Abstract
The 3rd European Workshop on Plant Chromatin (EWPC) was held on August 2013 in Madrid, Spain. A number of different topics on plant chromatin were presented during the meeting, including new factors mediating Polycomb Group protein function in plants, chromatin-mediated reprogramming in plant developmental transitions, the role of histone variants, and newly identified chromatin remodeling factors. The function of interactions between chromatin and transcription factors in the modulation of gene expression, the role of chromatin dynamics in the control of nuclear processes and the influence of environmental factors on chromatin organization were also reported. In this report, we highlight some of the new insights emerging in this growing area of research, presented at the 3rd EWPC.
Keywords: chromatin, epigenetics, plants, development, chromatin reprogramming
Introduction
Following the two previous editions, which took place in Zurich and Versailles,1,2 a large representation of European researchers working on plant chromatin met for the 3rd European Workshop on Plant Chromatin (3rd EWPC). The workshop was held at the Centro de Biotecnología y Genómica de Plantas (CBGP, Madrid, Spain), and served as a platform to discuss recent progress in several aspects of chromatin structure and dynamics in plants.
The structural dynamics of chromatin is essential for an appropriate control of numerous biological processes during the life cycle of plants. Plant chromatin is continuously remodeled to promote changes in gene expression and in that way orchestrate developmental programs and respond to a fluctuating environment. Besides its role in transcriptional control, chromatin dynamics is also involved in the modulation of other cellular processes, such as genome stability, and DNA replication and repair. The modulation of chromatin organization during these processes occurs through the covalent modifications of histones and DNA, and also by the change of nucleosome position, the destabilization nucleosomes or the substitution of canonical histones by histone variants. During the 3rd EWPC, a number of topics related to chromatin structure and function were discussed. New components of plant chromatin remodeling complexes, including Polycomb group (PcG) proteins, required to mediate the role of chromatin in transcription and chromosomal organization, were presented. The essential role of epigenetic reprogramming in plant developmental transitions was also discussed. In addition, several reports dealt with the role of histone variants and novel modifiers of histones in chromatin dynamics. The relevance of the interactions between transcription factor-mediated regulation of gene expression and chromatin remodeling processes was also emphasized in a number of contributions. The idea that all nuclear processes involving DNA take place in the context of chromatin is changing our perception of how these processes are regulated. Finally, a number of chromatin responses to environmental signals in plants were covered in different communications.
This review summarizes some of the topics reported during the meeting, highlighting new insights emerging in this growing area of research. We apologize to those groups whose work is not reported or fully discussed here due to space limitations.
PcG Proteins: Old Fellows with New Mates
PcG proteins are key transcriptional regulators with important functions in developmental regulation in plants.3 Trithorax group (TrxG) proteins also play central roles in the transcriptional regulation of plant development by antagonizing the function of PcG proteins.4 Both PcG and TrxG proteins form complexes that catalyze the modification of specific histone residues, leading to chromatin-mediated changes in gene activity.
A number of laboratories reported the identification of proteins that work together with these complexes in the regulation of gene expression, providing novel and exciting insights into PcG mechanism of action. J. Goodrich (Edinburgh, UK) presented a novel TrxG member, SUPPRESSOR OF POLYCOMB 12 (SOP12), identified through a genetic screen for suppressors of mutants in the Arabidopsis thaliana PcG gene CURLY LEAF (CLF). SOP12 encodes a highly conserved plant-specific Harbinger transposase nuclease-like protein that has lost transposase activity and antagonizes POLYCOMB REPRESSOR COMPLEX 2 (PRC2) function. The same gene was independently identified by F. Turck’s lab (Cologne, Germany) in a screen for suppressors of mutants in another PcG gene, LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), and was named ANTAGONIST OF LHP1 1 (ALP1).5 These observations suggest that ALP1/SOP12 interacts with multiple PcG members.
Although PcG components are well conserved in eukaryotes,6 plant-specific proteins involved in this pathway might have appeared during evolution to cover specific developmental requirements in plants. S. Farrona (Düsseldorf, Germany) presented the PWWP-DOMAIN INTERACTOR OF POLYCOMBS 1 (PWO1) protein as a new component of the PcG protein pathway. PWO1 belongs to a small plant-specific subfamily of PWWP proteins, containing a structural module characteristic of chromatin regulators.7 The analysis of pwo1 mutants showed that this gene interacts with CLF, revealing an important function of PWO1 in the regulation of plant development. PWO1 might be a putative regulator of PRC2 recruitment to chromatin and the binding of PWO1-PWWP is modulated by histone modifications. Analyses of PWO1 complex(es) in planta is expected to provide further insights into the role of PWO1 and PcG proteins on plant transcriptional regulation.
Several chromatin-modifying activities have been shown to modulate the expression of master genes of flowering.8,9 In fact, the study of vernalization, the requirement for a prolonged exposure to cold to induce flowering, in Arabidopsis has become a paradigm of epigenetic regulation of gene expression in plants. Vernalization is mediated by silencing of the floral repressor FLOWERING LOCUS C (FLC) through a conserved PRC2-dependent mechanism.10 J. Questa (Norwich, UK) presented data supporting the hypothesis that modulation of chromatin silencing mechanisms and non-coding RNA regulation through cis polymorphisms could be a general mechanism underlying adaptation to changing environments.11 The cold-induced nucleation of chromatin silencing and the maintenance of this silenced state upon return of plants to warm conditions have a central role in this response. Moreover, the long non-coding RNA COOLAIR is also involved in the modulation of this process accelerating FLC downregulation during cold.10 A detailed understanding of the molecular basis for the vernalization response is likely to provide working models for a variety of other plant biological processes that require a cellular memory.
