263–74005312 | - | 0.22 | 1.98E-07 | ||||
| 0q44on0oRTyiSnoQTc | SH3TC2 | chr5:148384360–148384409 | - | 0.1 | 5.29E-11 | ||||
| WVElhxxViAlxN9F9ec | RASGEF1C | chr5:179564876–179564899: 179564687–179564712 | - | 0.13 | 4.15E-07 | ||||
| 01Xd110F6gx9ITdQHQ | RNF39 | chr6_mcf_hap5:1420068–1420117 | - | 0.16 | 6.29E-07 | ||||
| 0cVOsh0SeXuuB.p1eU | PRRC2A | chr6_dbb_hap3:2890088–2890137 | + | 0.35 | 2.22E-07 | ||||
| Tqg7v6gLQi6yj6_nqY | C6orf132 | chr6:42070437–42070486 | - | 0.23 | 3.15E-10 | ||||
| HqI33tUcoB5g_xRIqI | C6orf176 | chr6:166338009–166338058 | - | 0.11 | 1.15E-08 | ||||
| cyxSgtKKIa_cDsvc3o | LOC441208 | chr7:32768986–32769035 | + | 0.14 | 3.61E-07 | ||||
| cyxSgtKKIa_cDsvc3o | LOC441208 | chr7:32768986–32769035 | + | 0.15 | < 2.00E-16 | ||||
| 3WfZR.WSd6.5x7nndc | C7orf52 | chr7:100813863–100813912 | - | 0.15 | 4.28E-09 | 0.15 | 1.03E-08 | 0.14 | 1.43E-09 |
| K_N4laRVOKE5IkXkig | CNTFR | chr9:34552159–34552166: 34552028–34552069 | - | 0.23 | < 2.00E-16 | 0.23 | 3.99E-14 | 0.23 | < 2.00E-16 |
| uJdPSKuSnCZOhiSaug | GPSM1 | chr9:139252547–139252596 | + | 0.25 | 2.95E-08 | ||||
| 90NSdfgVdSVn0l6k0c | ENTPD2 | chr9:139942751–139942800 | - | 0.09 | 6.35E-08 | ||||
| 3Z2Vqi1Jrte6J5ZTpU | IL15RA | chr10:6019449–6019498 | - | 0.08 | 8.68E-07 | 0.08 | 6.27E-08 | ||
| ZIRQ6g1Q6oLY56590U | IGF2 | chr11:2159459–2159460: 2156712–2156759 | - | 0.1 | 9.90E-14 | 0.09 | 1.45E-12 | 0.1 | < 2.00E-16 |
| Wl36k3qhE6w57l_Egs | CALCA | chr11:14988290–14988339 | - | 0.23 | 8.30E-09 | 0.23 | 5.73E-09 | 0.22 | 5.94E-09 |
| x9f4JKzfk6VFquC1eU | TMEM223 | chr11:62558209–62558258 | - | 0.28 | 6.22E-07 | ||||
| 9ooIoDrhULuqvuvSgI | SLC22A6 | chr11:62744143–62744192 | - | 0.18 | 3.65E-07 | 0.18 | 9.29E-07 | 0.18 | 9.39E-08 |
| Z9IldVfociaepInc.BI | CNIH2 | chr11:66051236–66051285 | + | 0.35 | 9.53E-07 | 0.36 | 4.88E-15 | ||
| HFwXl7tTpgV3E1wXlo | SMARCC2 | chr12:56558210–56558259 | - | 0.18 | 8.81E-08 | 0.19 | 4.36E-08 | 0.18 | 6.14E-07 |
| QO_wesnjwsy9XkVfeo | LOC220115 | chr13:53161055–53161104 | + | 0.12 | 8.32E-09 | ||||
| c70LXLcyj6S.A5.HVU | OLFM4 | chr13:53626107–53626156 | + | 0.09 | 4.37E-07 | 0.09 | 1.59E-07 | ||
| 36JWb571RKl2v.XB_c | SPSB3 | chr16:1826791–1826840 | - | -0.89 | 9.17E-07 | ||||
| BloWN0NHoOuep7agcE | TNRC6A | chr16:24834917–24834966 | + | 0.13 | 9.96E-09 | 0.13 | 1.69E-08 | 0.13 | 9.43E-09 |
| QLR0VHu.euUKd_KlUc | FAM64A | chr17:6354072–6354121 | + | 0.1 | 1.33E-07 | ||||
| EdOgRNRCeCkhClZIJ0 | RPL19 | chr17:37360385–37360427 | + | 0.7 | 6.11E-08 | ||||
| x6gXnXotNXeQEiJUi4 | RBFOX3 | chr17:77303806–77303842: 77231875–77231887 | - | 0.57 | 2.85E-09 | ||||
| iIoD3lFP9UreSVdSeo | RBFOX3 | chr17:77303839–77303888 | - | 0.3 | 3.77E-07 | ||||
| f4oYbqlTz8YVp9fc6U | FN3K | chr17:80708373–80708422 | + | 0.22 | 1.29E-10 | 0.21 | 7.08E-10 | 0.23 | 1.34E-14 |
| x3S.SNd5j.Pi6At_Z4 | MGC70870 | chr17_gl000205_random: 119141–119190 |
+ | 0.31 | 3.30E-07 | ||||
| oCVdUCZaWHZ7dnqh38 | NFIC | chr19:3463515–3463564 | + | 0.1 | 1.67E-08 | ||||
| QudxDtFe.RtNJF3qhU | OLFM2 | chr19:9964866–9964915 | - | 0.54 | 9.61E-07 | 0.57 | 1.20E-08 | ||
| 9RSiJQISr7l8VSiUVc | NACC1 | chr19:13251492–13251541 | + | 0.49 | 7.50E-11 | 0.49 | 1.49E-08 | 0.5 | < 2.00E-16 |
| uCKpSOEVwJKeNqQ6is | RAB3A | chr19:18307891–18307940 | - | 0.93 | 1.71E-12 | ||||
| l3roFeVfFJfjpJqDes | POU2F2 | chr19:42595686–42595735 | - | 0.09 | 6.52E-08 | ||||
