Abstract
Previously, spontaneous rifampin resistance mutations were isolated in cluster I of the rpoB gene, resulting in amino acid replacements (Q469R, H482R, H482Y, or S487L) in the Bacillus subtilis RNA polymerase β subunit (W. L. Nicholson and H. Maughan, J. Bacteriol. 184:4936-4940, 2002). In this study, each amino acid change in the β subunit was observed to result in its own unique spectrum of effects on growth and various developmental events, including sporulation, germination, and competence for transformation. The results thus establish the important role played by the RNA polymerase β subunit, not only in the catalytic aspect of transcription, but also in the regulation of major developmental events in B. subtilis.
The antibiotic rifampin (RIF) is a potent inhibitor of prokaryotic transcription initiation (33) and has long been used both to study bacterial transcription and as a highly clinically effective drug, particularly for the treatment of the infectious diseases tuberculosis and leprosy, caused by Mycobacterium tuberculosis and Mycobacterium leprae, respectively (22). Resistance to RIF (Rifr) arises from mutations in the rpoB gene encoding the β subunit of RNA polymerase (11), and the majority of Rifr mutations occur within a short (<100 bp) region within rpoB, designated cluster I in Escherichia coli (11, 28). Cluster I homologues have also been studied in a number of bacteria outside the enteric paradigm, including M. tuberculosis (7, 13, 24, 29, 31, 34, 36), Streptomyces spp. (8, 9), and Bacillus subtilis (2, 10, 16, 25).
Several lines of evidence indicate profound fundamental connections between RIF resistance, RNA polymerase structure and function, and global gene expression. First, RIF has long been known to specifically block transcription initiation but not elongation (33). In the recently elucidated three-dimensional structure of RNA polymerase, the RIF binding site, including cluster I, was localized to the β subunit in the DNA-RNA channel, ∼12 Å downstream from the active site; RIF binding apparently physically blocks initiation when the nascent transcript is 2 to 3 nucleotides (nt) long (4, 15).
Second, in addition to the Rifr phenotype, additional effects on gene expression have been noted in bacteria carrying mutations in rpoB cluster I. For example, binding of guanosine tetraphosphate (ppGpp) to RNA polymerase has long been recognized as an important modulator of global gene expression during growth, stationary phase, and the stringent response (reviewed in references 27 and 37). Although the exact location of the ppGpp binding site on RNA polymerase is at present unknown, it may reside in close proximity to the RIF binding site, because (i) in E. coli, Rifr mutations were isolated in rpoB which alleviated the toxic and growth-inhibitory effects of artificially induced ppGpp overproduction (32); (ii) certain mutations in cluster I of the B. subtilis rpoB gene confer both Rifr and hypersensitivity of B. subtilis RNA polymerase to the transcription termination factor NusG (10); and (iii) in Streptomyces lividans and Streptomyces coelicolor, Rifr mutants have been isolated which mimic postexponential phase- and stringent response-mediated antibiotic production, even in relA mutants that are unable to synthesize ppGpp (8, 9). From the analysis of a large number of Rifr mutant Streptomyces spp., a spectrum of stringent response mimicry was noted; certain amino acid changes within cluster I of rpoB led to strong mimicry of the stringent response and to elevated antibiotic production, whereas other mutant alleles exhibited weak or unapparent physiological effects (8, 9).
Third, we recently reported that for B. subtilis different physiological environments can lead to differences in the spectrum of spontaneous Rifr mutations in rpoB (16). We noted in reviewing the literature that the spectrum of spontaneous Rifr mutations obtained from vegetative cells of B. subtilis (10) was very different from that obtained from Rifr clinical specimens of M. tuberculosis taken from human tuberculosis cases (7), and we demonstrated that spores of B. subtilis 168 exhibited a spectrum of spontaneous Rifr mutations that was not only distinct from that of vegetative B. subtilis cells but also resembled the spectrum seen in clinical M. tuberculosis isolates (16). Thus, it appeared that the cell's physiological or developmental state could alter the spectrum of spontaneous Rifr mutations occurring in cluster I of rpoB.