PcG-mediated regulation plays important roles during seed germination, which requires silencing of seed developmental genes in A. thaliana.12 W.-H. Shen (Strasbourg, France) presented evidence of transcriptional gene repression mediated by PcG proteins in the control of this process. His group identified members of the ALFIN-LIKE family, nuclear proteins that bind H3K4me3, as novel PRC1-associated factors. The characterization of these PRC1-interacting proteins led to the conclusion that complexes containing these PcG proteins are required during germination for the switch from the H3K4me3-associated active chromatin to the H3K27me3-associated repressive transcription state of seed developmental genes.13 F. Bratzel (Heidelberg, Germany) reported that VAL B3-domain transcription factors and AtBMI1 protein complexes also participate in the PcG-mediated repression of the seed maturation program after germination to allow vegetative development.14 AtBMI-mediated H2A ubiquitination initiates the repression of the seed maturation program and subsequent activity of PRC2 deposits H3K27me3 to stably maintain repression. Moreover, PRC1 and PRC2 regulate each other’s activities, showing that PcG repression of seed maturation genes follows unexpected rules that place PRC1-mediated H2Aub and gene repression upstream of PRC2-mediated H3K27me3 deposition and maintenance (Fig. 1). These observations challenge the current view on interdependency of PRC complexes and uncover different PcG mechanisms mediating distinct levels of gene repression.14
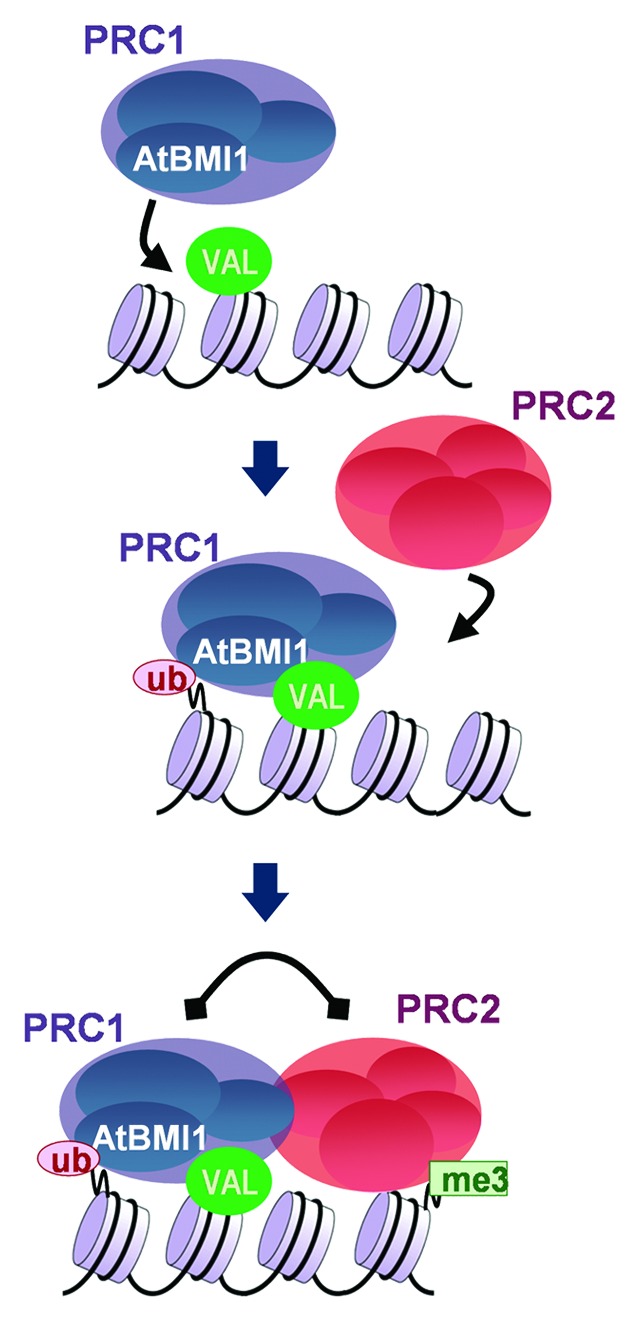
Figure 1. PcG protein regulation of seed maturation genes. Interplay between PRC1 and PRC2 represses seed maturation genes following unexpected rules that challenge the traditional views on PcG protein function. Courtesy of M. Calonje (Seville, Spain).
Additional partners of PcG proteins were presented. Screening by yeast two-hybrid revealed that BPC6 interacted with PRC complex members (A. Hecker, Tuebingen, Germany). This interaction was verified by a set of in vitro and in vivo methods that propose the formation of a novel complex that might be involved in the recruitment of PRC components to DNA in a sequence specific manner.
As potential part of PRC1, LHP1 is involved in chromatin-mediated gene repression.15 Although LHP1 is encoded by a single-copy gene,16 lhp1 loss-of-function mutants display mild phenotypes compared with those associated with full or partial loss-of-function of other PRC1 components. This suggests that PRC1 can repress most targets without LHP1 or that other unknown components have a redundant function with LHP1. F. Turck (Cologne, Germany) described a screen for genetic enhancers and suppressors of lhp1 that identified two ANTAGONISTS OF LHP1 (ALPs) and four ENHANCERS OF LHP1 (ELPs) so far. As mentioned earlier, SOP12/ALP1 is related to Harbinger transposases.5 ELPs fall in two distinct categories and are either related to DNA replication or to telomere function. Further characterization of these loci will provide new insights on the function of LHP1.
I. del Olmo (Madrid, Spain) presented evidence of interplay between the DNA replication machinery and the PcG protein complexes in the repression of flowering through a mechanism involving epigenetic gene silencing. A mutation in EARLY IN SHORT DAYS 7 (ESD7), a gene encoding the catalytic subunit of the DNA polymerase ε complex in Arabidopsis, esd7–1, causes similar pleiotropic phenotypic alterations to those displayed by PRC1 and PRC2 mutants. ESD7 interacts genetically and physically with LHP1.17 Furthermore, ESD7 and several genes encoding PRC2 components interact synergistically. These observations open new perspectives in the study of possible interactions between PcG complexes and DNA polymerases and may help to unveil how epigenetic information at pivotal loci in developmental control is transmitted within cell lineages from one cell to its progeny.
The activity of PcG proteins also appears to be required, together with DNA methylation, for imprinted gene expression, as shown by the loss of PHERES (PHE) parent of origin expression in medea mutants.18 Genomic imprinting is a phenomenon where only one of the two alleles of a particular gene is expressed—either the maternally or the paternally inherited allele. However, knowledge about the effect of H3K27me3 on imprinting is limited to individual genes. In the laboratory of C. Köhler (Uppsala, Sweden) global profiling techniques have been used in specific cell types (endosperm) to understand how genomic imprinting is regulated. J. Moreno showed the results obtained to set up the technology required to analyze this histone modification in a parent-specific allele and in genome-wide manner. This approach includes the use of Isolation of Nuclei TAgged in specific Cell Types (INTACT) methods19 based on the PHE promoter. Preliminary results suggest the advantages of the approach to obtain allele-specific levels of H3K27me3 and, therefore, parent of origin H3K27me3 profiles are expected to be available soon.