| T3DouuhUIkzS5yQDpI | UBE2V1 | chr20:48700666–48700677: 48699413–48699451 | - | 0.19 | 8.02E-09 | ||||
| uXOOCKm1HogpVesKUk | KIAA1647 | chr22:18958144–18958171 | + | 0.12 | 6.51E-07 | ||||
| i1_RF4d7R0Jf3UUpV0 | SERPIND1 | chr22:21141675–21141724 | + | 0.1 | 4.00E-07 | ||||
| Z5K0omXVEuL9VRHadc | RRP7A | chr22:42907975–42908024 | - | 0.16 | 4.44E-15 | ||||
| EU_ve.pMFc8DrmSJ4M | LOC389834 | chrUn_gl000218:51064–51113 | - | 0.14 | 1.87E-07 | ||||
| lt1Hf71dcOcPygIhR4 | CHIC1 | chrX:72903733–72903782 | + | 0.08 | 4.14E-06 | ||||
| WnUxEZ5faxUURBNQuk | L1CAM | chrX:153127445–153127494 | - | 0.77 | 1.28E-05 | ||||
| HLEukIKIKKaSqkOEnQ | L1CAM | chrX:153128160–153128209 | - | 0.08 | 2.23E-05 | ||||
| fKg7fXeNt9dKDIdCzk | RPS4Y1 | chrY:2712151–2712200 | + | 1.48 | < 2.00E-16 | ||||
| Q8jADkAqS017VJIV90 | NLGN4Y | chrY:16953254–16953303 | + | 0.06 | 1.27E-08 | ||||
| 6tUwTEFxS.3kFYCVKk | AL833666 | chrY:21724080–21724129 | - | 0.18 | 3.20E-10 | ||||
Light gray boxes indicate non-significant results. Dark gray boxes indicate that no comparison was made (X-chromosome-linked genes were compares to females only whereas Y-chromosome-linked genes were compared with males only). Annotation data for each probe obtained using the Bioconductor package illuminaHumanv4.db.39
Given previous evidence that KS is associated with skewed XCI in peripheral blood,14 we next assessed allelic patterns of DNA methylation in the proximity of a polymorphic repeat (CAG)n in the androgen receptor (AR) gene. Although the prefrontal cortex was characterized by subtle allelic imbalance of XCI (7.5% skewing), this did not exceed the range of normal skewing observed in our previous analysis of healthy individuals,40 and there was no evidence of allelic skewing in the cerebellum (0.4% skewing). We were also interested in examining the expression and DNA methylation status of X-linked genes believed to escape XCI in females.41 Between 5 and 15% of genes on the X-chromosome are thought to escape XCI in healthy females,42,43 and it has been suggested that these loci may play an important role in the KS phenotype.44,45 Although genes escaping XCI might be expected to show consistently high expression levels in females compared with males, many loci actually show variable levels of expression in females46 with evidence of tissue-specificity.45,47
Although the 47,XXY patient is an outlier for the expression of many genes thought to escape XCI, we find no consistent pattern of altered transcription; some loci are upregulated and others downregulated, with many showing changes specific to either male or female comparison groups (Table S2). There is also considerable evidence for tissue-specific differences in the expression of these genes, concurring with data from a study of genes escaping XCI in 41,XXY mice compared with karyotypically normal male and female mice.45 One of the loci showing noticeably higher expression in the 47,XXY patient compared with both females and males across both brain regions is the gene encoding Eukaryotic Translation Initiation Factor 1AX (EIF1AX) (Fig. S8), which has been suggested as a possible candidate gene for Turner syndrome (45,XO).48 Browseable tracks for viewing within the Integrative Genomics Viewer (IGV) (http://www.broadinstitute.org/igv/home) showing 47,XXY-associated changes in DNA methylation and gene expression for other loci are downloadable from our laboratory website (http://epigenetics.iop.kcl.ac.uk/XXY).