It has been well established that the developmental cycle of B. subtilis, consisting of postexponential-phase gene expression, sporulation, dormancy, and germination, is controlled in large part at the transcriptional level (reviewed in references 19 to 21). Prior evidence suggested that at least one mutation in rpoB cluster I can affect development. Rothstein et al. (25) isolated a Rifr B. subtilis mutant which exhibited a temperature-sensitive sporulation phenotype, and the mutation responsible, rfm2103, was later identified as causing the amino acid change H482Y in cluster I of the B. subtilis RNA polymerase β subunit (2). With a more extensive collection of cluster I rpoB mutations causing Rifr in hand (16), we initiated a systematic set of investigations to determine the effects of cluster I mutations on various aspects of the B. subtilis developmental cycle. We report in this communication that mutations in rpoB cluster I exert a number of both position-specific and allele-specific alterations in the expression of global regulons, such as those controlling growth, competence, sporulation, and germination.
The Rifr rpoB alleles studied in this work arose spontaneously from either vegetative cells or spores of B. subtilis strain 168, and their nucleotide and deduced amino acid sequences have been described (16). To ensure that the phenotypes observed in this study resulted directly from the rpoB alleles tested, we PCR amplified all alleles from the spontaneous Rifr mutants (12, 16) and transferred them by transformation (3, 16, 30) into the common host background of B. subtilis strain MH5636, a generous gift from Marion Hulett (23). Strain MH5636 carries an engineered rpoC gene which expresses a 6-histidine-tagged version of the RNA polymerase β′ subunit, which does not appear to interfere with normal RNA polymerase functioning (reference 23 and our unpublished observations). In all cases, the presence of the correct rpoB alleles in MH5636 was confirmed by PCR amplification and sequencing (12, 16).
Growth rates of Rifr mutants.
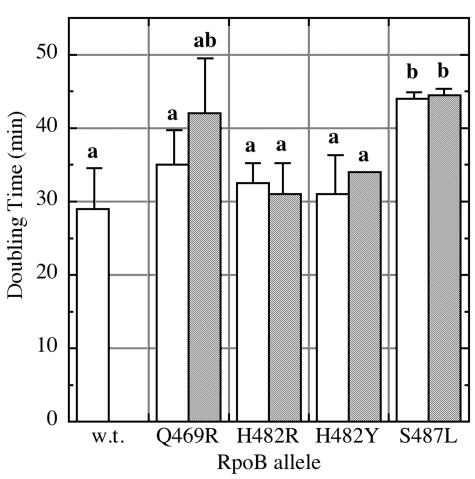
The effect of rpoB mutations on the exponential growth rate was determined by the cultivation of wild-type strain WN547 and all of its Rifr derivatives (Table 1) in Luria-Bertani (LB) medium (14) in triplicate at 37°C and by calculation of the average doubling times of the cultures. The wild-type parental strain and the Rifr mutants carrying the Q469R, H482R, and H482Y rpoB alleles exhibited doubling times ranging from 29 to 35 min (Fig. 1), which were not significantly different at a P value of 0.01 by analysis of variance (ANOVA). However, strain WN761, carrying the S487L mutation, grew significantly more slowly, with an average doubling time of 43 min (Fig. 1). The doubling times of strains carrying the rpoB mutations H482R, H482Y, and S487L were unaffected by the addition of RIF (50 μg/ml) to the LB medium (Fig. 1). However, the average doubling time of the strain carrying the Q469R mutation increased from 35 to 42 min in LB medium plus RIF, to more closely resemble the slower growth rate characteristic of the S487L-bearing strain (Fig. 1). Thus, it appeared that mutations at three closely spaced positions within rpoB cluster I exhibited three distinct phenotypes: (i) mutations H482R and H482Y did not affect the growth rate, (ii) mutation S487L caused a RIF-independent reduction in the growth rate, and (iii) mutation Q469R caused a RIF-dependent reduction in the growth rate (Fig. 1).
TABLE 1.