In addition to regulation of developmental programs, PcG proteins appear to be also involved in responses to environmental stresses.20 V. Gaudin (Versailles, France) presented data on the characterization of LIF2, an RNA-binding protein that interacts with LHP1.21 Besides a role in the maintenance of cell determination in floral meristems, LIF2 is involved in defense pathways. These data highlight new roles of PcG complexes in these responses and a new trade-off between growth and defense in plants.
Epigenetic Reprogramming in Plant Developmental Transitions
A number of developmental transitions take place during the life cycle of plants. For instance, somatic-to-reproductive switch occurs upon differentiation of the spore mother cells that will undergo meiosis. This transition leads to the generation of the haploid gametophytes that give rise to gametes. The results presented by C. Baroux (Zurich, Switzerland) showed that in the female megaspore mother cell in A. thaliana this cell fate switch is accompanied by a large-scale epigenetic reprogramming. This resetting probably creates a distinct transcriptional and epigenetic status that distinguishes these cells from the surrounding somatic tissue.22 Chromatin decondensation, depletion of linker histones, changes in the levels of histone variants as well as alterations in histone covalent modification landscapes are characteristic of this transition and have been proposed to be involved in the establishment of the competence for post meiotic gametophyte pluripotency (Fig. 2). These observations lead to the conclusion that, as in animal primordial germ cells, the somatic-to-reproductive transition in plant species is likely to be associated with large-scale epigenetic reprogramming.23
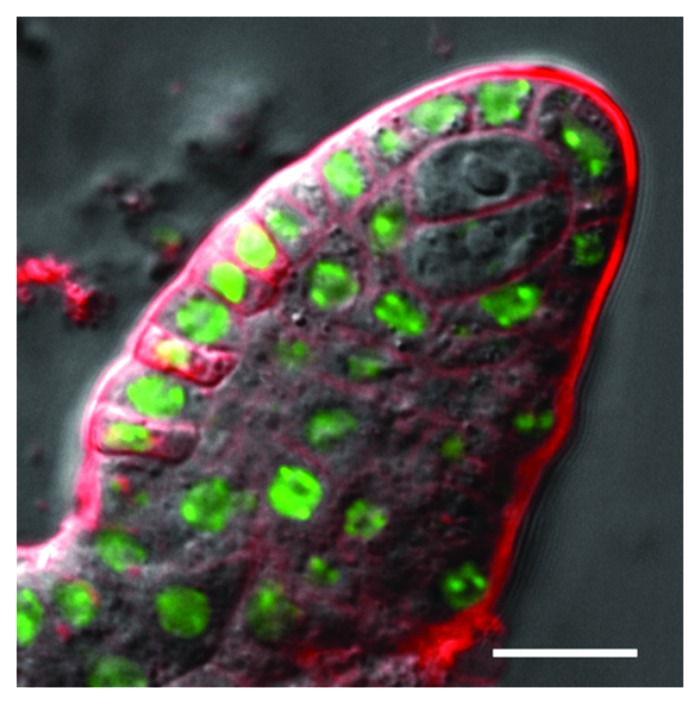
Figure 2. H1 eviction marks the somatic-to-reproductive fate transition in Arabidopsis ovule primordia. H1.1-GFP (green) is present in all nucellar cells of the ovule primordium but is evicted upon differentiation of megaspore mother cells (MMC). This image shows an ago9–4 ovule primordium producing ectopic cells engaged in an ameiotic gametophytic fate,64 showing H1 eviction like wild type MMCs. Scale bar: 10 µm. Courtesy of C. Baroux (Zurich, Switzerland).
Histone Variants: Small Changes Make a Big Difference
Recent observations underscore the pivotal role that histone variants play in the regulation of gene expression in the context of a number of plant biological processes. For instance, the SWR1 complex (SWR1-C), widely conserved in eukaryotic organisms, is required to exchange histone H2A by the histone variant H2A.Z, and this substitution has implications on the expression levels of underlying genes.24 In yeast, SWR1-C shares four subunits with the NuA4 complex (NuA4-C), which has histone acetyltransferase activity and is involved in the acetylation of histones, including H4 and H2A.Z.25 The functional relationship between these two complexes suggests that acetylation mediated by a putative Arabidopsis NuA4-C may also have a role in the regulation of flowering time. A. Mouriz (Madrid, Spain) presented data concerning the characterization of the role of the NuA4-C in the control of development in Arabidopsis. The characterization of mutants affected in subunits of the AtNuA4-C suggests that the function of this complex is required for a proper control of the floral transition.
Plant development is highly responsive to environmental factors, including temperature. However, although H2A.Z has a prevalent role in the thermosensory pathway in plants, how temperature is sensed by plants remains unknown.26 Ongoing experiments in the laboratory of P. Wigge (Cambridge, UK) are addressing this question. S. Cortijo showed that although H2A.Z is important for activation of temperature responsive genes, this histone variant does not seem to be required for the repression of these genes upon prolonged exposure to high temperature. The nature of this shutdown mechanism remains unidentified, and the putative role for other chromatin remodeling complexes in the transcriptional response to warm temperature will have to be analyzed in future experiments.
Chromatin dynamics participate in every DNA-templated process, including transcription, replication, recombination and repair.27 Chromatin remodeling activities are necessary to allow the access of DNA repair enzymes to genomic sites where DNA damage has occurred. O. Mittelsten Scheid (Vienna, Austria) presented evidence showing that the SWR1-C is required for DNA repair in Arabidopsis.28 Moreover, swr1 mutants are less effective in homologous recombination. Defects in fertility and alterations in meiosis suggest that SWR1-C is also necessary for proper meiosis.
Variants of the linker histone H1 also seem to mediate gene expression changes in response to environmental factors.29 In Arabidopsis there are two canonical histone H1 variants, H1.1 and H1.2, and one stress-inducible, H1.3,30 particularly under low light conditions. The results presented by K. Rutowicz (Warsaw, Poland) indicated that while H1.1 and H1.2 proteins showed a general pattern of expression, H1.3 was only ubiquitously expressed upon exposure to stress conditions. She also showed that the chromatin binding properties of H1.3 are different from those of the other histone H1 variants, suggesting that H1.3 is involved in plant adaptation to stress by facilitating chromatin changes that mediate phenotypic plasticity. Future studies will unveil how the relative ratios of H1 variants could mediate gene expression changes upon exposure to stress conditions.
