Abstract
An expanding body of evidence supports a role for gut microbes in the etiology of cancer. Previously, the focus was on identifying individual bacterial species that directly initiate or promote gastrointestinal malignancies; however, the capacity of gut microbes to influence systemic inflammation and other downstream pathways suggests that the gut microbial community may also affect risk of cancer in tissues outside of the gastrointestinal tract. Functional contributions of the gut microbiota that may influence cancer susceptibility in the broad sense include (1) harvesting otherwise inaccessible nutrients and/or sources of energy from the diet (i.e., fermentation of dietary fibers and resistant starch); (2) metabolism of xenobiotics, both potentially beneficial or detrimental (i.e., dietary constituents, drugs, carcinogens, etc.); (3) renewal of gut epithelial cells and maintenance of mucosal integrity; and (4) affecting immune system development and activity. Understanding the complex and dynamic interplay between the gut microbiome, host immune system, and dietary exposures may help elucidate mechanisms for carcinogenesis and guide future cancer prevention and treatment strategies.
Keywords: Microbiota, Gut microbiome, Gut microbial community, Cancer prevention
1 Introduction
Representing a vast ecosystem, the indigenous bacteria in the human gut have various physiological effects and carry out multiple metabolic functions that can influence the health of the host. Bacteria are hypothesized to benefit the host in many ways. These favorable effects include (1) facilitating the metabolic conversion and uptake of beneficial dietary components; (2) producing beneficial fermentation end products that affect intestinal pH and interact with gut mucosa epithelial cells; (3) excluding pathogens by competing for attachment sites within the gut mucosa; (4) interacting with the intestinal immune system and contributing to the regulation of immune function; (5) transforming or excreting toxic substances; and (6) generating fecal bulk that decreases transit time and dilutes toxic substances [127]. Laboratory studies of the phenotypic differences between germfree and conventional animals illustrate the importance of the normal microbiota for overall host health [128, 142]; germfree animals tended to have a lower body temperature, smaller lymph nodes, lack of deconjugation of bilirubin and bile acids, an absence of urease and β-glucuronidase activities, and lower organ weights [20, 139].
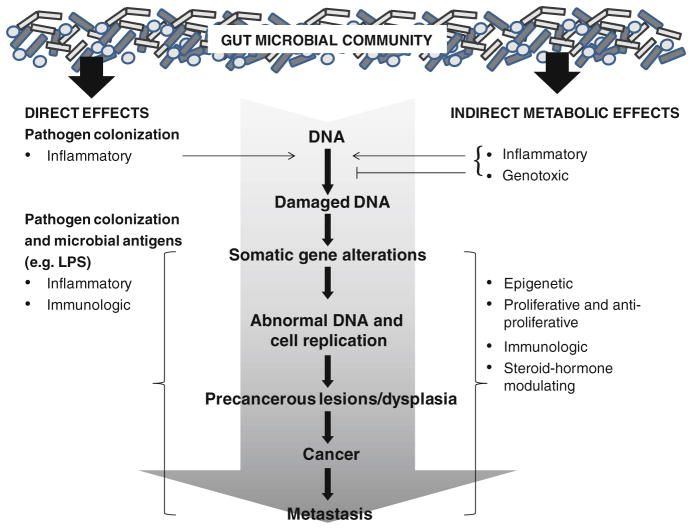
Carcinogenesis has been associated with the microbiome through direct and indirect routes (Fig. 1). Direct pathways include colonization of epithelia by pathogens or direct interaction with the innate immune system via bacterial antigenic particles with pattern recognition receptors (PRR, e.g. toll-like receptor). Indirect pathways include bacterial production of carcinogens and chemoprotective factors from exogenous sources, such as diet, or from endogenous sources, such as compounds resulting from human metabolism (e.g., bile acids and steroid hormones). We present below epidemiologic and experimental evidence for associations of the gut microbiome, diet, and cancer.
Fig. 1.
Direct and indirect mechanisms by which the gut microbial community may influence cancer risk. Direct colonization of gut epithelium by pathogens, as well as effects of microbial antigens (e.g., lipopolysaccharide—LPS), contribute to inflammation and altered immune function. Indirectly, microbial metabolites of exogenous substrates (i.e., dietary constituents) and endogenous host compounds (i.e., steroid hormones, bile acids, etc.) can affect the carcinogenesis continuum within the colon, as well as in other tissues via systemic effects
2 Molecular Characterization of the Gut Microbial Community
Bacteria colonize throughout the gastrointestinal tract, and to a great extent, bacteria in fecal samples reflect the bacterial composition in the lumen of the large intestine [49, 129]. The adult human intestine is host to a diverse community of microorganisms, including more than 800 species of bacteria [119]. However, the gut microbial community is distributed predominantly among two bacterial divisions, the Bacteroidetes and the Firmicutes, and one Archaeal species, Methanobacter brevii [49, 62, 119, 143]. A metagenomic analysis of the gut community also suggests that there is a core microbiome that individuals share; however, at the bacterial species level, large variation in gut microbial composition between individuals is observed [49, 62, 119, 143]. Therefore, the task of identifying particular bacteria associated with a specific phenotype in humans can be difficult. Conventional culture techniques for isolating and identifying active bacteria are arduous and time consuming. Furthermore, quantifying bacteria with these techniques is limited because it is estimated that approximately 40–60 % of mammalian bacterial species from the intestine cannot be cultured with conventional techniques [20, 140].
Because of the problems inherent in conventional culture techniques, studies of gut microbial communities have turned to molecular sequence-based approaches to identify intestinal bacterial species [5, 53, 65, 92, 150]. Bacterial DNA and RNA can be identified regardless of whether the bacterium itself can be cultured. For phylogenetic-based approaches, the 16S ribosomal RNA (rRNA) gene is ideal, because it contains regions of the DNA that are conserved across bacterial species, as well as sequences that are unique to a specific bacterial species. Furthermore, the relationship between rRNA content and growth rate in enteric bacteria is well established, and rRNA content per cell varies with growth rate under different nutrient conditions; thus, 16S rRNA content can be used as an estimate of microbial biomass [112, 124, 131] and the physiologically active bacteria. These molecular assays can be used to focus at the domain level (i.e., Eubacteria and Archae), the phylum level (i.e., Bacteroidetes and Firmicutes), the functional group level (i.e., sulfate-reducing bacteria), or the species level (i.e., Clostridia sp.).
Comparative omics technologies provide an opportunity to link microbial community structure and function to human health and disease. In a recent study using a metagenomic approach to catalog the genes in the microbiomes from 124 individuals, Qin et al. [119] identified 3.3 million bacterial sequences. This approach was used to putatively categorize humans into three classes, or enterotypes, based on the composition and functional potential of their gut microbiome [9]. This is intriguing because it suggests that the underlying physiology of the gut microbiome and thus, how the human host is influenced by the microbiome, varies in a potentially predictable way. However, the presence of a gene does not necessarily imply that it is actively being expressed and shaping microbiome–host interactions. Functional metagenomic approaches need to be integrated with other approaches to assess which of these genes are actively expressed (metatranscriptomics) and translated to functioning proteins (metaproteomics). The integration of these ‘omics technologies can also assess the presence of the metabolic pathways, such as sulfate reduction, nitrate reduction, secondary bile acid formation, and others that interact with diet to influence human health. They can also be used to measure the direct effects of pathogens that may promote carcinogenesis in epithelial cells. Evidence for the influence of the gut microbiome as a direct or indirect agent of carcinogenesis has been evident in the epidemiologic literature (see below). Coupled with studies of in vitro systems, mouse models, and controlled human interventions, we can start to understand the mechanisms associated with the gut microbiome which influence human health and risk of disease. However, until we can sample the gut microbiome in a prospective fashion, it will be hard to understand truly the causal effect of the gut microbiome on disease outcomes [86].
3 Direct Effects of the Gut Microbiome in Cancer Development
3.1 Gut Microbes as Infectious Agents
It is now clear that infectious agents are important to the development of specific cancer types; cervical, anal, penile, oropharyngeal, liver, and stomach cancers, along with certain types of lymphomas, have well-established infectious etiologies [23, 42]. Approximately 20 % of the total worldwide cancer burden is attributable to known infectious agents, and this proportion is expected to increase over time [23, 153]. The majority of known infection-associated cancers are caused by viral agents, such as the link between cervical cancer and oncogenic human papillomavirus alpha types, or between liver cancer and hepatitis C and B viruses [116]. However, bacteria, and in particular microbes found in the gut, have also been implicated as carcinogenic agents [96].