We also looked at the expression of genes located in the pseudoautosomal regions (PAR) 1 and 249 (http://www.genenames.org/genefamilies/PAR), which are represented by three copies in individuals with a 47,XXY karyotype. Again, although often an outlier for transcription at these loci (Table S3), the observed pattern in the 47,XXY individual is heterogeneous, differing across tissues and between male and female comparison groups. Some PAR genes (e.g., SLC25A6) are clearly upregulated in the cerebellum but not the prefrontal cortex, while others (e.g., DHRSX) are upregulated across both tissues. Other genes such as GTPBP6, for example, are consistent outliers for reduced expression in the 47,XXY patient suggesting that transcription is not always positively correlated with copy number in the PAR. Further work is needed to explore the regulatory mechanisms influencing expression of loci on the extra X-chromosome, and the processes involved in controlling dosage compensation. Another region of interest in KS is the X transposed region (XTR) on the Y chromosome,50 created by a 3.5 Mb duplication from Xq21.3 to Yp11.2 during hominin evolution.51-53 In Figure S4 the allele frequency plot for the 46,XY male (B) shows three bands across both chromosomal regions, whereas the 47,XXY patient (A) is characterized by four bands, most likely resulting from cross hybridization of microarray probes. Although epigenetic deregulation of the Protocadherin 11 X-linked (PCDH11X) and Y-linked (PCDH11Y) genes in these regions are of great interest in KS and neuropsychiatric disease,54-56 DNA methylation array probes in the vicinity of these genes were excluded during our stringent quality control steps due to cross-reactivity57 and could not be assessed in this study (see the “Materials and Methods” section). Because it is plausible that the XTR contains sequence and epigenetic differences that are important in KS and schizophrenia, future studies should utilize methods that can unambiguously profile variation in this region.
In summary, this study identifies widespread transcriptomic and epigenomic changes in the prefrontal cortex and cerebellum associated with a 47,XXY karyotype. Although our findings are based on data from only a single 47,XXY individual, and it will be important to confirm the observed patterns samples from additional patients, this study represents the first detailed molecular characterization of brain tissue from an individual with a 47,XXY karyotype. The patient, who was comorbid for schizophrenia, was found to have a notably reduced cerebellum mass and was characterized by considerable locus-specific changes in DNA methylation and gene expression, with many of these differences being autosomal and tissue-specific. Strikingly, global DNA methylation, assessed via the interrogation of LINE-1 and Alu repetitive elements, was significantly altered in the 47,XXY patient in a tissue-specific manner. Finally, we find evidence for alterations in gene expression at loci believed to normally escape XCI in females.
Materials and Methods
Samples and nucleic acid isolation
Post-mortem brain samples were obtained from the MRC London Neurodegenerative Diseases Brain Bank (http://www.kcl.ac.uk/iop/depts/cn/research/MRC-London-Neurodegenerative-Diseases-Brain-Bank/MRC-London-Neurodegenerative-Diseases-Brain-Bank.aspx). Patients (n = 49) were approached in life for written consent for brain banking. All samples were dissected by a trained neuropathologist, snap-frozen and stored at -80°C following legal and ethical guidelines. The time between death and removal of the brain was recorded as post-mortem interval (PMI). The samples used in this study comprised of prefrontal cortex (PFC, n = 49) and cerebellum samples (CER, n = 48), from 23 schizophrenia patients (including the 47,XXY patient) and 26 unaffected controls. All schizophrenia patients were diagnosed pre-mortem by psychiatrists in the UK using standardized diagnostic criteria. Demographic information about the samples is summarized in Table S4. Genomic DNA was extracted from each tissue sample using a standard phenol-chloroform extraction and tested for purity and degradation using spectrophotometry and gel electrophoresis, respectively. RNA was extracted using a standard Trizol extraction method and purified using an RNeasy Mini Kit with DNase I digestion (Qiagen), according to manufacturer’s instructions. RNA was tested for degradation and purity using an Agilent 2100 Bioanalyzer and RNA 6000 Nano kit (Agilent Technologies). All samples were randomized with respect to gender and disease status throughout all stages of the project to avoid potential batch effects.