Dynamic deposition of histone H3 variants accompanies developmental remodeling of the Arabidopsis transcriptome.31 M. Benoit (Aubière, France) highlighted the role of histone H3 variants and histone chaperones in the dynamics of heterochromatin organization in Arabidopsis cotyledons during early stages of post-germination development. During this period, the transcript levels of histone H3 variant genes and components of some histone chaperones involved in their deposition display significant variations. Moreover, the organization of 180 bp repeats is altered in mutants affected in genes encoding subunits of these chaperones. Possible changes in transcription, nucleosome and histone modifications associated with this reorganization of heterochromatin will need to be assessed to further understand the dynamics of heterochromatin organization.
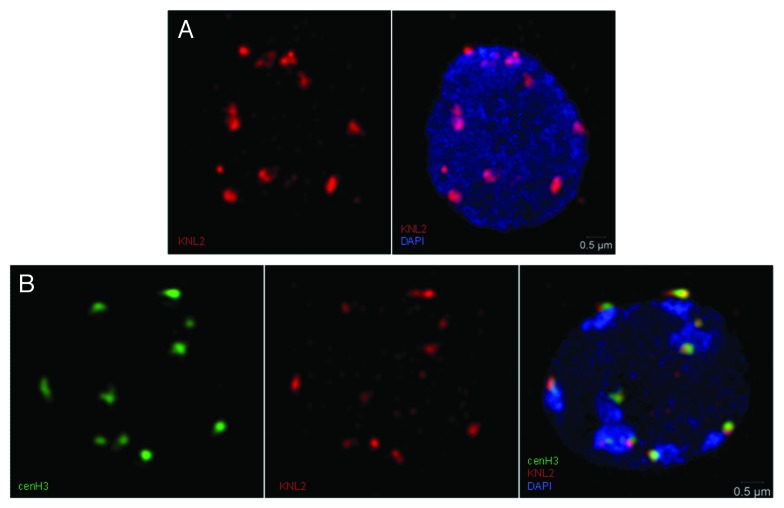
The centromeric histone H3 variant (cenH3) is necessary for the assembly and proper function of kinetochores during mitosis and meiosis.32 In Arabidopsis, knockdown of cenH3 causes a reduction of growth and fertility.33 Deposition of cenH3 onto centromeric chromatin is a complex process that involves the recognition of centromeres, loading of the newly synthesized histone and maintenance of the incorporated cenH3. I. Lermontova (Gatersleben, Germany) presented results showing that the Arabidopsis KINETOCHORE NULL2 (KNL2) is part of the machinery involved in the assembly of cenH3. The KNL2 protein is associated with centromeres and colocalizes with cenH3 (Fig. 3). Loss-of-function of KNL2 results in reduced levels of the cenH3 transcript, decreased cenH3 deposition and defects in mitosis and meiosis, as well as markedly reduced fertility. The observations reported suggest that KNL2 influences the epigenetic environment required for centromere function.34
Figure 3. KNL2 colocalizes with cenH3. (A) Localization of KNL2 in meristematic interphase nucleus of A thaliana (in red) and DAPI counterstaining (blue). (B) Colocalization of cenH3 (left, green) and KNL2 (middle, red) in meristematic interphase nucleus of A. thaliana. Overlay and DAPI counterstaining (right). Courtesy of I. Lermontova (Gatersleben, Germany).
New Players on the Posttranslational Modifications of Plant Histones
Histone modifications influence a number of biological processes related to DNA function, including DNA replication and repair, gene expression control and chromosome segregation.35 Specifically, methylation of lysine (K) residues on histone tails conferred by SET domain proteins influences chromatin structure and is involved in maintenance of transcriptional states of euchromatic genes, suppression of the activity of transposable elements (TEs), and compaction of heterochromatin.36 Although chromatin structure and histone modifications change during the cell cycle, few cell cycle-related chromatin modifiers have been characterized. R. B. Aalen (Oslo, Norway) described the identification and characterization of a SET domain protein needed to coordinate cell divisions in the Arabidopsis root meristem (Fig. 4). The corresponding gene is expressed in the quiescent center (QC) and the surrounding stem cells of the root stem cell niche, and is needed for normal cell division activity in the meristematic zone (MZ). Each cell in the MZ goes through a limited number of cell divisions with a pattern of synchronized replication of sister cells, which may facilitate symmetric root development. The identified SET domain protein displays H3K36 monomethyltransferase activity, and this histone modification is suggested to be used as a counting system for synchronization of MZ cell divisions.
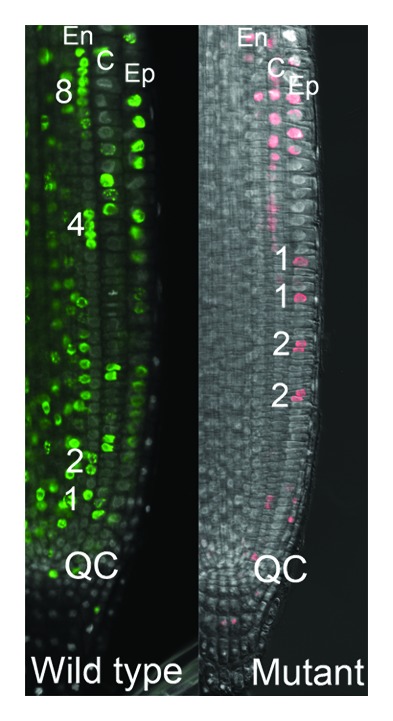
Figure 4. Coordinated replication in root meristem cell files is dependent on a SET-domain gene. Wild type (Col-0) and SET-domain gene mutant root meristems stained 1 h with the thymidine analog EdU to visualize DNA-replicating nuclei (in green and pink, respectively). The wild type image exemplifies a coordinated pattern, in this case along an endodermal (En) cell file, with a replicating singlet (1) nearest the quiescent center (QC), followed by a duplet (2), quartet (4) and octet (8) of co-replicating daughter, granddaughter and great-granddaughter cells. This coordinated replication pattern is distorted in the SET-domain gene mutant. C, cortex; Ep, epidermis. Courtesy of R. B. Aalen (Oslo, Norway); images by Robert Kumpf.