Helicobacter pylori, considered a class I carcinogen by the International Agency for Research on Cancer (IARC), is an established cause of gastric cancer and MALT lymphoma and accounts for approximately 5.5 % of cancers worldwide [72, 116]. Chronic carriage of Salmonella typhi, the causative agent of typhoid fever, is hypothesized to be linked with gallbladder cancer [90]. Additionally, several species of bacteria have been identified as potential candidates associated with colorectal cancer; these include Streptococcus bovis (also known as S. gallolyticus), Fusobacterium nucleatum, H. pylori, Coriobacterialies, and enterotoxigenic Bacteroides fragilis (ETFB) [27, 31, 63, 100].
Most of the studies linking potential infectious bacterial agents to carcinomas of the gut have used a case–control design. Although case–control studies are efficient for studying rare diseases, they are unable to establish the temporal relationship between infection and cancer [99]. This methodological concern is important to the interpretation of case–control study results. For cancer, the microenvironment around the tumor becomes more anaerobic, and possibly, more susceptible to infection as the cancer progresses [68]. Therefore, it is unclear if the malignant tumor creates an ideal environment for infection with specific bacterial agents, or if bacterial infection precedes the carcinoma and acts to drive carcinogenesis via inflammation or other pathways. Furthermore, some bacterial species may be important in early carcinogenesis but may not be able to tolerate the new tumor environment as the cancer develops; these potentially important agents would be missed in case–control studies of cancer in the gut. Thus, prospective studies and studies of precursors to cancer, such as colorectal adenomas, are important to determining causality in the relationship between bacterial agents and cancer.
In addition to determining the temporal relationship between bacterial agents and cancer, mechanisms by which infectious agents may promote carcinogenesis should be established. Each of the bacterial species discussed briefly above are hypothesized to follow a more traditional model of microbial carcinogenesis by promoting cancer directly at the site of infection. In this model, the microbe would infect the gut mucosa, resulting in a chronic, local inflammatory response which triggers cellular proliferation, cytokine production, and oxidative DNA damage due to an increase in reactive oxygen species [37]. Over time, DNA damage would accumulate in the infected cells, as well as adjacent cells, with mutations in tumor suppressor genes and oncogenes driving morphologic changes that eventually progress to a malignant tumor. Although this model is certainly important for H. pylori and the development of gastric cancer [51], increasing evidence points toward the potential for gut microbes to affect carcinogenesis at anatomic sites beyond the gastrointestinal tract and to have complex interactions with diet [36, 123]. For example, some researchers hypothesize that H. pylori and high salt intake may act synergistically to promote gastric cancer [147]. Further, recent studies have shown that polyamine catabolism contributes to ETFB-induced colon tumorigenesis in mice [63] (see dietary polyamine section below). Diet may also serve as a potential source of infection for possible carcinogenic agents. For example, high red meat intake is associated with an increased risk of colorectal cancer [114], and it is possible that this relationship is mediated by potentially carcinogenic bacterial contaminants of red meat products, such as S. bovis [84].
3.2 Gut Microbial Antigenic Particles Associated with Inflammation
Lipopolysaccharide (LPS, also known as endotoxin) is a bacterial cell wall component in gram-negative bacteria that is associated with low-grade, chronic inflammation in obesity [29, 30, 38] and colorectal cancer [28, 56]. LPS acts through toll-like receptor-4 (TLR-4), a PRR associated with innate immunity, which triggers TGF-β-mediated pathways [1, 97]. This leads to the expression of various genes that promote neoplasia, including those of growth factors and inflammatory mediators. Serum LPS-binding protein (LBP), a protein that binds LPS upon activation of TLR-4, is correlated with circulating concentrations of LPS [117], and a recent prospective study showed that polymorphisms in the LBP gene were associated with increased colorectal cancer risk [33]. To date, no studies have evaluated the association of these mutations to the distribution of gram-negative bacteria in the gut microbiome and cancer risk.
4 Indirect Effects of the Gut Microbiome in Cancer Development and Prevention
Host diet influences the amount and types of bacteria present in the gut, and gut microbial metabolism of dietary compounds affects the production of both protective and harmful metabolites. Therefore, the interaction between dietary intake and the commensal gut bacteria may ultimately influence cancer risk in humans. Cancers arise as a consequence of genomic and epigenomic instability. This instability permits the accumulation of genetic and epigenetic alterations that transform normal, healthy cells into cancer cells. Microbial metabolites may influence the development of microsatellite unstable (MSI) or chromosomal unstable (CIN) tumors through direct genotoxic effects on DNA, as well as by modulating DNA repair systems and by modulating epigenetic mechanisms through histone acetylation or CpG Island methylation [98, 137]. Groups of bacteria with unique metabolism—such as chemolithoheterotrophs that use inorganic compounds as an electron acceptor and organic carbon sources for growth and organoheterotrophs that use organic carbon for both respiration and growth—have been associated with cancer. We describe below several examples of metabolism unique to bacteria that support mechanisms by which the gut microbiome can 1) produce metabolites from exogenous sources (i.e., diet) that may influence tumorigenesis or 2) alter exposure to circulating levels of endogenous compounds, such as steroid hormones or bile acids, that influence tumorigenesis (Table 1).
Table 1.
Summary of the potential impact on cancer risk for gut microbial metabolism of specific exogenous and endogenous compounds
| Bacterial metabolism | Substrate sources | Bacterial species | Potential impact on cancer |
|---|---|---|---|
| Exogenous substrates | |||
| Sulfate reduction to produce hydrogen sulfide (H2S) | Dietary protein, especially sulfur-containing amino acids, and inorganic sulfur sources (SO4 in water) | Sulfate-reducing bacteria (SRB) | H2S has cytotoxic and genotoxic effects |
| Nitrate reduction to nitrite resulting in N-nitroso compounds (NOC) | Meat, particularly red meat | Multiple gut bacterial species | NOC can form DNA adducts |
| Polyamine production | Ornithine | Multiple gut bacterial species | Polyamines associated with increased inflammatory microenvironments |
| Flavonoid metabolism: daidzein to equol | Soybeans | Enterococcus faecium strain EPI1, Lactobacillus mucosae strain EPI2, Finegoldia magna strain EPI3, and Veillonella sp. | Equol production is associated with lower risks of breast and prostate cancer in high-soy- consuming populations |
| Metabolism of glucosinolates to isothiocyanates (ITC) | Cruciferous vegetables, such as broccoli, cabbage, kale, and brussels sprouts | Escherichia coli, Bacteroides thetaiotaomicron, Enterococcus faecalis, Enterococcus faecium, Peptostreptococcus sp., and Bifidobacterium sp. | ITC have anti-carcinogenic properties, including causing cell cycle arrest and inducing apoptosis |
| Endogenous substrates | |||
| Production of secondary bile acids (SBA) deoxycholic acid and lithocholic acid | Primary bile acids: cholic acid and chenodeoxycholic acid | Multiple Clostridium sp. | SBA may have tumor-promoting activity, and some studies associate high fecal SBA levels with increased risk of colorectal cancer |
| Metabolism of endogenous estrogens | Estradiol (E2), estrone 16αhydroxyestrone, 16-oxoestradiol, 15α-hydroxyestrone | Numerous reactions carried out by a wide variety of bacteria including Clostridium paraputrificum, Bacteroides sp., Eubacterium lentum | Higher circulating concentrations of E2 associated with breast cancer risk |
4.1 Gut Microbial Metabolism of Exogenous Substrates Associated with Carcinogenesis
4.1.1 Sulfate Reduction
Hydrogen sulfide (H2S) is produced by sulfate-reducing bacteria (SRB) and has been shown to have both cytotoxic and genotoxic effects in cell culture studies [13, 44, 75]. For example, using a modified comet assay, Attene-Ramos et al. [13] showed that H2S resulted in genomic DNA damage. Sulfide has also been shown to prevent the oxidation of butyrate by colonic epithelial cells, thereby reducing ATP formation and energy harvest [34]. This lowers the absorption of ions, mucus formation, and cellular detoxification. Roediger et al. [121, 122] reported decreased fatty acid oxidation in colonocytes exposed to H2S, and there is evidence that sulfide alters cellular redox potential which, in turn, alters cell proliferation [44].
The role of SRB in inflammatory bowel disease and colorectal cancer has been evaluated in several epidemiologic and clinical studies [15, 60, 80, 118, 121, 122, 130]. Genomic instability associated with sporadic colon cancer and ulcerative colitis (UC), a risk factor for colon cancer, is hypothesized to result in part from H2S exposure [14]. In a population-based study (n = 55), the distribution of SRB varied between different ethnic groups and the prevalence of SRB was associated with the diets of groups having higher rates of colon cancer [115]. UC has been linked to increased inflammation possibly associated with H2S generated by SRB [52]. Furthermore, when analyzed by culture methods, fecal samples from patients with UC showed higher concentrations of SRB than feces from patients without UC [61].