Global DNA methylation assay
Bisulfite-PCR pyrosequencing was used to assess the methylation status of LINE-1 and Alu repeats as a proxy of global DNA methylation levels, as described previously.30,31 Samples were run on the Pyromark Q24 pyrosequencer (Qiagen) according to manufacturer’s instructions. DNA methylation levels for each sample were calculated as the average of the three interrogated CpG sites on each assay. Fully methylated and fully unmethylated control samples were included in all procedures to act as assay controls.
Genome-wide DNA methylation array processing
500ng of genomic DNA from each sample was treated with sodium bisulfite in duplicate, using the EZ-96 DNA methylation kit (Zymo Research) following the manufacturer’s standard protocol. Duplicates were pooled and the samples (PFC n = 46 and CER n = 46) were assessed using the Illumina Infinium HumanMethylation450 BeadChip (Illumina Inc.) run on the HiScan System (Illumina Inc.). All samples were randomized with respect to gender and disease status to avoid batch effects, and processed on eight BeadChips.
Methylomic data processing and analysis
Signal intensities were extracted using Illumina GenomeStudio software (Illumina Inc.) and imported into R58 using the methylumi and minfi packages.59,60 Multi-dimensional scaling plots of variable probes on the X- and Y-chromosome were used to check concordance between predicted and reported sex for each individual (see the “Results and Discussion” section). The comparison of non-CpG SNP probes on the array confirmed that the PFC and CER were sourced from the same individual where expected. Raw β values of CpG probes within brain region-specific differentially methylated regions (DMRs) (extracted from ref. 61) were used to confirm that the predicted and reported brain region corresponded for each sample. Probes containing a SNP with MAF > 5% within 10 bp of the CG target site based on the Illumina annotation data (n = 35 413) and non-CG probes (n = 65) were removed. Further stringent data quality control and processing steps were conducted using the dasen function in the wateRmelon package as previously described.62 The pfilter function was used to filter data by beadcount and detection P value to stringently control for poor quality probes (PFC n = 5623 probes and CER n = 10 417 probes removed across all samples). Prior to statistical analyses cross-reactive probes co-hybridizing to the sex-chromosomes, as previously identified,57 were removed. The pnorm function used to identify differentially methylated CpG sites in the 47,XXY patient and the comb-p package63 was used to identify 500bp regions of 2 or more adjacent differentially-methylated probes. The identified regions of differential DNA methylation were compared with copy number variation (CNV) data of the 47,XXY patient to screen for overlaps with any large genomic aberrations.
Genome-wide expression array processing
An amount of 100 ng RNA from each sample (PFC n = 47 and CER n = 48) was biotinylated and amplified using the Illumina TotalPrep RNA Amplification kit (Life Technologies) to produce cRNA. cRNA was quantitated using a NanoDrop NO-1000 (Thermo Fisher Scientific) and RediPlate 96 RiboGreen RNA Quantitation Kit (Life Technologies). Genome-wide expression was assessed using the Illumina HumanHT-12 v4 Expression BeadChip (Illumina Inc.) according to manufacturer’s instruction.
Expression data processing and analysis
Signal intensities for each probe were extracted using Illumina GenomeStudio software (Illumina Inc.) and imported into R using the lumi package within Bioconductor.64 Initial quality control checks using functions within lumi identified clear outlying samples, which were removed from subsequent analyses (PFC n = 5, CER n = 4). The sex of the samples was checked by comparing the sex predicted by the expression levels of the XIST gene with the reported sex for each individual (see the “Results and Discussion” section). Probes targeting transcripts of genes in the vicinity of brain region-specific DMRs61 were used to confirm that the predicted brain region corresponded with the reported region for each sample. Remaining samples were processed using the lumi64 and MBCB65 Bioconductor packages in R. During processing, probes with a detection P value > 0.01 across all samples were considered non-detectable and removed from subsequent analysis. The ComBat function within the sva package in R66 was used to adjust the data to remove batch effects. The pnorm function was used to identify differentially expressed transcripts in the 47,XXY sample. Genes identified as differentially expressed were compared with the CNV data of the 47,XXY patient to record overlaps with large genomic aberrations.