H3 phosphorylation also has a crucial role in transcription, apoptosis, DNA repair and cell cycle-dependent chromosome condensation and segregation.37 E. Tomastikova (Olomouc, Czech Republic) presented evidence that at least two members of the Aurora family of kinases in Arabidopsis are involved in the site-specific phosphorylation of histone H3. The activity of both kinases is influenced by other posttranscriptional modifications present in histone H3. Moreover, the influence that many chromatin marks appear to have on the activity of these Aurora kinases suggests a role for these kinases in the control not only of chromosome segregation but also of transcriptional activation or repression.
CenH3 is essential for chromosome segregation in eukaryotes and also undergoes covalent posttranslational modifications. Interested in understanding the role of cenH3 phosphorylation in plants during the cell cycle, D. Demidov (Gatersleben, Germany) showed that certain Aurora kinases interact with cenH3 in vivo and that cenH3 is phosphorylated in vitro by these kinases in specific positions. Targeted mutation of the phosphorylated residues in cenH3 caused phenotypic alterations in Arabidopsis, indicating that regulated phosphorylation of specific cenH3 residues is necessary for proper plant development.
Unveiling Novel Interactions between Plant Chromatin and Transcription Factors
Chromatin dynamics underlies gene silencing and activation mechanisms required for plant developmental programs.38 A. Pajoro (Wageningen, The Netherlands) presented evidence that flower development is associated with dynamic changes in chromatin accessibility and DNA binding of transcription factors (TF). MADS-domain TFs play essential roles in establishing the identities of floral organs39 and interact with chromatin remodeling proteins during flower development.40 The characterization of the dynamics of chromatin accessibility and DNA-binding of MADS-domain TFs lead to the conclusion that binding of these transcriptional regulators to DNA is largely independent of chromatin accessibility, and that chromatin becomes more accessible after TF DNA-binding. This suggests that MADS-domain proteins can act as ‘pioneer factors’ that promote changes in chromatin accessibility to regulate gene expression.41 The SAND domain-containing proteins ULTRAPETALA1 (ULT1) and ULT2 have overlapping functions in regulating shoot and floral stem cell function, with ULT1 playing the main role.42 ULT1 antagonizes the activity of PcG proteins by repressing the deposition of repressive histone marks at target loci.43 C. Carles (Grenoble, France) showed that ULT1 physically interacts with a number of chromatin modifying proteins and these interactions suggest possible mechanisms mediating the role of ULT1 in transcriptional activation. Moreover, ULT1 also interacts with TFs, suggesting that this protein could link chromatin remodeling with transcriptional activation. Future studies are likely to unveil new interactions between TFs of different families with a variety of chromatin remodeling complexes and will contribute to reveal how molecular mechanisms underlying gene expression control work in plants and other eukaryotes.
Plant Chromatin Complexes Modulating other Aspects of Transcription and Chromosomal Organization
Modifications of either DNA or histones have fundamental effects on chromatin structure and function and, thus, influence processes such as transcription, replication or recombination.44 The heterodimeric complex SPT4/SPT5 is a transcript elongation factor that directly interacts with RNA polymerase II (RNAP II) to regulate mRNA synthesis in the chromatin context.45 A. Pfab (Regensburg, Germany) reported the characterization of the Arabidopsis SPT4/SPT5 complex, demonstrating that it is conserved in plants.46 SPT4-RNAi plants display auxin signaling phenotypes with developmental defects and severely reduced growth. Consistently, auxin-related genes, most strikingly AUX/IAAs, are downregulated in these plants. He showed that SPT5 colocalizes with transcribing RNAP II in euchromatic regions and is found over the entire transcription unit of RNAP II-transcribed genes. In SPT4-RNAi plants, elevated levels of RNAP II and SPT5 were detected within transcribed regions, indicating transcript elongation defects in these plants.
C. Oliver (Madrid, Spain) presented data revealing the participation of AGO4 in the Arabidopsis meiotic process. AGO4 is part of the RNA induced transcriptional silencing complex (RISC), which binds to repeat associated siRNAs (ra-siRNAs), and is guided by ra-siRNAs to the target DNA involving RNA-directed DNA methylation (RdDM).47 The reduced fertility of ago4 mutants has been associated to defects in flower development, but a detailed analysis of specialized cellular division in pollen mother cells of ago4 Arabidopsis mutants unveiled a variety of meiotic abnormalities that could contribute to the partial sterility displayed by the mutants. Therefore, AGO4 could be involved in the silencing of genes involved in the key meiotic process of homologous chromosome segregation as recently described in mice.
Paramutation is another phenomenon dependent on epigenetic regulation in which in trans interactions between two alleles result in meiotically stable transcriptional silencing of one of them. M. Stam (Amsterdam, The Netherlands) reported on the collaborative project performed in her laboratory and that of V. Chandler (Arizona, USA) to understand the molecular basis underlying paramutation of the b1 gene, a maize locus controlling anthocyanin pigmentation.48 Paramutation at the b1 locus requires non-coding transcribed tandem repeats located 100 kb upstream of the gene. The RdDM pathway is necessary for paramutation mediated by endogenous as well as transgenic tandem repeats. In fact, the tandem repeats generate small RNAs, but these siRNAs seem not sufficient for paramutation. Moreover, extensive repeat DNA methylation is not required for paramutation by transgenes. There is, however, a correlation between the level of silencing and the extent of DNA methylation in the endogenous repeats.49
Chromatin dynamics also have an essential role in the silencing of rDNA,50 although the epigenetic mechanisms mediating the transcriptional control of these rDNA loci are not yet fully understood. Nucleolins are nucleolar proteins with histone chaperone activity involved both in RNA Polymerase I (RNAP I)-mediated transcription of rDNA and RNAP II transcription. There are two nucleolin homologs in Arabidopsis (AtNUCs)51 that are being characterized in the laboratory of J. Saez-Vasquez (Perpignan, France). These proteins appear to act antagonistically to modulate chromatin dynamics of rDNA genes and interact with the proteasome to control the chromatin status of rDNA genes.
The chromatin landscapes of origins of replication (ORIs) in plants were also discussed during the workshop. To understand the regulation of ORIs it is essential to unveil their link to transcriptional activation and epigenetic landscapes. J. Sequeira-Mendes (Madrid, Spain) compared the genome-wide distribution of ORIs in Arabidopsis52 with the profiles obtained for a number of chromatin marks. In synchronized Arabidopsis cultured cells, ORIs tend to be enriched in particular chromatin marks and some histone variants while other chromatin covalent modifications are depleted in ORIs. In addition, the analysis of ORIs is being performed at the whole organism level, revealing that only some of the ORIs were shared with those found in cultured cells. Moreover, ORIs appear to be frequently associated with particular features, although they are not associated with a single chromatin signature but rather with a range of signatures that contribute to the specification of different ORIs, revealing that plasticity in their chromatin environment is characteristic.
