Abstract
Workers in our laboratory have previously identified the staphylococcal respiratory response AB (SrrAB), a Staphylococcus aureus two-component system that acts in the global regulation of virulence factors. This system down-regulates production of agr RNAIII, protein A, and toxic shock syndrome toxin 1 (TSST-1), particularly under low-oxygen conditions. In this study we investigated the localization and membrane orientation of SrrA and SrrB, transcription of the srrAB operon, the DNA-binding properties of SrrA, and the effect of SrrAB expression on S. aureus virulence. We found that SrrA is localized to the S. aureus cytoplasm, while SrrB is localized to the membrane and is properly oriented to function as a histidine kinase. srrAB has one transcriptional start site which results in either an srrA transcript or a full-length srrAB transcript; srrB must be cotranscribed with srrA. Gel shift assays of the agr P2, agr P3, protein A (spa), TSST-1 (tst), and srr promoters revealed SrrA binding at each of these promoters. Analysis of SrrAB-overexpressing strains by using the rabbit model of bacterial endocarditis demonstrated that overexpression of SrrAB decreased the virulence of the organisms compared to the virulence of isogenic strains that do not overexpress SrrAB. We concluded that SrrAB is properly localized and oriented to function as a two-component system. Overexpression of SrrAB, which represses agr RNAIII, TSST-1, and protein A in vitro, decreases virulence in the rabbit endocarditis model. Repression of these virulence factors is likely due to a direct interaction between SrrA and the agr, tst, and spa promoters.
Staphylococcus aureus is a gram-positive coccus that is responsible for a wide variety of community-acquired and hospital-acquired infections. S. aureus may cause relatively minor skin infections, but more severe infections, such as endocarditis, osteomyelitis, and toxic shock syndrome, may result from the coordinated expression of virulence factors (25).
Several global regulators of virulence in S. aureus have been described. One of these systems, the accessory gene regulator system (agr), has been well described in vitro. agr consists of two divergently transcribed promoters; promoter 2 directs transcription of RNAII, which encodes the AgrBDCA quorum-sensing two-component system, while promoter 3 directs transcription of RNAIII, which is the effector molecule of the agr locus (24, 28). In vitro studies have revealed that RNAIII expression is highest during the postexponential and stationary phases of growth, during which RNAIII enhances expression of exotoxins and represses expression of surface-associated virulence factors. Although these in vitro studies demonstrated that agr plays a role in the global regulation of virulence factors, several in vivo studies of agr expression have shown that RNAIII is repressed in animal infection models (11, 36), as well as during human lung infection (10). Recent evidence concerning the role of agr in vivo has indicated that agr is expressed by S. aureus intracellularly (26, 33).
Other global regulators of virulence include the staphylococcal accessory gene regulator (sar), as well as the staphylococcal accessory element (sae). SarA has been shown to activate transcription of extracellular proteins, such as α- and β-hemolysins, toxic shock syndrome toxin 1 (TSST-1), staphylococcal enterotoxin B, and cell surface proteins, including fibronectin binding protein. SarA also represses protease activity (3). Gel shift assays with SarA have indicated that its effects are mediated by agr-dependent mechanisms, as well as agr-independent mechanisms (4, 5, 13, 27, 34). The saeRS locus encodes a two-component system that controls exoprotein synthesis at the transcriptional level (7, 9). This system has been shown to enhance transcription of α- and β-hemolysins and coagulase, while disruption of sae has no effect on agr or sarA transcription (8).