Genome-wide CNV detection
200ng of genomic DNA from each prefrontal cortex sample were genotyped using the Illumina HumanOmniExpress BeadChip (Illumina Inc.). All samples were randomized with respect to gender and disease status to avoid batch effects. Illumina GenomeStudio was used to call genotypes (using the HumanOmniExpress-12v1_C.egt cluster file) with the default GenCall cut-off of 0.15. To compare the sex predicted by the genetic data with the reported sex for each individual previously published recommendations were followed.67 PLINK was used to assess the heterozygosity rate of the probes on the X-chromosome68 (see the “Results and Discussion” section). Autosomal CNVs were called using PennCNV.69
PCR-based sex-typing assay
A PCR-based sex-typing assay was performed as described previously.70 In brief, the X and Y amelogenin (AMELX) sequences were amplified, with amplicons distinguished on the basis of size; the X-chromosome produces a 977bp amplicon, whereas the Y-chromosome produces a 788bp amplicon.
X-Chromosome Inactivation assay
The allelic X-Chromosome Inactivation (XCI) ratios of both tissues of the 47,XXY sample were determined by assessing DNA methylation in the proximity of a polymorphic repeat (CAG)n in the human androgen receptor (AR) gene, as described previously.71,72 In brief, 50 ng of genomic DNA was incubated with HpaII, MspI or water in triplicate. The digestion product was amplified using fluorescently labeled primers flanking the polymorphic repeat (CAG)n. An ABI3130 (Life Technologies) was used to separate the fluorescently labeled amplification products and quantify the peak heights of each allele. The XCI ratio was then calculated as previously described.40
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by a UK Medical Research Council (MRC) project grant to J.M. R.P. was funded by an MRC PhD studentship. We thank Annet Damhuis at the Royal Devon and Exeter NHS Trust for the technical assistance.
Glossary
Abbreviations:
- KS
Klinefelter syndrome
- XCI
X-Chromosome inactivation
- LINE-1
long interspersed nucleotide element-1
- PMI
post-mortem interval
- CER
cerebellum
- PFC
prefrontal cortex
- DMR
differentially methylated regions
- DE
differentially expressed
- CNV
copy number variation
- PAR
pseudoautosomal regions
References
- 1.Wikström AM, Dunkel L. Klinefelter syndrome. Best Pract Res Clin Endocrinol Metab. 2011;25:239–50. doi: 10.1016/j.beem.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Frühmesser A, Kotzot D. Chromosomal variants in klinefelter syndrome. Sex Dev. 2011;5:109–23. doi: 10.1159/000327324. [DOI] [PubMed] [Google Scholar]
- 3.Klinefelter HF, Reifenstein E. C., Albright, F. Syndrome Characterized by Gynecomastia, Aspermatogenesis without A-Leydigism, and Increased Excretion of Follicle-Stimulating Hormone. Am J Clin Dermatol. 1942;2:615–27. [Google Scholar]
- 4.Boada R, Janusz J, Hutaff-Lee C, Tartaglia N. The cognitive phenotype in Klinefelter syndrome: a review of the literature including genetic and hormonal factors. Dev Disabil Res Rev. 2009;15:284–94. doi: 10.1002/ddrr.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Rijn S, Aleman A, Swaab H, Kahn R. Klinefelter syndrome (karyotype 47,XXY) and schizophrenia-spectrum pathology. Br J Psychiatry. 2006;189:459–60. doi: 10.1192/bjp.bp.105.008961. [DOI] [PubMed] [Google Scholar]
- 6.DeLisi LE, Maurizio AM, Svetina C, Ardekani B, Szulc K, Nierenberg J, Leonard J, Harvey PD. Klinefelter syndrome (XXY) as a genetic model for psychotic disorders. Am J Med Genet B Neuropsychiatr Genet. 2005;135B:15–23. doi: 10.1002/ajmg.b.30163. [DOI] [PubMed] [Google Scholar]
- 7.Fales CL, Knowlton BJ, Holyoak KJ, Geschwind DH, Swerdloff RS, Gonzalo IG. Working memory and relational reasoning in Klinefelter syndrome. J Int Neuropsychol Soc. 2003;9:839–46. doi: 10.1017/S1355617703960036. [DOI] [PubMed] [Google Scholar]
- 8.Warwick MM, Lawrie SM, Beveridge A, Johnstone EC. Abnormal cerebral asymmetry and schizophrenia in a subject with Klinefelter syndrome (XXY) Biol Psychiatry. 2003;53:627–9. doi: 10.1016/S0006-3223(02)01484-1. [DOI] [PubMed] [Google Scholar]