Telomeres are nucleoprotein structures present at the ends of eukaryotic chromosomes with an essential role in chromosome and genome stability. Classically considered to be heterochromatic regions, telomeres have been recently shown to contain also euchromatin features.53 M. Fojtova (Brno, Czech Republic) presented results suggesting a link between telomere maintenance and DNA methylation. Compromised in vivo telomerase action appears to be involved in the telomere shortening in hypomethylated A. thaliana plants.54
Plant Chromatin Responses to Stress/Environmental Factors
The regulation of gene expression patterns at the level of chromatin plays a central role in the capacity of plants to adapt to fluctuations in the environment. TEs provide an example of transcriptional alterations in response to environmental changes. TEs are repressed in both plant and animal genomes through the epigenetic inheritance of repressed chromatin and expression states. Stress situations lead to a transient transcriptional activation of endogenous repeats that are regulated by transcriptional gene silencing (TGS).55 O. Mittelsten Scheid (Vienna, Austria) discussed results obtained by her group indicating that TEs exploit defense mechanisms in plants.56 ONSEN retrotransposons are not expressed in wild type plants, but are activated upon heat stress.57 Efficient activation of ONSEN elements is accompanied by loss of nucleosomes and heterochromatin decondensation, and requires heat shock factors which also bind long-terminal repeats (LTR) in the mobile element. These observations suggest that ONSEN exploits for its amplification molecular mechanisms that plants use to respond to stressful conditions.
A number of other TEs and heterochromatic regions in A. thaliana are activated upon heat stress.58 L. López-González (Clermont-Ferrand, France) presented a genetic strategy aimed at identifying genes involved in the silencing of these TEs and in the release of silencing following the exposure to high temperature. A number of suppressors and enhancers were isolated following this screening. Among them, one of the mutants turned out to be affected in a gene encoding a member of the MEDIATOR (MED) complex that regulates transcription by RNAP II.59 As for other mutations in genes encoding MED subunits, this mutant displays developmental alterations such as a delay in flowering time. Consistent with this phenotype, master genes of flowering were misregulated in the mutant, suggesting a role for this protein in both transposon silencing and transcriptional control of euchromatic genes.
Epigenetic regulation of TEs is not exclusive of A. thaliana and analysis of other species may shed the light on its evolutionary trends. B. Pietzenuk (Cologne, Germany) presented results dealing with the regulation of TEs in A. lyrata, a close relative of A. thaliana. Previous reports suggested less efficient TE silencing in A. lyrata compared with A. thaliana.60,61 This is now studied at the genome-wide level by TE RNA-sequencing and will elucidate how this process is differentially regulated in these two relatives. In addition, a number of TE families are transcriptionally induced upon heat stimulus in A. thaliana and A. lyrata. Some of these elements are species-specific while others are common, suggesting that the response of several TE families is evolutionary conserved and might be part of the strategy for TE survival.
Besides the silencing of transposable elements, the activity of chromatin remodeling complexes is also necessary to maintain an inactive state of some euchromatic loci during normal growth. I. Mozgova (Uppsala, Sweden) discussed the role of the CHROMATIN ASSEMBLY FACTOR 1 (CAF1), a histone chaperone mediating DNA replication-coupled histone deposition, in regulation of stress-responsive genes in Arabidopsis. Variations in environmental cues such as light also cause massive changes in the status of chromatin, as discussed by F. Barneche (Paris, France). A rapid reprogramming of gene expression takes place upon switching the Arabidopsis seedling from dark to light conditions. This is an excellent system to analyze the impact of chromatin states on transcriptional reprogramming and to unveil the effect of light changes in chromatin dynamics on responsive genes. Global profiling studies revealed changes in the levels of histone marks such as ubiquitinated histone H2B (H2Bub) associated with light in a number of genes, which frequently impacts rapid modulation of gene expression in response to this environmental factor.62
The identification of novel components of chromatin has unveiled new aspects of chromatin participation in the response of plants to stress. DEK is an ubiquitous protein that in mammalian cells colocalizes with chromatin enriched in acetylated histone H4.63 In A. thaliana there are several members of this family of proteins with architectural roles in chromatin. S. Waidmann (Vienna, Austria) described the characterization of one of the AtDEK proteins and its role in plant stress responses. This AtDEK protein interacts with histones in vivo, and also with other chromatin components. Binding of this protein to chromatin influences the expression of specific genes, although how the activity of AtDEK is regulated will be analyzed in future studies.
Proteomic approaches are contributing significantly to better understand how chromatin modulates changes in gene expression in response to a number of stresses. D. Pflieger (Evry, France) presented one example based on phosphoproteomics of chromatin in Arabidopsis plants exposed to a simulated biotic stress. Her group has identified differentially phosphorylated proteins bound to chromatin. Some of them were known MAPK substrates while others were putative targets of MAPK, as well as of other classes of kinases, and could represent key factors in the response of Arabidopsis plants to biotic stress. Further efforts will be required to establish the function of these proteins in gene expression dynamics during pathogen challenges.
What is Next?
In coming years, researchers on the plant chromatin field will sustain efforts to identify and characterize new factors involved in the deposition of chromatin marks, their recognition, and in translating these modifications into signals mediating the control of gene expression and other biological processes, such as DNA replication and repair, and chromosome organization and stability. A deep understanding of the function of these chromatin remodeling factors will also require the identification of the protein complexes in which they are present, as well as the targets they act upon. We are beginning to see how interactions between different chromatin marks, rather than a particular modification, are likely to influence the functional dynamics of chromatin. Further studies will probably unveil more examples of how the combined action of different chromatin remodeling activities and their interactions with sequence-specific transcription factors regulate nuclear processes that involve DNA. Hopefully, technical and methodological advances will ease the path to generate integrative models that can explain how plant chromatin dynamics is modulated in response to developmental cues or a changing environment. The use of methods that allow the purification of chromatin from specific cell types will contribute to avoid the problems associated with the analysis of plant material containing different tissues with potentially distinct behaviors. The combination of these techniques with the use of next generation sequencing approaches will provide powerful tools to gather valuable information concerning chromatin function in the context of a variety of biological processes. A new perspective of chromatin dynamics is likely to emerge from the application of bioinformatics tools and mathematical modeling to these data sets.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all participants in the 3rd EWPC for their contributions and exciting discussions during the meeting, and particularly to those contributors that presented their results in the workshop and allowed us to discuss their unpublished results in this report. We acknowledge the financial support of INIA (Project AC2012–00056), MICINN (project BIO2010–15589 to JAJ and MP), CBGP, and INRA.