Although much is known about the in vitro activities of these global regulators of virulence, we know very little about how S. aureus responds to the in vivo infection environment. S. aureus is known to regulate TSST-1 production in response to NaCl, sucrose, magnesium, oxygen, temperature, and carbon dioxide (2, 15, 29, 35, 38). In particular, several authors have described the ability of S. aureus to regulate TSST-1 production in response to oxygen (15, 29, 38). There are no known systems by which S. aureus responds to oxygen. For this reason, workers in our laboratory began to search for an oxygen-responsive virulence regulator in S. aureus. We searched the S. aureus genome for homologs of the Bacillus subtilis ResDE two-component system, which has been described as a regulator of anaerobic respiration and aerobic respiration (21). We designated this putative staphylococcal two-component system the staphylococcal respiratory response AB (SrrAB). Analysis of the srrAB locus revealed two predicted open reading frames that overlap by 20 bp. srrA was predicted to encode a 28-kDa, 241-amino-acid response regulator, while srrB was predicted to encode a 66-kDa, 583-amino-acid histidine kinase. Creation of an srrB null mutant, as well as subsequent complementation of srrAB in trans, revealed that expression of agr RNAIII was inversely related to expression of SrrAB. Expression of TSST-1 in the srrB null mutant was decreased, particularly under microaerobic conditions. Overexpression of SrrAB resulted in repression of TSST-1, even under aerobic conditions. Expression of protein A in the srrB null mutant was upregulated in microaerobic conditions and decreased in aerobic conditions. When SrrAB was overexpressed, protein A production was restored to nearly wild-type levels. Overexpression of srrAB in trans also appeared to upregulate expression of the truncated chromosomal copy of srrAB. We concluded that SrrAB acts in the global regulation of staphylococcal virulence factors and may repress virulence factors under low-oxygen conditions (37). Following our description of the identification of SrrAB, another group determined the role of SrrAB in regulating metabolic processes and energy transduction in response to oxygen availability by using a proteomics approach (32).
As our studies have indicated that SrrAB plays a role in virulence factor regulation in response to oxygen availability, we performed this study in order to investigate the mechanisms by which SrrAB acts. Here we report on the subcellular localization and transcriptional regulation of SrrAB, the mechanism by which SrrAB may regulate agr RNAIII, TSST-1, protein A, and SrrAB expression, and the effect of SrrAB on the virulence of S. aureus in a rabbit endocarditis model.
MATERIALS AND METHODS
Strains and growth conditions.
The strains and plasmids used in this study are described in Table 1. Escherichia coli was grown in Luria-Bertani medium with appropriate antibiotic selection. S. aureus was grown in Todd-Hewitt medium (Difco Laboratories, Sparks, Md.) with appropriate antibiotic selection. All strains were grown in a laboratory aerobic atmosphere with shaking.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source and/or reference |
|---|---|---|
| E. coli BL21(DE3) | B F−dcm ompT hsdS (rB− mB−) gal λ (DE3) | Stratagene |
| S. aureus strains | ||
| MN8 | tstH | 1 |
| RN4220 | 8325-4 rK− mK+ | 16 |
| MN3050 | RN4220(pJMY10) tstH+ | 37 |
| MN4011 | RN4220 srrBΩpJMY2 Emr (pJMY11) tstH+srrAB+ | 37 |
| DU5875 | 8325-4 Δspa::Tcr, Tcr | T. Foster (20) |
| DU5875(pJMY11) | DU5875(pJMY11), Tcr Cmr | This study |
| RN6390 | 8325-4 | R. Novick (23) |
| RN6390(pJMY10) | 8325-4(pJMY10), Cmr | This study |
| RN6390(pJMY11) | 8325-4(pJMY11), Cmr | This study |
| Plasmids | ||
| pJMY10 | 805-bp Cmr cassette from pIJ002 cloned into the PstI and ScaI sites of pCE107, tstH, 5.4 kb, Cmr | 37 |
| pJMY11 | 2,712-bp srrAB operon cloned into the XmaI site of pJMY10, tstH srrAB, 8.2 kb, Cmr | 37 |
| pJMY21 | 402-bp portion of srrB (SrrB*) cloned into NheI-BamHI sites of pET28b | This study |
| pJMY22 | 726-bp full-length srrA cloned into NheI-BamHI sites of pET28b | This study |
Generation of anti-SrrA and anti-SrrB* antibodies.