SRB are often members of the normal gut microbiota, and diet can influence their distribution and activity. Dietary protein, especially sulfur-containing amino acids, and inorganic sulfur sources (SO4 in water) contribute to H2S production [46]. In a controlled feeding study, Magee et al. [95] showed that H2S was significantly related to the amount of meat protein consumed. In mice, inorganic sulfate consumption enhanced sulfate reduction [46], and sulfonated proteins (e.g., mucins) enhanced sulfate reduction and inhibited methanogenesis in a continuous culture inoculated with fecal slurry [59]. These studies show that SRB are required for sulfate reduction to occur and that diet can alter their abundance. Most SRB form a fairly phylogenetically discreet group found in the delta subdivision of the delta Proteobacteria. One exception is Desulfotomaculum, which is found in the Clostridium subdivision of the gram-positive bacteria [71]. The functional genes, dissimilatory (bi) sulfite reductase (dsrA) [154] and the adenosine-5′-phosphosulfate reductase (apsA), are key enzymes in the sulfate reduction pathway [45, 55].
4.1.2 Nitrate Reduction
Epidemiologic studies have suggested an increased risk of colon cancer associated with red and processed meat consumption [114]. Numerous constituents in red and processed meats may contribute to this increased risk [17], including protein and other nitrogenous residues which allow for increased gut bacterial production of N-nitroso compounds (NOC) [22]. Nitrate can be reduced endogenously to nitrite via nitrate reductase produced by the gut bacteria, and nitrite can interact with organic compounds to form NOC. Many classes of NOC have been identified in feces, including nitrosamines, nitrosamides, and nitrosoguanidine [25]. NOC can form DNA adducts which induce mutations. For example, it has been shown that some NOC are alkylating agents that induce GC to AT transitions at the second base of codon 12 or 13 of the K-ras gene—this is a common mutation found in colorectal tumors with K-ras mutations [25]. More recently, transcriptomic analysis of colon biopsies was compared in inflammatory bowel disease patients diagnosed with UC and irritable bowel syndrome patients without inflammation [66]. The investigators associated gene expression levels with fecal NOC in all study participants (cases and controls) and, using network analysis, found chromatin modification linked to altered regulation of 11 histone genes. This suggested that epigenetic mechanisms may be relevant to NOC-induced carcinogenesis.
Diet can influence NOC concentrations. Meat consumption increases the amount of nitrogenous residues in the colon [136], and in a controlled feeding study in eight men, there was a dose–response between intake of meat and fecal concentrations of NOC [73]. Fecal water genotoxicity correlated with colonic gene expression changes in pro-carcinogenic pathways, including DNA damage repair, cell cycle, and apoptosis pathways in a 7-day dietary intervention with red meat [67]. Additional controlled feeding studies in men showed that, while heme iron increased fecal NOC, protein sources low in heme (i.e., white meat and protein from vegetable sources) did not increase fecal NOC [22, 39]. The independent effect of heme on NOC suggested that chemical catalysis, in addition to bacterial N-nitrosation, may be responsible for the dose-dependent effect of red meat on increasing endogenous intestinal N-nitrosation [39]. The addition of broccoli, brussels sprouts, or green peas to a high red meat diet had no effect on mean levels of fecal NOC, but the addition of soy statistically significantly suppressed fecal NOC [74].
The importance of gut bacteria in N-nitrosation has been demonstrated by the fact that N-nitrosation does not occur in germfree rats given nitrate as a nitrosating agent, but it does occur in rats harboring a conventional gut microbiota [103]. A number of facultative and anaerobic bacteria are able to catalyze the formation of NOC via nitrate reductase, and dissimilatory nitrate reduction is carried out by a number of bacteria distributed across bacterial groups. The narG gene is responsible for the reduction of nitrate to nitrite. Interestingly, recent studies in humans observed an increased risk of colorectal cancer in people ingesting more than three servings of red meat per week and who had a polymorphism in the nucleotide excision repair pathway [79]. However, the combined effect of degree of gut microbial formation of NOC and variation in host DNA repair mechanisms on colorectal cancer risk has not been investigated.
4.1.3 Polyamine Production
Polyamine exposure has been linked to inflammation in colonic mucosa and subsequent colon cancer risk [58]. Ornithine is converted to the polyamine putrescine, which is a precursor of spermidine and spermine. While this pathway is important to normal growth, the polyamines can also be oxidized to produce reactive oxygen species contributing to a chronic inflammatory microenvironment. An increased flux of polyamines into epithelium up-regulates eukaryotic ornithine decarboxylase (ODC), a key regulatory enzyme involved in polyamine synthesis and up-regulated in colon cancer. This may favor colon cancers that have upregulated the MAPK signaling pathway downstream from K-ras mutations found in CIN tumors [76, 89, 105].
Although polyamines are produced endogenously, both polyamines from diet, as well as those generated by microbial metabolism of dietary precursors, influence levels to which gut epithelial cells are exposed. Gerner et al. [57] showed that the efficacy of chemopreventive treatments can be influenced by modulation of dietary putrescine; however, modulation of the gut microbiome is a chemoprevention avenue that has not received attention. A common chemoprevention treatment in familial adenomatous polyposis is to block ODC with difluoromethylornithine (DFMO) and nonsteroidal anti-inflammatory drugs [58]. DFMO acts on eukaryotic production of polyamines; however, it may be less effective in altering the supply of bacterially produced putrescine. Given the phylogenetic and structural diversity of bacterial amino acid decarboxylases [91], there are multiple, diverse pathways by which bacteria produce polyamines and influence lumenal polyamine concentrations. Thus, while DFMO may influence eukaryotic production of polyamines, bacterial production may not be influenced. For example, DFMO has been shown to be effective in altering growth rates in H. pylori, but not those of other enterics, such as E. coli and C. rodentium [16]. Identifying new approaches for reducing polyamine production by gut microbes or altering the gut microbial community may be another chemoprevention strategy in high-risk patient populations.
4.1.4 Flavonoid Metabolism
Epidemiologic studies have shown that the consumption of foods of plant origin is associated with lower risk of several cancers [152]. Flavonoids are polyphenolic compounds and are the most abundant phytonutrients in the human diet. Categorized into six major subgroups, they have various cancer-impeding activities, such as reducing DNA damage via antioxidant properties or interacting with inflammation pathways (reviewed in [138]). High inter-individual variation in excretion and circulating concentrations and the extent of metabolism is probably a reflection of variation in the gut microbiome.
Probably the most extensively studied flavonoid with regard to bacterial metabolism is the soy isoflavone daidzein. Studies have shown that approximately 30–50 and 80–90 % of the population are able to metabolize daidzein to equol [54, 88] and O-desmethylangolensin (ODMA) [6, 81], respectively. Several in vitro studies suggest that equol is more biologically active than its precursor daidzein. For example, equol has been shown to be more estrogenic [101], is a more potent antioxidant than daidzein [7, 120, 144, 145], and has a higher effective free fraction in serum than both genistein and 17β-estradiol [110]. This has led to increased interest in equol producers as potential “responders” to soy consumption; over 10 years ago, Setchell et al. [133] hypothesized that the failure to “bacteriotype” individuals for their ability to produce equol in previous intervention studies of soy or isoflavone supplements could explain the variable results seen in such studies. Some, although not all, studies have shown a lower risk of breast cancer and prostate cancers associated with equol production (reviewed in [87]) and favorable associations, in terms of breast cancer risk, between equol production and circulating concentrations of steroid hormones, urinary estrogen metabolites, and mammographic breast density [10, 48, 54, 113].
Human intestinal bacteria are responsible for the production of equol and ODMA [11, 32]. Certain bacteria have been identified that are capable of carrying out discrete steps in the pathway to equol production [135], but other work also suggests that a consortium of bacteria consisting of Enterococcus faecium strain EPI1, Lactobacillus mucosae strain EPI2, Finegoldia magna strain EPI3, and an as yet undescribed species related to Veillonella sp. may be involved in equol production [43].
4.1.5 Metabolism of Glucosinolates from Brassica
Consumption of cruciferous or Brassica vegetables has been shown to be inversely associated with risk of some cancers [85]. Isothiocyanates (ITC), the hydrolysis products of glucosinolates, have been shown to have anti-carcinogenic properties both in vitro and in vivo (reviewed in [111]). The biologic effects of ITC are diverse, including interaction with multiple signaling pathways important to carcinogenesis as well as cross talk between pathways. The inhibitory activity of ITC against tumorigenesis is inferred by its ability to modulate Phase 1 and 2 biotransformation enzyme activities, thereby affecting several processes related to chemical carcinogenesis, such as the metabolism and DNA binding of carcinogens [70]. In vitro studies have also indicated that ITC cause cell cycle arrest and induce apoptosis [109].