Bacterial strains used for this study
| Strain | Genotype and/or phenotype | Source or descriptiona (reference) |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| MH5636 (WN547) | trpC2 pheA1 cat 6His-rpoC; Cmr | Marion Hulett (23) |
| WN729 | trpC2 rpoB-S487L; Rifr | Spontaneous Rifr mutant of 168 (16) |
| WN736 | trpC2 rpoB-Q469R; Rifr | Spontaneous Rifr mutant of 168 (16) |
| WN737 | trpC2 rpoB-H482R; Rifr | Spontaneous Rifr mutant of 168 (16) |
| WN738 | trpC2 rpoB-H482Y; Rifr | Spontaneous Rifr mutant of 168 (16) |
| WN758 | trpC2 pheA1 rpoB-Q469R cat 6His-rpoC; Rifr Cmr | 1.78-kb rpoB-Q469R PCR product from WN736→MH5636; Rifr |
| WN759 | trpC2 pheA1 rpoB-H482R cat 6His-rpoC; Rifr Cmr | 1.78-kb rpoB-H482R PCR product from WN737→MH5636; Rifr |
| WN760 | trpC2 pheA1 rpoB-H482Y cat 6His-rpoC; Rifr Cmr | 1.78-kb rpoB-H482Y PCR product from WN738→MH5636; Rifr |
| WN761 | trpC2 pheA1 rpoB-S487L cat 6His-rpoC; Rifr Cmr | 1.78-kb rpoB-S487L PCR product from WN729→MH5636; Rifr |
Arrows denote transfers of donor DNA into recipient strains by transformation.
FIG. 1.
Growth rates of isogenic strains carrying the indicated rpoB alleles in LB medium (white bars) or LB medium plus Rif (hatched bars). Data are expressed as average doubling times in minutes ± standard deviations (n = 3). Data that were not significantly different (P > 0.01 by ANOVA) were placed into the same group (groups are indicated by lowercase letters). w.t., wild type.
Competence of Rifr mutants.
The expression of competence for transformation in B. subtilis is the result of a complex network of environmental and quorum-sensing signal transduction pathways leading ultimately to transcriptional regulation of the genes encoding DNA uptake and processing proteins (reviewed in reference 6). In order to test the possible effects of rpoB mutations on competence, we cultivated the parent strain WN547 and all of its isogenic Rifr derivatives in the GM-I/GM-II competence medium system (3, 30) and tested them for the ability to be transformed by chromosomal DNA prepared from strain 168 to prototrophy for the pheA1 marker (Table 2). In a separate experiment, all strains were also tested for their efficiency of transformation to prototrophy for the trpC2 marker, using 50 ng of plasmid pTRP-H3, which carries part of the cloned trpC+ gene in plasmid pBR322 (1, 35), as donor DNA, with essentially identical results (data not shown).
TABLE 2.
Effect of rpoB mutations on competencea
| Strain (rpoB allele) | Doubling time in GM-I medium (min) | Optical density at t0 in GM-I medium (Klett units) | No. of PheA+ transformants (per ml) | Total CFU/ml | Transformation efficiency relative to wild type |
|---|---|---|---|---|---|
| WN547 (wild type) | 45 | 94 | 1,325 | 1.20 × 108 | 1.0 |
| WN758 (Q469R) | 45 | 86 | 34 | 2.06 × 108 | 0.015 |
| WN759 (H482R) | 57 | 130 | 8 | 1.24 × 108 | 0.006 |
| WN760 (H482Y) | 51 | 100 | 44 | 3.08 × 108 | 0.013 |
| WN761 (S487L) | 45 | 48 | 0 | 2.4 × 107 | <0.004 |
One milliliter of competent cells, prepared in GM-I or GM-II medium containing 50 μg of tryptophan and phenylalanine per ml, was transformed with 50 ng of chromosomal DNA purified from strain 168, and PheA+ transformants were selected.
As observed previously with LB medium (Fig. 1), strain-specific growth patterns of the Rifr mutants were also present with GM-I competence medium containing tryptophan and phenylalanine. Firstly, all strains immediately initiated exponential growth upon aeration in GM-I medium, with the notable exception of strain WN761(rpoB S487L), which lagged for ∼3 h before commencing exponential growth. The wild-type parent strain and the strains carrying the Q469R and S487L mutations grew in GM-I medium with doubling times of ∼45 min, whereas the strains carrying the H482R and H482Y mutations grew slightly more slowly in GM-I medium (Table 2). All strains grew in GM-I medium to optical densities of >86 Klett units before entering stationary phase, again with the notable exception of strain WN761 carrying the S487L mutation, which grew to an optical density of only 48 Klett units (Table 2). The mutant strains carrying rpoB alleles Q469R, H482R, and H482Y exhibited between 2 and 3 orders of magnitude lower transformation efficiencies for the pheA marker (Table 2) or the trpC marker (data not shown) than did the isogenic wild-type parental strain WN547, indicating that these mutations in rpoB interfered with the proper expression of competence in B. subtilis. Interestingly, in strain WN761 carrying the S487L rpoB allele, no Phe+ or Trp+ transformants were detected above background revertants (Table 2), indicating that the S487L Rifr rpoB mutation led to a profound defect in the expression of competence.