- 9.Rezaie R, Daly EM, Cutter WJ, Murphy DG, Robertson DM, DeLisi LE, Mackay CE, Barrick TR, Crow TJ, Roberts N. The influence of sex chromosome aneuploidy on brain asymmetry. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:74–85. doi: 10.1002/ajmg.b.30772. [DOI] [PubMed] [Google Scholar]
- 10.Skakkebæk A, Gravholt CH, Rasmussen PM, Bojesen A, Jensen JS, Fedder J, Laurberg P, Hertz JM, Ostergaard JR, Pedersen AD, et al. Neuroanatomical correlates of Klinefelter syndrome studied in relation to the neuropsychological profile. Neuroimage Clin. 2013;4:1–9. doi: 10.1016/j.nicl.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen D, Liu D, Liu H, Clasen L, Giedd J, Davatzikos C. Automated morphometric study of brain variation in XXY males. Neuroimage. 2004;23:648–53. doi: 10.1016/j.neuroimage.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Warwick MM, Doody GA, Lawrie SM, Kestelman JN, Best JJ, Johnstone EC. Volumetric magnetic resonance imaging study of the brain in subjects with sex chromosome aneuploidies. J Neurol Neurosurg Psychiatry. 1999;66:628–32. doi: 10.1136/jnnp.66.5.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giedd JN, Clasen LS, Wallace GL, Lenroot RK, Lerch JP, Wells EM, Blumenthal JD, Nelson JE, Tossell JW, Stayer C, et al. XXY (Klinefelter syndrome): a pediatric quantitative brain magnetic resonance imaging case-control study. Pediatrics. 2007;119:e232–40. doi: 10.1542/peds.2005-2969. [DOI] [PubMed] [Google Scholar]
- 14.Iitsuka Y, Bock A, Nguyen DD, Samango-Sprouse CA, Simpson JL, Bischoff FZ. Evidence of skewed X-chromosome inactivation in 47,XXY and 48,XXYY Klinefelter patients. Am J Med Genet. 2001;98:25–31. doi: 10.1002/1096-8628(20010101)98:1<25::AID-AJMG1015>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 15.Bruining H, van Rijn S, Swaab H, Giltay J, Kates W, Kas MJ, van Engeland H, de Sonneville L. The parent-of-origin of the extra X chromosome may differentially affect psychopathology in Klinefelter syndrome. Biol Psychiatry. 2010;68:1156–62. doi: 10.1016/j.biopsych.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehta A, Malek-Jones M, Bolyakov A, Mielnik A, Schlegel PN, Paduch DA. Methylation-specific PCR allows for fast diagnosis of X chromosome disomy and reveals skewed inactivation of the X chromosome in men with Klinefelter syndrome. J Androl. 2012;33:955–62. doi: 10.2164/jandrol.111.016030. [DOI] [PubMed] [Google Scholar]
- 17.Singer H, Walier M, Nüsgen N, Meesters C, Schreiner F, Woelfle J, Fimmers R, Wienker T, Kalscheuer VM, Becker T, et al. Methylation of L1Hs promoters is lower on the inactive X, has a tendency of being higher on autosomes in smaller genomes and shows inter-individual variability at some loci. Hum Mol Genet. 2012;21:219–35. doi: 10.1093/hmg/ddr456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerkel K, Schupf N, Hatta K, Pang D, Salas M, Kratz A, Minden M, Murty V, Zigman WB, Mayeux RP, et al. Altered DNA methylation in leukocytes with trisomy 21. PLoS Genet. 2010;6:e1001212. doi: 10.1371/journal.pgen.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davidsson J, Veerla S, Johansson B. Constitutional trisomy 8 mosaicism as a model for epigenetic studies of aneuploidy. Epigenetics Chromatin. 2013;6:18. doi: 10.1186/1756-8935-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lepage JF, Hong DS, Mazaika PK, Raman M, Sheau K, Marzelli MJ, Hallmayer J, Reiss AL. Genomic imprinting effects of the X chromosome on brain morphology. J Neurosci. 2013;33:8567–74. doi: 10.1523/JNEUROSCI.5810-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newbury DF, Mari F, Sadighi Akha E, Macdermot KD, Canitano R, Monaco AP, Taylor JC, Renieri A, Fisher SE, Knight SJ. Dual copy number variants involving 16p11 and 6q22 in a case of childhood apraxia of speech and pervasive developmental disorder. Eur J Hum Genet. 2013;21:361–5. doi: 10.1038/ejhg.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Priebe L, Degenhardt FA, Herms S, Haenisch B, Mattheisen M, Nieratschker V, Weingarten M, Witt S, Breuer R, Paul T, et al. Genome-wide survey implicates the influence of copy number variants (CNVs) in the development of early-onset bipolar disorder. Mol Psychiatry. 2012;17:421–32. doi: 10.1038/mp.2011.8. [DOI] [PubMed] [Google Scholar]