Glucosinolates are converted into ITC by either the plant myrosinases or bacterially produced thioglucosidases. Cooking cruciferous vegetables deactivates the plant myrosinases, and given that most cruciferous vegetables consumed by humans are cooked, gut bacteria play a critical role in converting glucosinolates to ITC. Previous studies have shown that certain species of bacteria, such as Escherichia coli, Bacteroides thetaiotaomicron, Enterococcus faecalis, E. faecium, Peptostreptococcus sp., and Bifidobacterium sp., isolated from the human gut or feces can convert glucosinolates into ITC and other derivatives [26, 50, 69]. Controlled feeding studies in humans have shown significant inter-individual differences in urinary ITC excretion after participants consumed the same amount of cruciferous vegetables that had been either heated or microwaved prior to consumption to remove the plant myrosinase activity [125, 134, 146]. Similar effects have been found in controlled feeding studies with rats [35, 126]. This suggests inter-individual differences in the activity or composition of the intestinal bacteria involved in ITC formation. In support of this hypothesis, we showed recently that the fecal bacteria from individuals who excrete higher amounts of ITC in their urine after a standard meal of cooked broccoli metabolize more glucoraphanin [93].
4.2 Gut Microbial Metabolism of Endogenous Substrates Associated with Carcinogenesis
4.2.1 Production of Secondary Bile Acids
The secondary bile acid (SBA), deoxycholic acid (DCA), a colonic bacterial transformation product, has been implicated in gallstone formation [141] and colorectal carcinogenesis [21, 106]. The primary bile acids, cholic acid (CA) and chenodeoxycholic acid, are synthesized in the liver from cholesterol, are conjugated with either glycine or taurine, and undergo enterohepatic circulation (EHC). Although EHC of bile acids between the liver, gallbladder, and intestines is approximately 95 % efficient, up to 5 % of bile acids escape EHC and are transformed by anaerobic bacteria in the colon to SBA, DCA and lithocholic acid (LCA). Diet can affect the amount of bile acids entering the colon, and studies suggest that high-fat diets may result in higher fecal SBA concentrations [40, 104]. SBA have been shown to have tumor-promoting actions in animal studies, and some studies in humans have shown increased risks of colorectal cancer associated with high fecal bile acid concentrations (reviewed in [104]). However, not all studies have shown such associations [107]. Associations between fecal bile acid concentrations and colon cancer are complex given that factors such as gut transit time and pH may also influence fecal SBA concentrations, and it has been suggested that fecal bile acid concentrations may overestimate the amount of DCA in the bile acid pool given that nearly all bile acids that escape EHC are converted to SBA before excretion [104]. Nonetheless, serum DCA levels, which may reflect the bile acid pool more accurately, also have been shown to be higher in patients with colon cancer than in healthy individuals [18, 19]. A potential mechanism for associations between DCA and colon cancer is that DCA may change the balance between apoptosis, proliferation, and differentiation in the intestinal epithelium [64], acting through interference of tumor suppression and enhancing stimulation of growth via cell signaling pathways.
The bacteria responsible for DCA formation have been identified and belong to the genus Clostridium [47, 83, 148, 149]. These bacteria are classified into two classes with either high or low 7α-hydroxylating activity which may explain some of the inter-individual variation in DCA concentrations. In addition, the bile acid inducible (bai) operon, involved in the bacterial 7α-dehydroxylation of CA to DCA, has been characterized [148].
4.2.2 Metabolism of Endogenous Estrogens
Breast cancer risk is associated with higher levels of circulating estrogens, such as estradiol (E2), estrone (E1), E1 sulfate (E1S), estriol (E3) and dehydroepiandrosterone (DHEA) [82], and 16α-hydroxyestrone (16α-OHE1) in urine [10]. Both genetic and environmental factors that alter circulating estrogen levels may influence the risk of breast cancer.
Estrogens circulate in the bloodstream either free or bound to protein, conjugated or unconjugated, and either interact with target tissue or are excreted. Estrogens are sulfated, glucuronidated, or methylated in the liver, and about 50 % are excreted in urine and the other half are excreted in bile and undergo EHC. In the colon, some of the compounds are excreted in feces and some are metabolized by the gut microbiome (reviewed in [108]). The gut microbiome can (1) increase the exposure to circulating hormones via deconjugation and (2) influence the composition (or types) of hormones in circulation via hydroxylation/dehydroxylation and methylation/demethylation. β-Glucosidases, β-glucuronidases, and sulfatases from a wide variety of gut bacteria hydrolyze hormone conjugates [41]. These unconjugated estrogens are reabsorbed into the bloodstream and excreted in urine or can undergo EHC again [132]. Adlercreutz and others showed over 30 years ago that when patients were given antibiotics, urinary estrogen excretion decreased suggesting that gut bacteria are important in regulating EHC of estrogens and therefore exposure [3, 4, 102]. Diet may also influence serum hormone concentrations either directly by binding of estrogens by dietary fiber [8] or indirectly by influencing gut microbial community metabolism of estrogens [2].
Many gut bacteria are capable of performing the initial step of hydrolyzing conjugated steroids [151], which enables further metabolism by intestinal bacteria to occur. Microbial metabolism of estrogens includes reduction, oxidation, and the generation of E2 from E1, as well as from E2-3-glucuronide, E1 from E2, and from estrone-3-sulfate (E1S), and E3 from 16α-OHE1. In addition, in vitro incubation showed conversion of E1, E2, and 16α-dehydroxyestrone to E3, 16-oxoestradiol to 16-epiestriol, and 15α-hydroxyestrone to 15α-hydroxyestradiol [77, 78, 94]. Ring- A reduction is catalyzed by enzymes from a wide variety of bacteria including Clostridium paraputrificum and Bacteroides sp., and reductive dehydroxylation of ring-D and oxidative reactions, such as dehydrogenation, is carried out by bacteria such as Eubacterium lentum [24].
We recently evaluated associations between gut microbial community composition and circulating steroid hormone concentrations in a cross-sectional study of 115 healthy premenopausal women, age 40–45 years [12]. The gut microbial community from fecal samples was measured by terminal restriction length polymorphism (TRFLP) analysis with two restriction enzymes, Alu I (predominantly Bacteroidetes) and Rsa I (predominantly Firmicutes), and quantitative PCR (qPCR) of the 16S rRNA genes of Bacteria, Bacteroides, Clostridia Cluster XIVa, and Archaea. Regression models were fit to assess associations between hormones and the gut microbial community structure. The outcomes measured were serum concentrations of E1, E1S, total and free E2, sex-hormone-binding globulin (SHBG), free E2:total E2 ratio, and exposures were gut microbial community multivariate axes from TRFLP analysis and qPCR. The final solution for NMS analysis of the Alu I TRFLP patterns had stable stress values of 16.65, after 400 iterations. The three axes cumulatively explained 79.7 %of the variation in both subsets of TRFLP data. Axes 1, 2, and 3 explained 26.2, 31.7, and 21.7 %, respectively. The final solution for NMS analysis of the Rsa I TRFLP patterns had stable stress values of 16.87, after 400 iterations. NMS analysis for Rsa I TRFLP explained a total of 82 % of the variation in the data for axis 1 (23 %), axis 2 (29 %), and axis 3 (29 %). E1 (p<0.004), E1S (p<0.017), E2, and free E2:total E2 ratio were significantly associated with the gut microbial community using ALU I (p<0.05). E1S, E2, and free E2:total E2 ratio were significantly associated with the gut microbial community described by Rsa I (p<0.05). E2 free E2:total E2 ratio, and SHBG were associated with Bacteroides. E1 and E1S were associated with Archaea (p<0.05). When adjusted for dietary factors and demographics, there was a significant association between E1S and either Archaea or Alu Axis I, free E2:total E2 and Alu Axis 2, and E2 and Rsa Axis I, suggesting that the composition of the gut microbiome may be a factor in determining concentrations and types of circulating steroid hormones. This study suggests that it is important to consider an exposure such as diet within the context of gut microbial community (Table 2).
Table 2.
Associations between serum hormone concentrations (outcome)a, the gut microbial community (exposure), and diet (exposure) in premenopausal women
| β-Coefficient | p value | |
|---|---|---|
| Estrone sulfate | ||
| Archaeab | −0.11 | 0.028 |
| Total fiber | −0.01 | 0.019 |
| Alu I Axis 1 | −0.18 | 0.017 |
| Total fiber | −0.01 | 0.094 |
| Total fat | −0.005 | 0.045 |
| Free estradiol: all estradiol | ||
| Alu I Axis 2 | 0.21 | 0.013 |
| Total fiber | −0.01 | 0.099 |
| Total estradiol | ||
| Rsa I Axis 2 | 0.17 | 0.028 |
| Total fiber | −0.011 | 0.053 |
Using stepwise GLM that considered inclusion at 0.10 level for hormones, percent body fat, and potentially confounding demographic factors
Adjustment for adiposity
5 Conclusions
Aspects of human health are influenced by the interaction of the gut microbiome, diet, and host physiology. We presented examples of microbially mediated pathways associated with cancer. These pathways involve both 1) direct contact of the pathogen with human host and 2) indirect effects of microbial metabolism of exogenous and endogenous substrates. These pathways alter inflammation, modify DNA leading to mutations, or influence epigenetics and gene silencing. Recent metagenomic studies of the gut microbiome have revealed the varied anaerobic metabolisms, both chemoheterotrophic and organoheterotrophic, involved in fermentation and the production of metabolites that are either beneficial or harmful to the host [9, 143]. In addition, these approaches have characterized differences in the composition of the microbial community associated with tumor and nontumor regions in the colon [100]. Inter-individual variation in cancer risk may therefore be associated with microbial biomass, composition, and function, and the interaction with host factors such as diet. Future studies need to consider the gut microbiome as a contributing functional unit in relation to host exposures in order to better understand both its impact and those of the exposure on cancer risk and to design appropriate prevention strategies.