Sporulation of Rifr mutants.
Sporulation in B. subtilis is another complex developmental process which relies on precise control of both the timing and compartmentalization of transcription (reviewed recently in references 5, 20, and 21). The notion that mutations in the B. subtilis RNA polymerase β subunit could affect cellular differentiation can be traced to the isolation and characterization of a Rifr rpoB mutation, designated rfm2103 (H482Y), which was observed to cause a temperature-sensitive defect in sporulation (25). To test the effect on sporulation of the various rpoB mutations used in this study, we grew wild-type parent strain WN547 and each of the isogenic Rifr strains at 37 and 48°C in liquid Schaeffer's sporulation medium (SSM) (26) or SSM plus RIF, and the efficiency of sporulation was quantified by determination of the ratio of heat-resistant (80°C, 10 min) spores (S) to total viable cells (V) from 24-h cultures, as described previously (18). The wild-type parental strain WN547 sporulated efficiently at 37°C in SSM (S/V = 0.79) (Table 3). Isogenic strains carrying the Q469R, H482Y, and S487L mutations sporulated at slightly lower efficiencies at 37°C than did the parent (with S/V values ranging from 0.4 to 0.6) in either SSM or SSM plus RIF (Table 3), but the sporulation frequencies of these mutants were not significantly different from the wild-type parent by ANOVA. In contrast, the strain carrying the H482R mutation consistently sporulated at a significantly lower frequency (P = 0.014) in either SSM (S/V = 0.13) or SSM plus RIF (S/V = 0.12) than did the wild-type parent (Table 3). Thus, it appeared that the H482R rpoB mutation interfered with, but did not abolish, efficient sporulation at 37°C.
TABLE 3.
Effect of rpoB mutations on sporulation at 37 and 48°Ca
| Strain | rpoB allele | Sporulation efficiency (S/V)b
|
|||
|---|---|---|---|---|---|
| 37°C
|
48°C
|
||||
| SSM | SSM + Rif | SSM | SSM + Rif | ||
| WN547 | Wild type | 0.79 | 0.24 | ||
| WN758 | Q469R | 0.47 | 0.52 | 0.22 | 0.23 |
| WN759 | H482R | 0.13 | 0.12 | 0.009 | 0.011 |
| WN760 | H482Y | 0.55 | 0.56 | 0.008 | 0.004 |
| WN761 | S487L | 0.55 | 0.53 | 0.69 | 0.67 |
Data are averages of two independent experiments which differed by <10%.
S/V, spore titer/viable titer, determined at 24 h.
To test whether the rpoB mutations conferred a temperature-sensitive sporulation phenotype, we also sporulated the above strains in either SSM or SSM plus RIF at 48°C and determined their sporulation efficiencies. We observed that the sporulation efficiency of the wild-type parental strain WN547 was slightly but not dramatically reduced at 48°C, to an S/V value of 0.24 (Table 3). The sporulation efficiency at 48°C was also slightly reduced, to an S/V value of ∼0.2, in the isogenic Rifr strain carrying the Q469R mutation, and no difference was observed when the Q469R mutant was sporulated in the presence or absence of RIF (Table 3). Sporulation of the Rifr strain carrying the S487L allele was also not impaired at 48°C; in fact, the S487L mutant appeared to sporulate slightly more efficiently at 48°C than at 37°C (Table 3). In sharp contrast, strains carrying rpoB alleles H482R and H482Y were severely impaired in sporulation at 48°C, with S/V values that were 10 to 100 times lower than those at 37°C (Table 3). These results confirmed the original observation that the rfm2103 allele, also an H482Y mutation, results in a temperature-sensitive sporulation phenotype (25). It is interesting that both the H482R and H482Y mutations caused a sporulation defect at 48°C, but only the H482R allele also demonstrated decreased sporulation efficiency at 37°C (Table 3). Therefore, the proper expression of sporulation regulons was altered in an allele-specific manner by amino acid substitutions in the β subunit at position H482. In none of the strains tested was there a difference observed in the frequency of sporulation in the presence or absence of RIF added to SSM at 50 μg/ml (Table 3).