- 23.Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–5. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 24.Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, D’arcy M, deBerardinis R, Frackelton E, Kim C, et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol Psychiatry. 2010;15:637–46. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geschwind DH, Boone KB, Miller BL, Swerdloff RS. Neurobehavioral phenotype of Klinefelter syndrome. Ment Retard Dev Disabil Res Rev. 2000;6:107–16. doi: 10.1002/1098-2779(2000)6:2<107::AID-MRDD4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Richter S, Dimitrova A, Maschke M, Gizewski E, Beck A, Aurich V, Timmann D. Degree of cerebellar ataxia correlates with three-dimensional mri-based cerebellar volume in pure cerebellar degeneration. Eur Neurol. 2005;54:23–7. doi: 10.1159/000087241. [DOI] [PubMed] [Google Scholar]
- 27.Laval SH, Dann JC, Butler RJ, Loftus J, Rue J, Leask SJ, Bass N, Comazzi M, Vita A, Nanko S, et al. Evidence for linkage to psychosis and cerebral asymmetry (relative hand skill) on the X chromosome. Am J Med Genet. 1998;81:420–7. doi: 10.1002/(SICI)1096-8628(19980907)81:5<420::AID-AJMG11>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 28.Blumenthal JD, Baker EH, Lee NR, Wade B, Clasen LS, Lenroot RK, Giedd JN. Brain morphological abnormalities in 49,XXXXY syndrome: A pediatric magnetic resonance imaging study. Neuroimage Clin. 2013;2:197–203. doi: 10.1016/j.nicl.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shear PK, Sullivan EV, Lane B, Pfefferbaum A. Mammillary body and cerebellar shrinkage in chronic alcoholics with and without amnesia. Alcohol Clin Exp Res. 1996;20:1489–95. doi: 10.1111/j.1530-0277.1996.tb01153.x. [DOI] [PubMed] [Google Scholar]
- 30.Bollati V, Galimberti D, Pergoli L, Dalla Valle E, Barretta F, Cortini F, Scarpini E, Bertazzi PA, Baccarelli A. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav Immun. 2011;25:1078–83. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Baccarelli A, Shu XO, Ji BT, Yu K, Tarantini L, Yang G, Li HL, Hou L, Rothman N, et al. Blood leukocyte Alu and LINE-1 methylation and gastric cancer risk in the Shanghai Women’s Health Study. Br J Cancer. 2012;106:585–91. doi: 10.1038/bjc.2011.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin W, Zhou X, Zhang M, Li Y, Miao S, Wang L, Zong S, Koide SS. Expression and function of the HSD-3.8 gene encoding a testis-specific protein. Mol Hum Reprod. 2001;7:811–8. doi: 10.1093/molehr/7.9.811. [DOI] [PubMed] [Google Scholar]
- 33.Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–7. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 34.Machinis K, Pantel J, Netchine I, Léger J, Camand OJ, Sobrier ML, Dastot-Le Moal F, Duquesnoy P, Abitbol M, Czernichow P, et al. Syndromic short stature in patients with a germline mutation in the LIM homeobox LHX4. Am J Hum Genet. 2001;69:961–8. doi: 10.1086/323764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda M, Aleksic B, Yamada K, Iwayama-Shigeno Y, Matsuo K, Numata S, Watanabe Y, Ohnuma T, Kaneko T, Fukuo Y, et al. Genetic evidence for association between NOTCH4 and schizophrenia supported by a GWAS follow-up study in a Japanese population. Mol Psychiatry. 2013;18:636–8. doi: 10.1038/mp.2012.74. [DOI] [PubMed] [Google Scholar]
- 36.Ho SK, Kovacević N, Henkelman RM, Boyd A, Pawson T, Henderson JT. EphB2 and EphA4 receptors regulate formation of the principal inter-hemispheric tracts of the mammalian forebrain. Neuroscience. 2009;160:784–95. doi: 10.1016/j.neuroscience.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Liebl DJ, Morris CJ, Henkemeyer M, Parada LF. mRNA expression of ephrins and Eph receptor tyrosine kinases in the neonatal and adult mouse central nervous system. J Neurosci Res. 2003;71:7–22. doi: 10.1002/jnr.10457. [DOI] [PubMed] [Google Scholar]
- 38.Dolga AM, Culmsee C. Protective Roles for Potassium SK/K(Ca)2 Channels in Microglia and Neurons. Front Pharmacol. 2012;3:196. doi: 10.3389/fphar.2012.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunning M, Lynch A, Eldridge M. IlluminaHumanv4.db: Illumina HumanHT12v4 annotation data (chip illuminaHumanv4). R package version 1180, 2013.