References
- 1.Köhler C, Gaudin V, Hennig L. Green chromatin dynamics in Zurich: meeting summary based on the European Workshop on Plant Chromatin 2009 in Zurich, Switzerland. Epigenetics. 2010;5:80–3. doi: 10.4161/epi.5.1.10376. [DOI] [PubMed] [Google Scholar]
- 2.Houba-Hérin N, Hennig L, Köhler C, Gaudin V. A fruitful chromatin harvest: meeting summary of the Second European Workshop on Plant Chromatin 2011 in Versailles, France. Epigenetics. 2012;7:307–11. doi: 10.4161/epi.7.3.19104. [DOI] [PubMed] [Google Scholar]
- 3.Hennig L, Derkacheva M. Diversity of Polycomb group complexes in plants: same rules, different players? Trends Genet. 2009;25:414–23. doi: 10.1016/j.tig.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Köhler C, Hennig L. Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr Opin Genet Dev. 2010;20:541–7. doi: 10.1016/j.gde.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 5.Hartwig B, James GV, Konrad K, Schneeberger K, Turck F. Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 2012;160:591–600. doi: 10.1104/pp.112.200311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharif J, Endo TA, Ito S, Ohara O, Koseki H. Embracing change to remain the same: conservation of polycomb functions despite divergence of binding motifs among species. Curr Opin Cell Biol. 2013;25:305–13. doi: 10.1016/j.ceb.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Stec I, Nagl SB, van Ommen GJ, den Dunnen JT. The PWWP domain: a potential protein-protein interaction domain in nuclear proteins influencing differentiation? FEBS Lett. 2000;473:1–5. doi: 10.1016/S0014-5793(00)01449-6. [DOI] [PubMed] [Google Scholar]
- 8.Jarillo JA, Piñeiro M, Cubas P, Martínez-Zapater JM. Chromatin remodeling in plant development. Int J Dev Biol. 2009;53:1581–96. doi: 10.1387/ijdb.072460jj. [DOI] [PubMed] [Google Scholar]
- 9.He Y. Chromatin regulation of flowering. Trends Plant Sci. 2012;17:556–62. doi: 10.1016/j.tplants.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Song J, Irwin J, Dean C. Remembering the prolonged cold of winter. Curr Biol. 2013;23:R807–11. doi: 10.1016/j.cub.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 11.Coustham V, Li P, Strange A, Lister C, Song J, Dean C. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science. 2012;337:584–7. doi: 10.1126/science.1221881. [DOI] [PubMed] [Google Scholar]
- 12.Holdsworth MJ, Bentsink L, Soppe WJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 13.Molitor AM, Bu Z, Yu Y, Shen W-H. Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 2014;10:e1004091. doi: 10.1371/journal.pgen.1004091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C, Bratzel F, Hohmann N, Koch M, Turck F, Calonje M. VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr Biol. 2013;23:1324–9. doi: 10.1016/j.cub.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 15.Molitor A, Shen WH. The polycomb complex PRC1: composition and function in plants. J Genet Genomics. 2013;40:231–8. doi: 10.1016/j.jgg.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Gaudin V, Libault M, Pouteau S, Juul T, Zhao G, Lefebvre D, Grandjean O. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development. 2001;128:4847–58. doi: 10.1242/dev.128.23.4847. [DOI] [PubMed] [Google Scholar]
- 17.del Olmo I, López-González L, Martín-Trillo MM, Martínez-Zapater JM, Piñeiro M, Jarillo JA. EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 2010;61:623–36. doi: 10.1111/j.1365-313X.2009.04093.x. [DOI] [PubMed] [Google Scholar]
- 18.Köhler C, Page DR, Gagliardini V, Grossniklaus U. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet. 2005;37:28–30. doi: 10.1038/ng1495. [DOI] [PubMed] [Google Scholar]
- 19.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Lucia F, Gaudin V. Epigenetic control by plant polycomb proteins: New perspectives and emerging roles in stress response. From plant genomics to plant biotechnology, (Woodhead Publishing, Elsevier, Cambridge, UK 2013) 2013; In Palmiro Poltronieri NB, Corrado Fogher, Editors:31-48. [Google Scholar]
- 21.Latrasse D, Germann S, Houba-Hérin N, Dubois E, Bui-Prodhomme D, Hourcade D, Juul-Jensen T, Le Roux C, Majira A, Simoncello N, et al. Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS One. 2011;6:e16592. doi: 10.1371/journal.pone.0016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baroux C, Raissig MT, Grossniklaus U. Epigenetic regulation and reprogramming during gamete formation in plants. Curr Opin Genet Dev. 2011;21:124–33. doi: 10.1016/j.gde.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 23.She W, Grimanelli D, Rutowicz K, Whitehead MW, Puzio M, Kotlinski M, Jerzmanowski A, Baroux C. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development. 2013;140:4008–19. doi: 10.1242/dev.095034. [DOI] [PubMed] [Google Scholar]
- 24.Deal RB, Henikoff S. Histone variants and modifications in plant gene regulation. Curr Opin Plant Biol. 2011;14:116–22. doi: 10.1016/j.pbi.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu PY, Lévesque N, Kobor MS. NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem Cell Biol. 2009;87:799–815. doi: 10.1139/O09-062. [DOI] [PubMed] [Google Scholar]
- 26.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–47. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 27.MacAlpine DM, Almouzni G. Chromatin and DNA replication. Cold Spring Harb Perspect Biol. 2013;5:a010207. doi: 10.1101/cshperspect.a010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosa M, Von Harder M, Cigliano RA, Schlögelhofer P, Mittelsten Scheid O. The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell. 2013;25:1990–2001. doi: 10.1105/tpc.112.104067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JM, To TK, Nishioka T, Seki M. Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 2010;33:604–11. doi: 10.1111/j.1365-3040.2009.02076.x. [DOI] [PubMed] [Google Scholar]
- 30.Wierzbicki AT, Jerzmanowski A. Suppression of histone H1 genes in Arabidopsis results in heritable developmental defects and stochastic changes in DNA methylation. Genetics. 2005;169:997–1008. doi: 10.1534/genetics.104.031997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wollmann H, Holec S, Alden K, Clarke ND, Jacques PE, Berger F. Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 2012;8:e1002658. doi: 10.1371/journal.pgen.1002658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravi M, Shibata F, Ramahi JS, Nagaki K, Chen C, Murata M, Chan SW. Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 2011;7:e1002121. doi: 10.1371/journal.pgen.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lermontova I, Koroleva O, Rutten T, Fuchs J, Schubert V, Moraes I, Koszegi D, Schubert I. Knockdown of CENH3 in Arabidopsis reduces mitotic divisions and causes sterility by disturbed meiotic chromosome segregation. Plant J. 2011;68:40–50. doi: 10.1111/j.1365-313X.2011.04664.x. [DOI] [PubMed] [Google Scholar]