Germination of Rifr mutants.
The germination and outgrowth of B. subtilis spores are yet more complex developmental processes, the success of which is dependent upon correct timing and compartmentalization of transcription. The success of germination, however, relies on the products of two sets of genes: the expression of one gene set is necessary during germination itself and the other is needed during the previous round of sporulation (reviewed in references 5, 17, and 19). In order to test if rpoB alleles conferring Rifr exerted effects on spore germination, we performed the following experiment. All Rifr mutants were sporulated at 37°C in either SSM or SSM plus RIF, and spores were purified by buffer washing and heat shock (18) to remove vegetative cells. Serial 10-fold dilutions of spores were plated in duplicate on LB medium plus RIF (or on LB medium in the case of the wild-type parent strain WN547) and incubated for 24 h at 37 and 48°C, and the resulting titers were compared (Fig. 2).
FIG. 2.
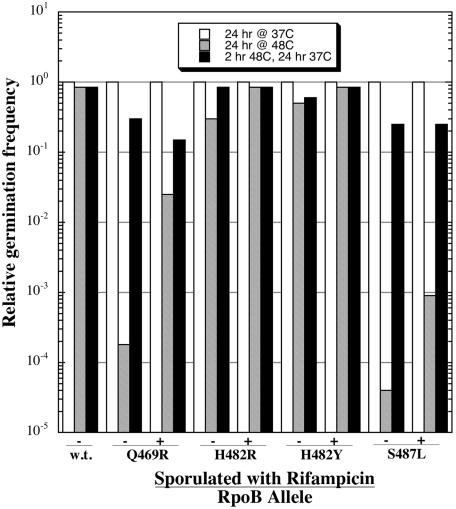
Spore germination of Rifr mutants and their wild-type (w.t.) parental strain. Germinated spore titers after 24 h at 48°C (hatched bars) or after a shift from 24 h at 48°C to an additional 24 h at 37°C (black bars) are depicted, normalized to the germinated spore titers of parallel plates of the same strain after 24 h of incubation at 37°C (white bars). Plus and minus signs indicate that spores were produced by growth in SMM in the presence or absence, respectively, of RIF (50 μg/ml). Strains are identified by the amino acid alteration in rpoB. Results are from two independent experiments performed in duplicate, from which the data differed by <10%.
Spores of all strains germinated efficiently on LB medium plus RIF plates at 37°C, and spores of the wild-type parent strain WN547 germinated nearly as efficiently at 48°C as they did at 37°C (Fig. 2). Spores of Rifr strains harboring rpoB alleles H482R and H482Y also germinated well at either 37 or 48°C, regardless of whether they had been sporulated in the presence or absence of RIF (Fig. 2). In contrast, spores of the strain harboring the Q469R allele were temperature sensitive for germination, showing a titer that was nearly 4 orders of magnitude lower at 48°C than that at 37°C (Fig. 2). However, the temperature-sensitive germination defect of the Q469R mutant could be rescued to a nearly wild-type level simply by shifting the plates from 48 to 37°C and incubating them for an additional 24 h (Fig. 2). Interestingly, the preparation of Q469R spores in the presence of RIF resulted in an ∼2-log increase in germination efficiency at 48°C, which could be rescued still further, to near-wild-type germination efficiency, by shifting from 48 to 37°C (Fig. 2). Spores of the mutant carrying the S487L allele exhibited yet a third distinct germination phenotype, in that they were severely temperature sensitive for germination, having a >4 orders of magnitude lower titer at 48°C than at 37°C, regardless of whether the S487L mutant had been sporulated in the presence or absence of RIF (Fig. 2). Again, as seen with the Q469R mutant, the S487L mutant could also be rescued to nearly the wild-type level of germination efficiency by shifting the plates from 48 to 37°C and incubating them for an additional 24 h (Fig. 2). Thus, mutations conferring Rifr, located at three closely spaced positions within cluster I of rpoB, exerted three dramatically different effects on spore germination. Furthermore, the temperature-sensitive germination phenotype of Rifr mutants was observed to be reversible by shifting of the spores from the restrictive to the permissive temperature. At present, we do not know if these germination defects resulted from alterations in the expression of germination-essential genes expressed during germination, during the previous round of sporulation, or both.