- 40.Wong CC, Caspi A, Williams B, Houts R, Craig IW, Mill J. A longitudinal twin study of skewed X chromosome-inactivation. PLoS One. 2011;6:e17873. doi: 10.1371/journal.pone.0017873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Craig IW. Imprinting, Inactivation and the Behaviour Genetics of the X Chromosome. In: Petronis A, Mill J, eds. Brain, behavior and epigenetics. Heidelberg, Germany: Springer, 2011:127-8. [Google Scholar]
- 42.Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–67. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 44.Ferguson-Smith MA. Karyotype-Phenotype Correlations in Gonadal Dysgenesis and Their Bearing on the Pathogenesis of Malformations. J Med Genet. 1965;2:142–55. doi: 10.1136/jmg.2.2.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werler S, Poplinski A, Gromoll J, Wistuba J. Expression of selected genes escaping from X inactivation in the 41, XX(Y)* mouse model for Klinefelter syndrome. Acta Paediatr. 2011;100:885–91. doi: 10.1111/j.1651-2227.2010.02112.x. [DOI] [PubMed] [Google Scholar]
- 46.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 47.Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: an X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci U S A. 1999;96:7364–9. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lahn BT, Page DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–80. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 49.El-Mogharbel N. JAM G. X and Y Chromosomes: Homologous Regions. Encyclopedia of Life Sciences 2008:1-9. [Google Scholar]
- 50.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–37. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sargent CA, Briggs H, Chalmers IJ, Lambson B, Walker E, Affara NA. The sequence organization of Yp/proximal Xq homologous regions of the human sex chromosomes is highly conserved. Genomics. 1996;32:200–9. doi: 10.1006/geno.1996.0106. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz A, Chan DC, Brown LG, Alagappan R, Pettay D, Disteche C, McGillivray B, de la Chapelle A, Page DC. Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum Mol Genet. 1998;7:1–11. doi: 10.1093/hmg/7.1.1. [DOI] [PubMed] [Google Scholar]
- 53.Williams NA, Close JP, Giouzeli M, Crow TJ. Accelerated evolution of Protocadherin11X/Y: a candidate gene-pair for cerebral asymmetry and language. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:623–33. doi: 10.1002/ajmg.b.30357. [DOI] [PubMed] [Google Scholar]
- 54.Crow TJ. The XY gene hypothesis of psychosis: Origins and current status. Am J Med Genet B Neuropsychiatr Genet. 2013 doi: 10.1002/ajmg.b.32202. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ross NL, Wadekar R, Lopes A, Dagnall A, Close J, Delisi LE, Crow TJ. Methylation of two Homo sapiens-specific X-Y homologous genes in Klinefelter syndrome (XXY) Am J Med Genet B Neuropsychiatr Genet. 2006;141B:544–8. doi: 10.1002/ajmg.b.30339. [DOI] [PubMed] [Google Scholar]
- 56.Melchior L, Bertelsen B, Debes NM, Groth C, Skov L, Mikkelsen JD, Brøndum-Nielsen K, Tümer Z. Microduplication of 15q13.3 and Xq21.31 in a family with tourette syndrome and comorbidities. Am J Med Genet B Neuropsychiatr Genet. 2013 doi: 10.1002/ajmg.b.32186. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 57.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2013. [Google Scholar]
- 59.Hansen KD, Aryee, M. Minfi: Analyze Illumina's 450k methylation arrays. R package version 1.2.0., 2013.
- 60.Davis S, Du P, Bilke S, Triche T, Bootwalla M. Methylumi: Handle Illumina methylation data. R package version 2.2.0. 2012.
- 61.Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pidsley R, Y Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28:2986–8. doi: 10.1093/bioinformatics/bts545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 65.Allen JD, Chen M, Xie Y. Model-Based Background Correction (MBCB): R Methods and GUI for Illumina Bead-array Data. J Cancer Sci Ther. 2009;1:25–7. doi: 10.4172/1948-5956.1000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes G, et al. Quality control procedures for genome-wide association studies. Curr Protoc Hum Genet 2011; Chapter 1:Unit 1 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakahori Y, Hamano K, Iwaya M, Nakagome Y. Sex identification by polymerase chain reaction using X-Y homologous primer. Am J Med Genet. 1991;39:472–3. doi: 10.1002/ajmg.1320390420. [DOI] [PubMed] [Google Scholar]
- 71.Rosa A, Picchioni MM, Kalidindi S, Loat CS, Knight J, Toulopoulou T, Vonk R, van der Schot AC, Nolen W, Kahn RS, et al. Differential methylation of the X-chromosome is a possible source of discordance for bipolar disorder female monozygotic twins. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:459–62. doi: 10.1002/ajmg.b.30616. [DOI] [PubMed] [Google Scholar]
- 72.Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–39. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.