Acknowledgments
Supported by US NIH grants U01 CA162077, R01 DK084157 and Fred Hutchinson Cancer Research Center.
Abbreviations
- PRR
Pattern recognition receptors
- ETFB
Enterogenic Bacteroides fragilis
- LPS
Lipopolysaccharide
- TLR-4
Toll-like receptor -4
- LBP
LPS-binding protein
- MSI
Microsatellite unstable
- CIN
Chromosomal unstable
- SRB
Sulfate-reducing bacteria
- UC
Ulcerative colitis
- NOC
N-nitroso compounds
- ODC
Ornithine decarboxylase
- DFMO
Difluoromethylornithine
- ODMA
O-desmethylangolensin
- ITC
Isothiocyanates
- EHC
Enterohepatic circulation
- SBA
Secondary bile acids
- CA
Cholic acid
- DCA
Deoxycholic acid
- LCA
Lithocholic acid
- E2
Estradiol
- E1
Estrone
- E1S
Estrone-3-sulfate
- E3
Estriol
- DHEA
Dehydroepiandrosterone
- 16α-OHE1
16α-hydroxyestrone
- TRFLP
Terminal restriction length polymorphism
- SHBG
Sex-hormone-binding globulin
Contributor Information
Meredith A. J. Hullar, Cancer Prevention Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, M4-B402, PO Box 19024Seattle, WA 98109, USA
Andrea N. Burnett-Hartman, Cancer Prevention Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, M4-B402, PO Box 19024Seattle, WA 98109, USA
Johanna W. Lampe, Email: jlampe@fhcrc.org, Cancer Prevention Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, M4-B402, PO Box 19024Seattle, WA 98109, USA. Department of Epidemiology, School of Public Health, University of Washington, Seattle, WA 98195, USA
References
- 1.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 2.Adlercreutz H, Martin F. Biliary excretion and intestinal metabolism of progesterone and estrogens in man. J Steroid Biochem. 1980;13:231–244. doi: 10.1016/0022-4731(80)90196-x. [DOI] [PubMed] [Google Scholar]
- 3.Adlercreutz H, Martin F, Pulkkinen M, et al. Intestinal metabolism of estrogens. J Clin Endocrinol Metab. 1976;43:497–505. doi: 10.1210/jcem-43-3-497. [DOI] [PubMed] [Google Scholar]
- 4.Adlercreutz H, Martin F, Lehtinen T, et al. Effect of ampicillin administration on plasma conjugated and unconjugated estrogen and progesterone levels in pregnancy. Am J Obstet Gynecol. 1977;128:266–271. doi: 10.1016/0002-9378(77)90620-2. [DOI] [PubMed] [Google Scholar]
- 5.Amann RI, Binder BJ, Olson RJ, et al. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arai Y, Uehara M, Sato Y, et al. Comparison of isoflavones among dietary intake, plasma concentration and urinary excretion for accurate estimation of phytoestrogen intake. J Epidemiol. 2000;10:127–135. doi: 10.2188/jea.10.127. [DOI] [PubMed] [Google Scholar]
- 7.Arora A, Nair MG, Strasburg GM. Antioxidant activities of isoflavones and their biological metabolites in a liposomal system. Arch Biochem Biophys. 1998;356:133–141. doi: 10.1006/abbi.1998.0783. [DOI] [PubMed] [Google Scholar]
- 8.Arts CJM, Govers CARL, Van den Berg H, et al. In vitro binding of estrogens by dietary fiber and the in vivo apparent digestibility tested in pigs. J Steroid Biochem Mol Biol. 1991;38:621–628. doi: 10.1016/0960-0760(91)90321-u. [DOI] [PubMed] [Google Scholar]
- 9.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atkinson C, Skor HE, Dawn Fitzgibbons E, et al. Urinary equol excretion in relation to 2-hydroxyestrone and 16alpha-hydroxyestrone concentrations: an observational study of young to middle-aged women. J Steroid Biochem Mol Biol. 2003;86:71–77. doi: 10.1016/s0960-0760(03)00259-0. [DOI] [PubMed] [Google Scholar]
- 11.Atkinson C, Berman S, Humbert O, et al. In vitro incubation of human feces with daidzein and antibiotics suggests interindividual differences in the bacteria responsible for equol production. J Nutr. 2004;134:596–599. doi: 10.1093/jn/134.3.596. [DOI] [PubMed] [Google Scholar]
- 12.Atkinson C, Newton KM, Stanczyk FZ, et al. Daidzein-metabolizing phenotypes in relation to serum hormones and sex hormone binding globulin, and urinary estrogen metabolites in premenopausal women in the United States. Cancer Causes Control. 2008;19:1085–1093. doi: 10.1007/s10552-008-9172-3. [DOI] [PubMed] [Google Scholar]
- 13.Attene-Ramos MS, Wagner ED, Plewa MJ, et al. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9–14. doi: 10.1158/1541-7786.MCR-05-0126. [DOI] [PubMed] [Google Scholar]
- 14.Attene-Ramos MS, Wagner ED, Gaskins HR, et al. Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 2007;5:455–459. doi: 10.1158/1541-7786.MCR-06-0439. [DOI] [PubMed] [Google Scholar]
- 15.Balamurugan R, Rajendiran E, George S, et al. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 16.Barry DP, Asim M, Leiman DA, et al. Difluoromethylornithine is a novel inhibitor of Helicobacter pylori growth, CagA translocation, and interleukin-8 induction. PLoS ONE. 2011;6:e17510. doi: 10.1371/journal.pone.0017510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bastide NM, Pierre FH, Corpet DE. Heme iron from meat and risk of colorectal cancer: a meta-analysis and a review of the mechanisms involved. Cancer Prev Res (Phila) 2011;4:177–184. doi: 10.1158/1940-6207.CAPR-10-0113. [DOI] [PubMed] [Google Scholar]
- 18.Bayerdorffer E, Mannes GA, Richter WO, et al. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology. 1993;104:145–151. doi: 10.1016/0016-5085(93)90846-5. [DOI] [PubMed] [Google Scholar]
- 19.Bayerdorffer E, Mannes GA, Ochsenkuhn T, et al. Variation of serum bile acids in patients with colorectal adenomas during a one-year follow-up. Digestion. 1994;55:121–129. doi: 10.1159/000201136. [DOI] [PubMed] [Google Scholar]
- 20.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein C, Holubec H, Bhattacharyya AK, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol. 2011;85:863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bingham SA, Hughes R, Cross AJ. Effect of white versus red meat on endogenous N-nitrosation in the human colon and further evidence of a dose response. J Nutr. 2002;132:3522S–3525S. doi: 10.1093/jn/132.11.3522S. [DOI] [PubMed] [Google Scholar]
- 23.Blaser MJ. Understanding microbe-induced cancers. Cancer Prev Res. 2008;1:15–20. doi: 10.1158/1940-6207.CAPR-08-0024. [DOI] [PubMed] [Google Scholar]
- 24.Bokkenheuser VD, Winter J. Biotransformation of steroid hormones by gut bacteria. Am J Clin Nutr. 1980;33:2502–2506. doi: 10.1093/ajcn/33.11.2502. [DOI] [PubMed] [Google Scholar]
- 25.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 26.Brabban AD, Edwards C. Isolation of glucosinolate degrading microorganisms and their potential for reducing the glucosinolate content of rapemeal. FEMS Microbiol Lett. 1994;119:83–88. doi: 10.1111/j.1574-6968.1994.tb06871.x. [DOI] [PubMed] [Google Scholar]
- 27.Burnett-Hartman AN, Newcomb PA, Potter JD. Infectious agents and colorectal cancer: a review of Helicobacter pylori, Streptococcus bovis, JC virus, and human papillomavirus. Cancer Epidemiol Biomarkers Prev. 2008;17:2970–2979. doi: 10.1158/1055-9965.EPI-08-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cammarota R, Bertolini V, Pennesi G, et al. The tumor microenvironment of colorectal cancer: stromal TLR-4 expression as a potential prognostic marker. J Transl Med. 2010;8:112. doi: 10.1186/1479-5876-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cani PD, Neyrinck AM, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 30.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 31.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2011;22:299. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang YC, Nair MG. Metabolism of daidzein and genistein by intestinal bacteria. J Nat Prod. 1995;58:1892–1896. doi: 10.1021/np50126a014. [DOI] [PubMed] [Google Scholar]
- 33.Chen RFL, Wang Y, et al. LBP and CD14 polymorphisms correlate with increased colorectal carcinoma risk in Han Chinese. World J Gastroenterol. 2011;17:2326–2331. doi: 10.3748/wjg.v17.i18.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christl SU, Eisner HD, Dusel G, et al. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: a potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci. 1996;41:2477–2481. doi: 10.1007/BF02100146. [DOI] [PubMed] [Google Scholar]
- 35.Conaway CC, Getahun SM, Liebes LL, et al. Disposition of glucosinolates and sulforaphane in humans after ingestion of steamed and fresh broccoli. Nutr Cancer. 2000;38:168–178. doi: 10.1207/S15327914NC382_5. [DOI] [PubMed] [Google Scholar]