Collectively, the data presented in this communication indicate that alterations in closely linked amino acids residing within rpoB cluster I (Q469, H482, and S487) not only confer Rifr, but also exert a range of effects on major developmental events in B. subtilis. In our experience, the Q469R allele is the most frequently isolated spontaneous Rifr mutation from B. subtilis vegetative cells, while the S487L allele is the most frequently isolated mutation from spores; the H482Y and H482R alleles are isolated from both spores and vegetative cells at roughly equivalent frequencies (16). In addition to the Rifr phenotype, all the mutant strains described here had the following in common: they demonstrated 2- to 3-log lower transformation efficiencies than the isogenic wild-type parent (Table 2), and their sporulation efficiencies were not affected by the presence or absence of RIF in the sporulation medium (Table 3). Surprisingly, apart from these common phenotypes, each amino acid change in rpoB cluster I resulted in its own unique spectrum of effects.
Q469R.
Growth of the Q469R mutant in LB or GM-I medium was essentially indistinguishable from that of the isogenic wild-type parent strain, but this mutant was unique in that its growth rate was slower in LB medium plus RIF. The Q469R mutant sporulated as well as the wild type at either 37 or 48°C, but its germination at 48°C was severely impaired. Interestingly, the temperature-sensitive germination defect of the Q469R mutant was alleviated somewhat if spores of the strain were prepared in the presence of RIF (Fig. 2).
H482R and H482Y.
With codon 482 of rpoB, we had the opportunity to compare the physiological effects of replacing H with either a basic (R) or an aromatic (Y) amino acid. Considering the rather dramatic chemical differences the amino acid changes would be predicted to confer, both mutants behaved remarkably similarly. Both mutants grew at the same rate as the wild type in LB medium, and growth was not slowed in LB medium plus RIF. However, in GM-I medium, both H482R and H482Y mutants grew more slowly than, but to the same final density as, wild-type cells. Both mutants were severely defective in sporulation at 48°C. In the case of the H482Y mutant, our results confirmed the original observation of Rothstein et al. (25) that this rpoB allele caused a temperature-sensitive sporulation phenotype. Germination, in contrast, was not affected in either mutant. In fact, the only difference that we could detect between the two alleles was that the H482R, but not the H482Y, mutant had a slightly but significantly reduced sporulation efficiency at 37°C (Table 2). It is interesting that both mutations at H482 caused sporulation, but not germination, to be temperature sensitive, exactly opposite of the effect caused by the Q469R mutation.
S487L.
Unlike the other mutants, growth of the S487L mutant was slower than that of the wild type in LB medium. However, in contrast to the Q469R mutant, growth of the S487L mutant was not further slowed in LB medium plus RIF. The result for the growth of the S487L mutant in GM-I medium deserves some preliminary explanation. In the GM-I/GM-II transformation protocol, cultures are inoculated into a tube containing GM-I medium, incubated overnight at room temperature without aeration, and then introduced into a flask and aerated vigorously at 37°C (3). Thus, culture growth is initiated by an abrupt upshift in both temperature and oxygenation. Whereas the wild-type parent and other mutant strains responded to this upshift by immediately commencing exponential growth, the S487L mutant lagged for ∼3 h, indicating a problem in dealing with a temperature and/or oxygen upshift. However, once growth in GM-I medium commenced, the S487L mutant grew at the wild-type rate, but only to half the optical density of the other cultures, before entering stationary phase. This defective growth of the S487L mutant in the GM-I/GM-II system may contribute to its apparently complete inability to be transformed to either Phe+ or Trp+. Similar to the Q469R mutant, the S487L mutant sporulated well at either 37 or 48°C (in fact, it was perhaps somewhat better at sporulation than the wild type at 48°C) but was severely temperature sensitive for germination. Unlike the Q469R mutant, however, germination of the S487L mutant at 48°C was not improved by prior sporulation of the strain in the presence of RIF (Fig. 2).