Recombinant SrrA and SrrB* (the putative extracellular portion of SrrB, QYYFT. . . IEDTN) were produced by independently cloning the full-length srrA gene and a portion of the srrB gene into the pET28b expression vector (Novagen, Madison, Wis.). E. coli strains BL21(pJMY22) and BL21(pJMY21) containing the pET28b-SrrA and pET28b-SrrB* constructs, respectively, were grown in Luria-Bertani medium, and when an optical density at 600 nm of ∼0.5 was reached, they were induced with 200 μM isopropyl β-d-thiogalactopyranoside (Sigma Chemical Co., St. Louis, Mo.) and incubated for an additional 3 h. Cells were harvested by centrifugation, resuspended in the loading buffer of a His-Bind resin and buffer kit (Novagen), and lysed by ultrasonication. The histidine-tagged proteins were then purified by column chromatography by following the manufacturer's directions. The histidine tag was removed from SrrA by thrombin cleavage (thrombin cleavage capture kit; Novagen), and both proteins were purified by high-performance liquid chromatography (HPLC) (University of Minnesota Microchemical Facility). The purity and size of proteins were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the sequence was verified by N-terminal sequencing (Mayo Protein Core Facility). Anti-SrrA and anti-SrrB* antibodies were obtained by immunizing Dutch-Belted rabbits with recombinant SrrA (rSrrA) and recombinant His6-SrrB* (rSrrB*). Rabbits were given three subcutaneous injections of 40 μg of purified protein that was resuspended in phosphate-buffered saline (PBS) (5 mM NaPO4, 0.15 M NaCl; pH 7.2) and emulsified in Freund's incomplete adjuvant (GibcoBRL, Grand Island, N.Y.) at 2-week intervals. Development of sensitive and specific antibodies was assessed by Western analysis by using an S. aureus cell extract and the appropriate recombinant protein.
Subcellular localization of SrrA and SrrB.
S. aureus strains DU5875 and DU5875(pJMY11) were grown to the postexponential phase. Samples were pelleted, washed twice in PBS, and resuspended in 100 μl of lysis buffer (30 mM Tris [pH 8], 1 mM EDTA, 20% sucrose, 1 mg of lysostaphin per ml). Samples were incubated on ice for 10 min and then pelleted and resuspended in 100 μl of 100 mM Tris (pH 8). The bacteria were lysed by sonication, and cytoplasmic and membrane extracts were separated by ultracentrifugation (48,000 rpm in a Beckman Optima preparative ultracentrifuge with an SW50.1 rotor at 4°C for 2 h). Samples were subjected to SDS-PAGE and blotted for Western analysis with anti-SrrA and anti-SrrB* by using standard procedures (30).
Immunostaining with anti-SrrB*.
Cells were stained with a 3,3′-diaminobenzidine (DAB) substrate kit for peroxidase (Vector Laboratories, Inc., Burlingame, Calif.) used according to the manufacturer's instructions. Briefly, S. aureus strain MN8 was grown to the stationary phase, washed in PBS, diluted, and spotted onto slides. The cells were dried and fixed with 10% formalin, and endogenous peroxidases were inactivated with 1.5% H2O2. The cells were blocked with horse serum and then incubated with a 1:100 dilution of either anti-SrrB* or normal rabbit serum at 4°C overnight. The cells were washed and incubated with a 1:100 dilution of the secondary anti-rabbit peroxidase conjugate (Sigma) for 1 h at room temperature. Staining was detected with the chromogenic peroxidase substrate DAB. The cells were counterstained with hematoxylin and examined for brown DAB staining by using light microscopy.
Primer extension analysis.
Primer extension was conducted as described by Sambrook and Russell (30). Briefly, primers located approximately 100 bp downstream of the start codons for srrA and srrB were radiolabeled with [γ-33P]ATP and cleaned with Microspin G-25 columns (Amersham Pharmacia Biotech), ∼2.5 × 105 cpm was incubated with reverse transcriptase, and RNA was harvested from exponential-phase cultures of MN8 grown in the presence of 21% oxygen and 7% carbon dioxide. The same primers were used in a PCR-based sequencing reaction with radiolabeled terminators (dideoxynucleoside triphosphates [15]) by using a U.S. Biochemicals ThermoSequence radiolabeled terminator cycle sequencing kit and MN8 genomic DNA as the template. The PCR conditions were 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min. Products from both the primer extension analysis and PCRs were electrophoresed simultaneously on a denaturing polyacrylamide gel (QuickPoint rapid nucleic acid separation system; NOVEX/Invitrogen). Gels were dried and exposed to radiographic film.
Gel shifts.
Gel shift reactions were carried out by using a digoxigenin gel shift kit (Roche, Mannheim, Germany) according to the manufacturer's instructions. Briefly, approximately 100-bp regions upstream of the translational start sites of the agrB, agr RNAIII, spa, srr, and tst loci were PCR amplified as described in Table 2. DNA probes were purified by using a QIAquick nucleotide removal kit (Qiagen, Valencia, Calif.) and labeled with digoxigenin. His6-SrrA was purified from E. coli BL21(pJMY22) as described above and then immediately frozen in single-use aliquots in order to preserve the activity of SrrA. Gel shift reactions were carried out by incubating 1.2 nM labeled probe and 3 ng of poly(dI-dC) with no SrrA or with 0.3, 0.7, 1.4, or 2.8 μM SrrA for 15 min at room temperature. Samples were electrophoresed on a 5% nondenaturing polyacrylamide gel and blotted onto a positively charged nylon membrane. The blots were developed with an antidigoxigenin antibody and chemiluminescent detection, and images were detected with radiographic film. As a negative control for SrrA-DNA binding, 100 bp of the internal coding sequence of S. aureus rpsC was analyzed by the gel shift procedure as well.