- 36.Conterno L, Fava F, Viola R, et al. Obesity and the gut microbiota: does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr. 2011;6:241–260. doi: 10.1007/s12263-011-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creely SJ, McTernan PG, Kusminski CM, et al. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292:E740–E747. doi: 10.1152/ajpendo.00302.2006. [DOI] [PubMed] [Google Scholar]
- 39.Cross AJ, Pollock JR, Bingham SA. Haem, not protein or inorganic iron, is responsible for endogenous intestinal N-nitrosation arising from red meat. Cancer Res. 2003;63:2358–2360. [PubMed] [Google Scholar]
- 40.Cummings JH, Wiggins HS, Jenkins DJ, et al. Influence of diets high and low in animal fat on bowel habit, gastrointestinal transit time, fecal microflora, bile acid, and fat excretion. J Clin Invest. 1978;61:953–963. doi: 10.1172/JCI109020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dabek M, McCrae SI, Stevens VJ, et al. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. 2008;66:487–495. doi: 10.1111/j.1574-6941.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- 42.De Flora S, Bonanni P. The prevention of infection-associated cancers. Carcinogenesis. 2011;32:787–795. doi: 10.1093/carcin/bgr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Decroos K, Vanhemmens S, Cattoir S, et al. Isolation and characterisation of an equol-producing mixed microbial culture from a human faecal sample and its activity under gastrointestinal conditions. Arch Microbiol. 2005;183:45–55. doi: 10.1007/s00203-004-0747-4. [DOI] [PubMed] [Google Scholar]
- 44.Deplancke B, Gaskins HR. Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J. 2003;17:1310–1312. doi: 10.1096/fj.02-0883fje. [DOI] [PubMed] [Google Scholar]
- 45.Deplancke B, Hristova KR, Oakley HA, et al. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl Environ Microbiol. 2000;66:2166–2174. doi: 10.1128/aem.66.5.2166-2174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deplancke B, Finster K, Graham WV, et al. Gastrointestinal and microbial responses to sulfate-supplemented drinking water in mice. Exp Biol Med (Maywood) 2003;228:424–433. doi: 10.1177/153537020322800413. [DOI] [PubMed] [Google Scholar]
- 47.Doerner KC, Takamine F, LaVoie CP, et al. Assessment of fecal bacteria with bile acid 7 alpha-dehydroxylating activity for the presence of bai-like genes. Appl Environ Microbiol. 1997;63:1185–1188. doi: 10.1128/aem.63.3.1185-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan AM, Merz-Demlow BE, Xu X, et al. Premenopausal equol excretors show plasma hormone profiles associated with lowered risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:581–586. [PubMed] [Google Scholar]
- 49.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elfoul L, Rabot S, Khelifa N, et al. Formation of allyl isothiocyanate from sinigrin in the digestive tract of rats monoassociated with a human colonic strain of Bacteroides thetaiotaomicron. FEMS Microbiol Lett. 2001;197:99–103. doi: 10.1111/j.1574-6968.2001.tb10589.x. [DOI] [PubMed] [Google Scholar]
- 51.Ernst P. Review article: the role of inflammation in the pathogenesis of gastric cancer. Aliment Pharmacol Ther. 1999;13(Suppl 1):13–18. doi: 10.1046/j.1365-2036.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 52.Florin T, Neale G, Gibson GR, et al. Metabolism of dietary sulfate—absorption and excretion in humans. Gut. 1991;32:766–773. doi: 10.1136/gut.32.7.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank DN, Pace NR. Molecular-phylogenetic analyses of human gastrointestinal microbiota. Curr Opin Gastroenterol. 2001;17:52–57. doi: 10.1097/00001574-200101000-00010. [DOI] [PubMed] [Google Scholar]
- 54.Frankenfeld CL, McTiernan A, Aiello EJ, et al. Mammographic density in relation to daidzein-metabolizing phenotypes in overweight, postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2004;13:1156–1162. [PubMed] [Google Scholar]
- 55.Friedrich MW. Phylogenetic analysis reveals multiple lateral transfers of adenosine- 5′-phosphosulfate reductase genes among sulfate-reducing microorganisms. J Bacteriol. 2002;184:278–289. doi: 10.1128/JB.184.1.278-289.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukata M, Abreu MT. TLR4 signalling in the intestine in health and disease. Biochem Soc Trans. 2007;35:1473–1478. doi: 10.1042/BST0351473. [DOI] [PubMed] [Google Scholar]
- 57.Gerner EW. Impact of dietary amino acids and polyamines on intestinal carcinogenesis and chemoprevention in mouse models. Biochem Soc Trans. 2007;35:322–325. doi: 10.1042/BST0350322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerner EW, Meyskens FL., Jr Combination chemoprevention for colon cancer targeting polyamine synthesis and inflammation. Clin Cancer Res. 2009;15:758–761. doi: 10.1158/1078-0432.CCR-08-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gibson GR, Cummings JH, Macfarlane GT. Use of a three-stage continuous culture system to study the effect of mucin on dissimilatory sulfate reduction and methanogenesis by mixed populations of human gut bacteria. Appl Environ Microbiol. 1988;54:2750–2755. doi: 10.1128/aem.54.11.2750-2755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson GR, Cummings JH, Macfarlane GT. Growth and activities of sulfate-reducing bacteria in gut contents of healthy-subjects and patients with ulcerative-colitis. FEMS Microbiol Ecol. 1991;86:103–111. [Google Scholar]
- 61.Gibson GR, Macfarlane S, Macfarlane GT. Metabolic interactions involving sulfate-reducing and methanogenic bacteria in the human large-intestine. FEMS Microbiol Ecol. 1993;12:117–125. [Google Scholar]
- 62.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodwin AC, Destefano Shields CE, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci USA. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hague A, Elder DJ, Hicks DJ, et al. Apoptosis in colorectal tumour cells: induction by the short chain fatty acids butyrate, propionate and acetate and by the bile salt deoxycholate. Int J Cancer. 1995;60:400–406. doi: 10.1002/ijc.2910600322. [DOI] [PubMed] [Google Scholar]
- 65.Hayashi H, Sakamoto M, Benno Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol Immunol. 2002;46:535–548. doi: 10.1111/j.1348-0421.2002.tb02731.x. [DOI] [PubMed] [Google Scholar]
- 66.Hebels DG, Sveje KM, de Kok MC, et al. N-nitroso compound exposure-associated transcriptomic profiles are indicative of an increased risk for colorectal cancer. Cancer Lett. 2011;309:1–10. doi: 10.1016/j.canlet.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Hebels DG, Sveje KM, de Kok MC, et al. Red meat intake-induced increases in fecal water genotoxicity correlate with pro-carcinogenic gene expression changes in the human colon. Food Chem Toxicol. 2012;50:95–103. doi: 10.1016/j.fct.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 68.Hirayama A, Kami K, Sugimoto M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- 69.Holst B, Williamson G. A critical review of the bioavailability of glucosinolates and related compounds. Nat Prod Rep. 2004;21:425–447. doi: 10.1039/b204039p. [DOI] [PubMed] [Google Scholar]
- 70.Holst JJ, Deacon CF. Glucagon-like peptide 1 and inhibitors of dipeptidyl peptidase IV in the treatment of type 2 diabetes mellitus. Curr Opin Pharmacol. 2004;4:589–596. doi: 10.1016/j.coph.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 71.Hristova KR, Mau M, Zheng D, et al. Desulfotomaculum genus- and subgenus-specific 16S rRNA hybridization probes for environmental studies. Environ Microbiol. 2000;2:143–159. doi: 10.1046/j.1462-2920.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 72.Huang JQ, Sridhar S, Chen Y, et al. Meta-analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology. 1998;114:1169–1179. doi: 10.1016/s0016-5085(98)70422-6. [DOI] [PubMed] [Google Scholar]
- 73.Hughes R, Cross AJ, Pollock JR, et al. Dose-dependent effect of dietary meat on endogenous colonic N-nitrosation. Carcinogenesis. 2001;22:199–202. doi: 10.1093/carcin/22.1.199. [DOI] [PubMed] [Google Scholar]