- 34.Lermontova I, Kuhlmann M, Friedel S, Rutten T, Heckmann S, Sandmann M, Demidov D, Schubert V, Schubert I. Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant cenH3 deposition at centromeres. Plant Cell. 2013;25:3389–404. doi: 10.1105/tpc.113.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lauria M, Rossi V. Epigenetic control of gene regulation in plants. Biochim Biophys Acta. 2011;1809:369–78. doi: 10.1016/j.bbagrm.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Thorstensen T, Grini PE, Aalen RB. SET domain proteins in plant development. Biochim Biophys Acta. 2011;1809:407–20. doi: 10.1016/j.bbagrm.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Sawicka A, Seiser C. Histone H3 phosphorylation - a versatile chromatin modification for different occasions. Biochimie. 2012;94:2193–201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gentry M, Hennig L. Remodelling chromatin to shape development of plants. Exp Cell Res. 2014;321:40–6. doi: 10.1016/j.yexcr.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 39.Smaczniak C, Immink RG, Angenent GC, Kaufmann K. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development. 2012;139:3081–98. doi: 10.1242/dev.074674. [DOI] [PubMed] [Google Scholar]
- 40.Smaczniak C, Immink RG, Muiño JM, Blanvillain R, Busscher M, Busscher-Lange J, Dinh QD, Liu S, Westphal AH, Boeren S, et al. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc Natl Acad Sci U S A. 2012;109:1560–5. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pajoro A, Madrigal P, Muiño JM, Matus JT, Jin J, Mecchia MA, et al. Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 2014 doi: 10.1186/gb-2014-15-3-r41. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monfared MM, Carles CC, Rossignol P, Pires HR, Fletcher JC. The ULT1 and ULT2 trxG genes play overlapping roles in Arabidopsis development and gene regulation. Mol Plant. 2013;6:1564–79. doi: 10.1093/mp/sst041. [DOI] [PubMed] [Google Scholar]
- 43.Carles CC, Fletcher JC. The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 2009;23:2723–8. doi: 10.1101/gad.1812609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smolle M, Workman JL. Transcription-associated histone modifications and cryptic transcription. Biochim Biophys Acta. 2013;1829:84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hartzog GA, Fu J. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta. 2013;1829:105–15. doi: 10.1016/j.bbagrm.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dürr J, Lolas IB, Sørensen BB, Schubert V, Houben A, Melzer M, et al. The transcript elongation factor SPT4/SPT5 is involved in auxin-related gene expression in Arabidopsis. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku096. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell. 2012;23:251–64. doi: 10.1016/j.devcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stam M, Belele C, Ramakrishna W, Dorweiler JE, Bennetzen JL, Chandler VL. The regulatory regions required for B’ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics. 2002;162:917–30. doi: 10.1093/genetics/162.2.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Belele CL, Sidorenko L, Stam M, Bader R, Arteaga-Vazquez MA, Chandler VL. Specific tandem repeats are sufficient for paramutation-induced trans-generational silencing. PLoS Genet. 2013;9:e1003773. doi: 10.1371/journal.pgen.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benoit M, Layat E, Tourmente S, Probst AV. Heterochromatin dynamics during developmental transitions in Arabidopsis - a focus on ribosomal DNA loci. Gene. 2013;526:39–45. doi: 10.1016/j.gene.2013.01.060. [DOI] [PubMed] [Google Scholar]
- 51.Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, et al. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001225. doi: 10.1371/journal.pgen.1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costas C, de la Paz Sanchez M, Stroud H, Yu Y, Oliveros JC, Feng S, Benguria A, López-Vidriero I, Zhang X, Solano R, et al. Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol. 2011;18:395–400. doi: 10.1038/nsmb.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaquero-Sedas MI, Gámez-Arjona FM, Vega-Palas MA. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2011;39:2007–17. doi: 10.1093/nar/gkq1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogrocká A, Polanská P, Majerová E, Janeba Z, Fajkus J, Fojtová M. Compromised telomere maintenance in hypomethylated Arabidopsis thaliana plants. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1285. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14:267–74. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Cavrak VV, Lettner N, Jamge S, Kosarewicz A, Bayer LM, Mittelsten Scheid O. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1004115. Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Mittelsten Scheid O. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–29. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rigal M, Mathieu O. A “mille-feuille” of silencing: epigenetic control of transposable elements. Biochim Biophys Acta. 2011;1809:452–8. doi: 10.1016/j.bbagrm.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 59.Hentges KE. The Mediator complex: crucial functions in transcription with links to development and disease. Semin Cell Dev Biol. 2011;22:728. doi: 10.1016/j.semcdb.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 60.Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci U S A. 2011;108:2322–7. doi: 10.1073/pnas.1018222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ito H, Yoshida T, Tsukahara S, Kawabe A. Evolution of the ONSEN retrotransposon family activated upon heat stress in Brassicaceae. Gene. 2013;518:256–61. doi: 10.1016/j.gene.2013.01.034. [DOI] [PubMed] [Google Scholar]
- 62.Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, Colot V, Bowler C, Barneche F. Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 2012;8:e1002825. doi: 10.1371/journal.pgen.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu HG, Scholten I, Gruss C, Knippers R. The distribution of the DEK protein in mammalian chromatin. Biochem Biophys Res Commun. 2007;358:1008–14. doi: 10.1016/j.bbrc.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 64.Olmedo-Monfil V, Durán-Figueroa N, Arteaga-Vázquez M, Demesa-Arévalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature. 2010;464:628–32. doi: 10.1038/nature08828. [DOI] [PMC free article] [PubMed] [Google Scholar]