TABLE 2.
Primers used to amplify gel shift probesa
| Promoter | Target | PCR primers |
|---|---|---|
| agr P2 | 100 bp upstream of RNAII translational start site | 5′-TAAAATATTAAATACAAATTACATTT-3′ |
| 5′-ATTTTACACCACTCTCCTCA-3′ | ||
| agr P3 | 100 bp upstream of RNAIII translational start site | 5′-TCAACTATTTTCCATCACATC-3′ |
| 5′-ACATAAAAAAATTTACAGTTAAGA-3′ | ||
| spa (protein A) | 100 bp upstream of translational start site | 5′-ATTAATACCCCCTGTATGTA-3′ |
| 5′-AACTTACATTTAAATTTAATTATAA-3′ | ||
| srr (SrrAB) | 100 bp upstream of translational start site | 5′-ACAGGTCATACCTCCCAC-3′ |
| 5′-AGAATTTTTTCACAAAATTTTAG-3′ | ||
| tst (TSST-1) | 100 bp upstream of translational start site | 5′-ATGGTTAATTGATTCATTTAAA-3′ |
| 5′-ATTAGTAATTTTTTATTCATTTTT-3′ | ||
| rpsC (30S ribosomal protein S3) | 100 bp of coding sequence | 5′-TTCATACTGGTAAACCTGG-3′ |
| 5′-TTTGTACACGACGGAAT-3′ |
Each experimental probe was designed to incorporate approximately 100 bp upstream of the translational start site for each gene. The control probe consisted of 100 bp of the coding sequence of the rpsC gene. Probes were PCR amplified by using the primers.
Endocarditis model.
Rabbits were challenged with S. aureus in the acute infective endocarditis model as described elsewhere (19, 31). Briefly, New Zealand White rabbits were anesthetized with ketamine and xylazine, and the left carotid artery was exposed. To induce damage to the aortic valve, a catheter (diameter, 1.27 mm; Becton Dickinson and Co., Franklin Lakes, N.J.) was passed through the carotid artery, aorta, and aortic valve into the left ventricle. The catheter remained in place for 2 h, and then it was removed and the incision was closed. The rabbits were immediately challenged with an intravenous injection consisting of 1 ml (optical density at 600 nm, 0.6) of an S. aureus strain that had been grown to the stationary phase, washed twice, and resuspended in PBS. Animals were sacrificed at 96 h, and their aortic valves were examined for vegetations. Vegetations were harvested, homogenized, and plated onto Todd-Hewitt agar. S. aureus CFU were enumerated at 24 h. The experiments described here complied with all relevant federal guidelines and institutional policies regarding animal use.
Nucleotide sequence accession number.
The S. aureus MN8 srrAB locus sequence has been deposited in the GenBank database under accession number AF260326.
RESULTS
SrrAB structure.
Analysis of SrrB by the dense surface alignment method (http://www.sbc.su.se/∼miklos/DSA) (6) revealed the presence of two putative membrane-spanning regions in the N-terminal portion of the protein (residues 10 to 33 and 176 to 194). Further inspection of SrrAB revealed that SrrB contains a hydrophilic region that is approximately 130 amino acids long (from Q36 to N170) in the N-terminal half of the protein, between the two membrane-spanning regions. This hydrophilic region may be exposed to the extracellular environment.
In order to examine the three-dimensional structural domains of SrrAB in the context of other two-component systems, the SrrAB system was compared to structures deposited in the Conserved Domain Database (http://www.ncbi.nlm.nih.gov:80/Structure/cdd/cdd.shtml) at the National Center for Biotechnology Information. Examination of the putative response regulator SrrA showed that its structure was predicted to contain a winged helix-turn-helix DNA binding domain, which is also found in the E. coli response regulator OmpR (18). Examination of the putative transmembrane histidine kinase SrrB revealed phosphoacceptor, dimerization, and ATPase domains. Based on these comparisons with known two-component systems, we hypothesized that SrrA is located in the cytoplasm and binds DNA, while SrrB is located in the membrane. Furthermore, we predicted that the N-terminal 130-amino-acid hydrophilic region of SrrB is located extracellularly, while the C-terminal phosphoacceptor, dimerization, and ATPase domains are located intracellularly.