- 74.Hughes R, Pollock JR, Bingham S. Effect of vegetables, tea, and soy on endogenous N-nitrosation, fecal ammonia, and fecal water genotoxicity during a high red meat diet in humans. Nutr Cancer. 2002;42:70–77. doi: 10.1207/S15327914NC421_10. [DOI] [PubMed] [Google Scholar]
- 75.Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med (Maywood) 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- 76.Ignatenko NA, Gerner EW, Besselsen DG. Defining the role of polyamines in colon carcinogenesis using mouse models. J Carcinog. 2011;10:10. doi: 10.4103/1477-3163.79673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Järvenpää P. In vitro metabolism of catechol estrogens by human fecal microflora. J Steroid Biochem. 1990;35:289–292. doi: 10.1016/0022-4731(90)90286-2. [DOI] [PubMed] [Google Scholar]
- 78.Järvenpää P, Kosunen T, Fotsis T, et al. In vitro metabolism of estrogens by isolated intestinal micro-organisms and by human faecal microflora. J Steroid Biochem. 1980;13:345–349. doi: 10.1016/0022-4731(80)90014-x. [DOI] [PubMed] [Google Scholar]
- 79.Joshi AD, Corral R, Siegmund KD, et al. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30:472–479. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanazawa K, Konishi F, Mitsuoka T, et al. Factors influencing the development of sigmoid colon cancer. Bacteriologic and biochemical studies. Cancer. 1996;77:1701–1706. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1701::AID-CNCR42>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 81.Kelly GE, Joannou GE, Reeder AY, et al. The variable metabolic response to dietary isoflavones in humans. Proc Soc Exp Biol Med. 1995;208:40–43. doi: 10.3181/00379727-208-43829. [DOI] [PubMed] [Google Scholar]
- 82.Key TJ. Serum oestradiol and breast cancer risk. Endocr Relat Cancer. 1999;6:175–180. doi: 10.1677/erc.0.0060175. [DOI] [PubMed] [Google Scholar]
- 83.Kitahara M, Takamine F, Imamura T, et al. Assignment of Eubacterium sp. VPI 12708 and related strains with high bile acid 7alpha-dehydroxylating activity to Clostridium scindens and proposal of Clostridium hylemonae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol. 2000;50(Pt 3):971–978. doi: 10.1099/00207713-50-3-971. [DOI] [PubMed] [Google Scholar]
- 84.Knudtson LM, Hartman PA. Comparison of fluorescent gentamicin-thallous-carbonate and KF streptococcal agars to enumerate enterococci and fecal streptococci in meats. Appl Environ Microbiol. 1993;59:936–938. doi: 10.1128/aem.59.3.936-938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kristal AR, Lampe JW. Brassica vegetables and prostate cancer risk: a review of the epidemiological evidence. Nutr Cancer. 2002;42:1–9. doi: 10.1207/S15327914NC421_1. [DOI] [PubMed] [Google Scholar]
- 86.Lampe JW. The human microbiome project: getting to the guts of the matter in cancer epidemiology. Cancer Epidemiol Biomarkers Prev. 2008;17:2523–2524. doi: 10.1158/1055-9965.EPI-08-0792. [DOI] [PubMed] [Google Scholar]
- 87.Lampe J. Emerging research on equol and cancer. J Nutr. 2010;140:1369S–1372S. doi: 10.3945/jn.109.118323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lampe JW, Karr SC, Hutchins AM, et al. Urinary equol excretion with a soy challenge: influence of habitual diet. Proc Soc Exp Biol Med. 1998;217:335–339. doi: 10.3181/00379727-217-44241. [DOI] [PubMed] [Google Scholar]
- 89.Laukaitis CM, Gerner EW. DFMO: targeted risk reduction therapy for colorectal neoplasia. Best Pract Res Clin Gastroenterol. 2011;25:495–506. doi: 10.1016/j.bpg.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lazcano-Ponce EC, Miquel JF, Munoz N, et al. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J Clin. 2001;51:349–364. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 91.Lee J, Michael AJ, Martynowski D, et al. Phylogenetic diversity and the structural basis of substrate specificity in the beta/alpha-barrel fold basic amino acid decarboxylases. J Biol Chem. 2007;282:27115–27125. doi: 10.1074/jbc.M704066200. [DOI] [PubMed] [Google Scholar]
- 92.Ley RE, Bäckhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li F, Hullar MA, Beresford SA, et al. Variation of glucoraphanin metabolism in vivo and ex vivo by human gut bacteria. Br J Nutr. 2011;106:408–416. doi: 10.1017/S0007114511000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lombardi P, Goldin B, Boutin E, et al. Metabolism of androgens and estrogens by human fecal microorganisms. J Steroid Biochem. 1978;9:795–801. doi: 10.1016/0022-4731(78)90203-0. [DOI] [PubMed] [Google Scholar]
- 95.Magee EA, Curno R, Edmond LM, et al. Contribution of dietary protein and inorganic sulfur to urinary sulfate: toward a biomarker of inorganic sulfur intake. Am J Clin Nutr. 2004;80:137–142. doi: 10.1093/ajcn/80.1.137. [DOI] [PubMed] [Google Scholar]
- 96.Mager DL. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 2006;4:14. doi: 10.1186/1479-5876-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manco M, Putignani L, Bottazzo GF. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev. 2010;31:817–844. doi: 10.1210/er.2009-0030. [DOI] [PubMed] [Google Scholar]
- 98.Maneval ML, Eckert KA. Effects of oxidative and alkylating damage on microsatellite instability in nontumorigenic human cells. Mutat Res. 2004;546:29–38. doi: 10.1016/j.mrfmmm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 99.Mann CJ. Observational research methods. Research design II: cohort, cross sectional, and case-control studies. Emerg Med J. 2003;20:54–60. doi: 10.1136/emj.20.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS ONE. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Markiewicz L, Garey J, Adlercreutz H, et al. In vitro bioassays of non-steroidal phytoestrogens. J Steroid Biochem Mol Biol. 1993;45:399–405. doi: 10.1016/0960-0760(93)90009-l. [DOI] [PubMed] [Google Scholar]
- 102.Martin F, Peltonen J, Laatikainen T, et al. Excretion of progesterone metabolites and estriol in faeces from pregnant women during ampicillin administration. J Steroid Biochem. 1975;6:1339–1346. doi: 10.1016/0022-4731(75)90363-5. [DOI] [PubMed] [Google Scholar]
- 103.Massey RC, Key PE, Mallett AK, et al. An investigation of the endogenous formation of apparent total N-nitroso compounds in conventional microflora and germ-free rats. Food Chem Toxicol. 1988;26:595–600. doi: 10.1016/0278-6915(88)90230-x. [DOI] [PubMed] [Google Scholar]
- 104.McGarr SE, Ridlon JM, Hylemon PB. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol. 2005;39:98–109. [PubMed] [Google Scholar]
- 105.Meyskens FL, Jr, Gerner EW. Back to the future: mechanism-based, mutation-specific combination chemoprevention with a synthetic lethality approach. Cancer Prev Res (Phila) 2011;4:628–632. doi: 10.1158/1940-6207.CAPR-11-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mower HF, Ray RM, Shoff R, et al. Fecal bile acids in two Japanese populations with different colon cancer risks. Cancer Res. 1979;39:328–331. [PubMed] [Google Scholar]
- 107.Mudd DG, McKelvey ST, Norwood W, et al. Faecal bile acid concentration of patients with carcinoma or increased risk of carcinoma in the large bowel. Gut. 1980;21:587–590. doi: 10.1136/gut.21.7.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Munro IC, Harwood M, Hlywka JJ, et al. Soy isoflavones: a safety review. Nutr Rev. 2003;61:1–33. doi: 10.1301/nr.2003.janr.1-33. [DOI] [PubMed] [Google Scholar]
- 109.Myzak MC, Hardin K, Yan M, et al. Sulforaphane inhibits HDAC activity in prostate cancer cells, retards growth of PC3 xenografts, and inhibits HDAC activity in vivo. FASEB J. 2006;20:A150–A150. [Google Scholar]
- 110.Nagel SC, vom Saal FS, Welshons WV. The effective free fraction of estradiol and xenoestrogens in human serum measured by whole cell uptake assays: physiology of delivery modifies estrogenic activity. Proc Soc Exp Biol Med. 1998;217:300–309. doi: 10.3181/00379727-217-44236. [DOI] [PubMed] [Google Scholar]
- 111.Navarro SL, Li F, Lampe JW. Mechanisms of action of isothiocyanates in cancer chemoprevention: an update. Food Funct. 2011;2:579–587. doi: 10.1039/c1fo10114e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Neidhardt FC, Magasanik B. Studies on the role of ribonucleic acid in the growth of bacteria. Biochim Biophys Acta. 1960;42:99–116. doi: 10.1016/0006-3002(60)90757-5. [DOI] [PubMed] [Google Scholar]