Studies of developmental transcription in B. subtilis over the past 3 decades have concentrated mainly upon the regulatory roles of transcription accessory factors (i.e., repressors and activators) and the sigma subunit of RNA polymerase (20, 21); components of the RNA polymerase core enzyme have largely been considered catalytic rather than regulatory elements. However, the results reported here clearly establish the importance of the RNA polymerase β subunit for the properly regulated expression of major developmental regulons in B. subtilis. At present, the exact mechanism(s) involved is obscure, but it is certainly amenable to experimental elucidation. We are directing current experiments toward a definition of how various Rifr mutations in cluster I affect in vivo expression of the complete B. subtilis transcriptome and individual relevant genes during growth, competence development, sporulation, germination, and other physiological conditions. We predict that such studies will lead to the identification of key developmental genes whose expression is altered in rpoB mutants and which are thus dependent upon the regulatory function of rpoB. In addition, as can be seen in Table 1, we constructed our set of isogenic Rifr strains with future in vitro studies in mind by moving the various rpoB mutations into the background of B. subtilis strain MH5636, thus facilitating rapid purification of the histidine-tagged RNA polymerase holoenzyme containing Rifr β subunits for in vitro transcription and structure-function studies.
Acknowledgments
We thank Marion Hulett for the generous donation of strain MH5636.
This work was supported by a grant from the Arizona Agricultural Experiment Station (USDA Hatch) to W.L.N. H.M. was supported in this work by an NSF-IGERT fellowship and a Research Training Grant from the Department of Ecology and Evolutionary Biology.
REFERENCES
- 1.Band, L., H. Shimotsu, and D. J. Henner. 1984. Nucleotide sequence of the Bacillus subtilis trpE and trpD genes. Gene 27:55-65. [DOI] [PubMed] [Google Scholar]
- 2.Boor, K. J., M. L. Duncan, and C. W. Price. 1995. Genetic and transcriptional organization of the region encoding the beta subunit of Bacillus subtilis RNA polymerase. J. Biol. Chem. 270:20329-20336. [DOI] [PubMed] [Google Scholar]
- 3.Boylan, R. J., N. Mendelson, D. Brooks, and F. E. Young. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, E. A., N. Korzheva, A. Mustaev, K. Murakami, S. Nair, A. Goldfarb, and S. A. Darst. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901-912. [DOI] [PubMed] [Google Scholar]
- 5.Driks, A. 2002. Proteins of spore core and coat, p. 527-535. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 6.Dubnau, D., and C. M. Lovett, Jr. 2002. Transformation and recombination, p. 453-471. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 7.Garcia, L., M. Alonso-Sanz, M. J. Rebollo, J. C. Tercero, and F. Chaves. 2001. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis isolates in Spain and their rapid detection by PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 39:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, H., and K. Ochi. 2001. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistance mutations. Appl. Environ. Microbiol. 67:1885-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, H., Q. Zhang, and K. Ochi. 2002. Activation of antibiotic synthesis by specific mutations in the rpoB gene (encoding the β subunit) in Streptomyces lividans. J. Bacteriol. 184:3984-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingham, C. J., and P. A. Furneaux. 2000. Mutations in the β subunit of the Bacillus subtilis RNA polymerase that confer both rifampicin resistance and hypersensitivity to NusG. Microbiology 146:3041-3049. [DOI] [PubMed] [Google Scholar]
- 11.Jin, D. J., and C. A. Gross. 1988. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J. Mol. Biol. 202:45-58. [DOI] [PubMed] [Google Scholar]
- 12.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, B.-J., K.-H. Lee, B.-N. Park, S.-J. Kim, E.-M. Park, Y.-G. Park, G.-H. Bai, S.-J. Kim, and Y. H. Kook. 2001. Detection of rifampin-resistant Mycobacterium tuberculosis in sputa by nested PCR-linked single-strand conformation polymorphism and DNA sequencing. J. Clin. Microbiol. 39:2610-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller, J. 1972. Experiments in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Naryshkina, T., A. Mustaev, S. A. Darst, and K. Severinov. 2001. The β' subunit of Escherichia coli RNA polymerase is not required for interaction with initiating nucleotide but is necessary for interaction with rifampicin. J. Biol. Chem. 276:13308-13313. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson, W. L., and H. Maughan. 2002. The spectrum of spontaneous rifampin resistance mutations in the rpoB gene of Bacillus subtilis 168 spores differs from that of vegetative cells and resembles that of Mycobacterium tuberculosis. J. Bacteriol. 184:4936-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of bacterial spores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Sussex, England.