Subcellular localization of SrrA and SrrB.
In order to demonstrate the expression and subcellular localization of SrrAB, cell fractionation methods were used. Full-length SrrA and SrrB* were overexpressed and purified as described in Materials and Methods. rSrrA was isolated, purified by HPLC, and identified as a 34-kDa protein by SDS-PAGE. Its identity was confirmed by N-terminal sequencing. rSrrB* was isolated, purified by HPLC, and identified as an 18-kDa protein by SDS-PAGE (Fig. 1A). The sensitivity and specificity of the antibodies generated by immunizing rabbits with rSrrA or rSrrB* were evaluated by performing a Western analysis with the recombinant antigen and an S. aureus cell extract.
FIG. 1.
Subcellular localization of SrrA and SrrB. (A) rSrrA and rSrrB* were produced by independently cloning the full-length srrA gene and the putative extracellular portion of the srrB gene into the pET28b expression vector. E. coli strains BL21(pJMY22) and BL21(pJMY21) containing the pET28b-SrrA and pET28b-SrrB* constructs, respectively, were grown in Luria-Bertani medium, induced to express recombinant protein, and harvested by centrifugation. Recombinant protein was obtained as described in Materials and Methods. The recombinant histidine-tagged proteins were purified by column chromatography. The histidine tag was removed from SrrA by thrombin cleavage, and both proteins were purified by HPLC. rSrrA was present as an approximately 34-kDa protein on an SDS-polyacrylamide gel, while rSrrB* was present as an approximately 18-kDa protein. (B) S. aureus strains DU5875 and DU5875(pJMY11) were grown to the postexponential phase, lysed, and separated into membrane and cytoplasmic fractions as described in the text. Fractions were electrophoresed by SDS-PAGE and blotted for Western analysis. Cytoplasmic fractions are on the left and membrane fractions are on the right of each blot. (Left panel) Western blot with anti-SrrA (α-SrrA). SrrA was present as an approximately 34-kDa protein that was in the cytoplasmic fraction of strain DU5875(pJMY11). (Center panel) Western blot with anti-SrrB* (α-SrrB*). SrrB was present as an approximately 60-kDa protein that was in both the DU5875 and DU5875(pJMY11) membrane fractions. (Right panel) Western blot with normal serum. Numbers on the left are molecular masses, in kilodaltons.
The subcellular localization of SrrA and SrrB was determined by Western analysis. S. aureus strain DU5875 (spa::Tcr) was used in this assay in order to avoid interfering protein A bands. Strain DU5875 was transformed with pJMY11, an SrrAB overexpression vector, in order to ensure clear visualization of SrrA and SrrB. The anti-SrrA antibody identified SrrA as an approximately 34-kDa protein present in the cytoplasmic extract when SrrA was overexpressed. SrrA was not found in either of the membrane extracts. The anti-SrrB* antibody identified SrrB as an approximately 60-kDa protein in the membrane fractions of both DU5875 and DU5875(pJMY11). SrrB was not found in either of the cytoplasmic fractions. Western analysis with normal rabbit serum performed as a negative control resulted in a weak band in the DU5875 membrane fraction at approximately 50 kDa that was also present in the anti-SrrA and anti-SrrB* blots (Fig. 1B). These findings demonstrated that SrrA is found only in cytoplasmic extracts and that SrrB is found only in membrane extracts of S. aureus cells.
Immunostaining with anti-SrrB*.
S. aureus strain MN8 was stained with either anti-SrrB* or normal rabbit serum in order to determine if SrrB*, the predicted extracellular domain of SrrB, was present at the cell surface. SrrB* was detected by peroxidase-DAB staining, which imparted a brown-orange color. Counterstaining with hematoxylin stained cells light purple. S. aureus MN8 was stained by anti-SrrB* but not by normal rabbit serum (Fig. 2). As the cells were not permeabilized prior to antibody exposure, this finding indicates that the predicted extracellular domain of SrrB, SrrB*, is located extracellularly.
FIG. 2.