- 113.Nettleton JA, Greany KA, Thomas W, et al. The effect of soy consumption on the urinary 2:16-hydroxyestrone ratio in postmenopausal women depends on equol production status but is not influenced by probiotic consumption. J Nutr. 2005;135:603–608. doi: 10.1093/jn/135.3.603. [DOI] [PubMed] [Google Scholar]
- 114.Norat T, Bingham S, Ferrari P, et al. Meat, fish, and colorectal cancer risk: the European prospective investigation into cancer and nutrition. J Natl Cancer Inst. 2005;97:906–916. doi: 10.1093/jnci/dji164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O’Keefe SJD, Carrim Y, van der Merwe CF, et al. Differences in diet and colonic bacterial metabolism that might account for the low risk of colon cancer in native Africans compared with Americans. J Nutr. 2004;134:3526S–3527S. [Google Scholar]
- 116.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 117.Pastor Rojo O, Lopez San Roman A, Albeniz Arbizu E, et al. Serum lipopolysaccharide-binding protein in endotoxemic patients with inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:269–277. doi: 10.1002/ibd.20019. [DOI] [PubMed] [Google Scholar]
- 118.Pitcher MCL, Beatty ER, Gibson GR, et al. Sulfate-reducing bacteria—prevalence in active and inactive ulcerative-colitis. Gastroenterology. 1995;108:A894–A894. [Google Scholar]
- 119.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rimbach G, De Pascual-Teresa S, Ewins BA, et al. Antioxidant and free radical scavenging activity of isoflavone metabolites. Xenobiotica. 2003;33:913–925. doi: 10.1080/0049825031000150444. [DOI] [PubMed] [Google Scholar]
- 121.Roediger WEW. Decreased sulphur amino acid intake in ulcerative colitis. Lancet. 1998;351:1555. doi: 10.1016/s0140-6736(05)61120-8. [DOI] [PubMed] [Google Scholar]
- 122.Roediger WEW, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci. 1997;42:1571–1579. doi: 10.1023/a:1018851723920. [DOI] [PubMed] [Google Scholar]
- 123.Rooks MG, Garrett WS. Bacteria, food, and cancer. F1000 Biol Rep. 2011;3:12. doi: 10.3410/B3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rosset R, Julien J, Monier R. Ribonucleic acid composition of bacteria as a function of growth rate. J Mol Biol. 1966;18:308–320. doi: 10.1016/s0022-2836(66)80248-6. [DOI] [PubMed] [Google Scholar]
- 125.Rouzaud G, Rabot S, Ratcliffe B, et al. Influence of plant and bacterial myrosinase activity on the metabolic fate of glucosinolates in gnotobiotic rats. Br J Nutr. 2003;90:395–404. doi: 10.1079/bjn2003900. [DOI] [PubMed] [Google Scholar]
- 126.Rouzaud G, Young SA, Duncan AJ. Hydrolysis of glucosinolates to isothiocyanates after ingestion of raw or microwaved cabbage by human volunteers. Cancer Epidemiol Biomarkers Prev. 2004;13:125–131. doi: 10.1158/1055-9965.epi-085-3. [DOI] [PubMed] [Google Scholar]
- 127.Rowland IR. Toxicological implications of the normal microflora. In: Tannock GW, editor. Medical importance of the normal microflora. Kluwer Academic Publishers; Dordrecht: 1999. [Google Scholar]
- 128.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 130.Scanlan PD, Shanahan F, Clune Y, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. 2008;10:789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 131.Schaechter M, Maalow O, Kjelgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 132.Setchell KDR, Adlercreutz H. Mammalian lignans and phyto-oestrogens: recent studies on their formation, metabolism and biological role in health and disease. In: Rowland I, editor. Role of the gut flora in toxicity and cancer. Academic Press; NY: 1988. pp. 316–345. [Google Scholar]
- 133.Setchell KD, Brown NM, Lydeking-Olsen E. The clinical importance of the metabolite equol-a clue to the effectiveness of soy and its isoflavones. J Nutr. 2002;132:3577–3584. doi: 10.1093/jn/132.12.3577. [DOI] [PubMed] [Google Scholar]
- 134.Shapiro TA, Fahey JW, Wade KL, et al. Human metabolism and excretion of cancer chemoprotective glucosinolates and isothiocyanates of cruciferous vegetables. Cancer Epidemiol Biomarkers Prev. 1998;7:1091–1100. [PubMed] [Google Scholar]
- 135.Shimada Y, Yasuda S, Takahashi M, et al. Cloning and expression of a novel NADP(H)-dependent daidzein reductase, an enzyme involved in the metabolism of daidzein, from equol-producing Lactococcus strain 20–92. Appl Environ Microbiol. 2010;76:5892–5901. doi: 10.1128/AEM.01101-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Silvester KR, Cummings JH. Does digestibility of meat protein help explain large bowel cancer risk? Nutr Cancer. 1995;24:279–288. doi: 10.1080/01635589509514417. [DOI] [PubMed] [Google Scholar]
- 137.Soreide K. Proteinase-activated receptor 2 (PAR-2) in gastrointestinal and pancreatic pathophysiology, inflammation and neoplasia. Scand J Gastroenterol. 2008;43:902–909. doi: 10.1080/00365520801942141. [DOI] [PubMed] [Google Scholar]
- 138.Spencer JPE, Crozier A. Flavonoids and related compounds. CRC Press, Taylor & Francis Group; Boca Raton: 2012. [Google Scholar]
- 139.Tannock GW. The normal microflora: an introduction. In: Tannock GW, editor. Medical importance of the normal microfloraed. Kluwer Academic Publishers; Dordrecht: 1999. pp. 1–23. [Google Scholar]
- 140.Tannock GW, Munro K, Harmsen HJ, et al. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl Environ Microbiol. 2000;66:2578–2588. doi: 10.1128/aem.66.6.2578-2588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thomas LA, Veysey MJ, Bathgate T, et al. Mechanism for the transit-induced increase in colonic deoxycholic acid formation in cholesterol cholelithiasis. Gastroenterology. 2000;119:806–815. doi: 10.1053/gast.2000.16495. [DOI] [PubMed] [Google Scholar]
- 142.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 143.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Turner R, Baron T, Wolffram S, et al. Effect of circulating forms of soy isoflavones on the oxidation of low density lipoprotein. Free Radic Res. 2004;38:209–216. doi: 10.1080/10715760310001641854. [DOI] [PubMed] [Google Scholar]
- 145.Vedavanam K, Srijayanta S, O’Reilly J, et al. Antioxidant action and potential antidiabetic properties of an isoflavonoid-containing soyabean phytochemical extract (SPE) Phytother Res. 1999;13:601–608. doi: 10.1002/(sici)1099-1573(199911)13:7<601::aid-ptr550>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 146.Vermeulen M, Van den Berg R, Freidig AP, et al. Association between consumption of cruciferous vegetables and condiments and excretion in urine of isothiocyanate mercapturic acids. J Agric Food Chem. 2006;54:5350–5358. doi: 10.1021/jf060723n. [DOI] [PubMed] [Google Scholar]
- 147.Wang XQ, Terry PD, Yan H. Review of salt consumption and stomach cancer risk: epidemiological and biological evidence. World J Gastroenterol. 2009;15:2204–2213. doi: 10.3748/wjg.15.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wells JE, Hylemon PB. Identification and characterization of a bile acid 7alpha-dehydroxylation operon in Clostridium sp. strain TO-931, a highly active 7alpha-dehydroxylating strain isolated from human feces. Appl Environ Microbiol. 2000;66:1107–1113. doi: 10.1128/aem.66.3.1107-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wells JE, Berr F, Thomas LA, et al. Isolation and characterization of cholic acid 7alpha-dehydroxylating fecal bacteria from cholesterol gallstone patients. J Hepatol. 2000;32:4–10. doi: 10.1016/s0168-8278(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 150.Wilson KH, Blitchington RB. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Winter J, Bokkenheuser VD. Bacterial metabolism of natural and synthetic sex hormones undergoing enterohepatic circulation. J Steroid Biochem. 1987;27:1145–1149. doi: 10.1016/0022-4731(87)90201-9. [DOI] [PubMed] [Google Scholar]
- 152.World Cancer Research Fund. Food, nutrition, physical activity, and the prevention of cancer: a global perspective. American Institute for Cancer Research; Washington, D.C: 2007. [DOI] [PubMed] [Google Scholar]
- 153.Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 154.Zverlov V, Klein M, Lucker S, et al. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J Bacteriol. 2005;187:2203–2208. doi: 10.1128/JB.187.6.2203-2208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]