- 19.Paidhungat, M., and P. Setlow. 2002. Spore germination and outgrowth, p. 537-548. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 20.Perego, M., and J. A. Hoch. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p. 473-481. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 21.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 22.Plorde, J. J. 1994. Mycobacteria, p. 401-416. In K. J. Ryan (ed.), Sherris medical microbiology, 3rd ed. Appleton & Lang, East Norwalk, Conn.
- 23.Qi, Y., and F. M. Hulett. 1998. PhoP∼P and RNA polymerase sigma A holoenzyme are sufficient for transcription of Pho regulon promoters in Bacillus subtilis: PhoP∼P activator sites within the coding region stimulate transcription in vitro. Mol. Microbiol. 28:1187-1198. [DOI] [PubMed] [Google Scholar]
- 24.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 25.Rothstein, D. M., C. L. Keeler, and A. L. Sonenshein. 1976. Bacillus subtilis RNA polymerase mutants temperature-sensitive for sporulation, p. 601-616. In R. L. Losick and M. Chamberlin (ed.), RNA polymerase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schaeffer, P., J. Millet, and J.-P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider, D. A., W. Ross, and R. L. Gourse. 2003. Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 6:151-156. [DOI] [PubMed] [Google Scholar]
- 28.Severinov, K., M. Soushko, A. Goldfarb, and V. Nikiforov. 1993. Rifampicin region revisited: new rifampicin-resistant and streptolydigin-resistant mutants in the β subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 268:14820-14825. [PubMed] [Google Scholar]
- 29.Sintchenko, V., W. K. Chew, P. J. Jelfs, and G. L. Gilbert. 1999. Mutations in rpoB gene and rifabutin susceptibility of multidrug-resistant Mycobacterium tuberculosis strains isolated in Australia. Pathology 31:257-260. [DOI] [PubMed] [Google Scholar]
- 30.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniguchi, H., H. Aramaki, Y. Nikaido, Y. Mizuguchi, M. Nakamura, T. Koga, and S. Yoshida. 1996. Rifampicin resistance and mutation of the rpoB gene in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 144:103-108. [DOI] [PubMed] [Google Scholar]
- 32.Tedin, K., and H. Bremer. 1992. Toxic effects of high levels of ppGpp in Escherichia coli are relieved by rpoB mutations. J. Biol. Chem. 267:2337-2344. [PubMed] [Google Scholar]
- 33.Wehrli, W., F. Knusel, K. Schmid, and M. Staehelin. 1968. Interaction of rifamycin with bacterial RNA polymerase. Proc. Natl. Acad. Sci. USA 61:667-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams, D. L., L. Spring, M. Salfinger, T.-P. Gillis, and D. H. Persing. 1998. Evaluation of polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampicin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin. Infect. Dis. 26:446-450. [DOI] [PubMed] [Google Scholar]
- 35.Xue, Y., and W. L. Nicholson. 1996. The two major spore DNA repair pathways, nucleotide excision repair and spore photoproduct lyase, are sufficient for the resistance of Bacillus subtilis spores to artificial UV-C and UV-B but not to solar radiation. Appl. Environ. Microbiol. 62:2221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, B., H. Koga, H. Ohno, K. Ogawa, M. Fukuda, Y. Hirakata, S. Maesaki, K. Tomono, T. Tashiro, and S. Kohno. 1998. Relationship between antimycobacterial activities of rifampicin, rifabutin, and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 42:621-628. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X., P. Dennis, M. Ehrenberg, and H. Bremer. 2002. Kinetic properties of rrn promoters in Escherichia coli. Biochimie 84:981-996. [DOI] [PubMed] [Google Scholar]