Predicted extracellular domain of SrrB is present extracellularly. S. aureus strain MN8 was grown to the stationary phase, washed in PBS, diluted, and spotted onto slides. Cells were dried, blocked with horse serum, and then incubated with either anti-SrrB* or normal rabbit serum overnight. Cells were washed and incubated with the secondary anti-rabbit peroxidase conjugate. Staining was detected with the chromogenic peroxidase substrate DAB, which imparts a brown-orange color to cells. Cells were counterstained with hematoxylin and examined for brown DAB staining by light microscopy. (Top panel) S. aureus MN8 stained with anti-SrrB*, detected with DAB, and counterstained with hematoxylin. (Bottom panel) S. aureus MN8 stained with normal rabbit serum, detected with DAB, and counterstained with hematoxylin.
Primer extension analysis.
Primer extension analysis revealed that the transcriptional start site for srrA was 24 nucleotides upstream of the start codon at a G residue (Fig. 3). Although there was sufficient primer for the sequencing reaction, no primer extension product for srrB was observed. This result, combined with the sequence and Northern blot analyses described previously (37), indicates that srrA and srrB have the same promoter and are cotranscribed. Further inspection of the srrAB promoter revealed a Pribnow box, which was identified based on its position (−10) relative to the transcriptional start site, as well as its consensus sequence (TATAAT). A −35 promoter region was not identified by consensus sequence analysis, which is not an unexpected finding for staphylococcal promoters. A Shine-Delgarno sequence (GGAGGT) was identified at positions −10 to −15, which is consistent with its role in ribosome binding.
FIG. 3.
Primer extension analysis of the srrAB promoter. Primers located approximately 100 bp downstream of the start codons for srrA and srrB were radiolabeled with [γ-33P]ATP and incubated with reverse transcriptase and S. aureus MN8 RNA. The same primers were used in a sequencing reaction with radiolabeled terminators and S. aureus MN8 genomic DNA as the template. Products from both the primer extension analysis and PCRs were electrophoresed simultaneously on a denaturing polyacrylamide gel. Analysis revealed that the transcriptional start site for the srrAB operon is 24 nucleotides upstream of the srrA translational start site. No primer extension product for srrB was observed, indicating that srrB is not transcribed independent of srrA. A Pribnow box at position −10, a −35 region, and a ribosome-binding site (RBS) were also predicted, as indicated.
Gel shift assays.
Previous work (37) indicated that expression of SrrAB alters the production of agr RNAIII, protein A (spa), TSST-1 (tst), and SrrAB (srr). We conducted gel shift assays with the agr P2, agr P3, spa, tst, and srr promoters to investigate if the regulation of these proteins may be due to a direct interaction between SrrA and these promoters. Gel shift promoter probes were designed so that they consisted of the 100 bp upstream of the translational start site of each gene. Each gel shift assay consisted of six lanes, two control lanes and four lanes containing increasing concentrations of SrrA. The leftmost lane contained a DNA probe-only control, while the next four lanes contained the probe plus increasing concentrations of SrrA. The SrrA concentrations ranged from 0.3 to 2.8 μM and increased twofold from left to right. The rightmost lane contained labeled probe, 2.8 μΜ SrrA, and a 125-fold excess of unlabeled probe.
Gel shift assays with the agr P2 promoter resulted in the appearance of three shifted bands. Unlabeled competitor added at a 125-fold excess successfully competed with the shifted bands (Fig. 4A). A gel shift analysis with the agr P3 promoter probe revealed the presence of two shifted bands (Fig. 4B). Analysis of the tst promoter revealed the presence of two shifted bands in all lanes with SrrA. The second lane from the right also contained two more shifted bands, as indicated in Fig. 4C. Gel shift analysis of the spa promoter resulted in three bands, a lower band present in all lanes and two higher bands present in the third, fourth, and fifth lanes from the left (Fig. 4D). Gel shift analysis with the srr promoter revealed the presence of one shifted band (Fig. 4E). Gel shift analysis of the rpsC gene failed to produce shifted bands, indicating that SrrA does not bind staphylococcal DNA indiscriminately (Fig. 4F).
FIG. 4.
SrrA interacts with the agr, tst, srr, and spa promoters. Regions consisting of approximately 100 bp upstream of the translational start sites of the agrB, agr RNAIII, spa, srr, tst, and rpsC loci were PCR amplified as described in the text. DNA probes were purified and labeled. Gel shift reactions were carried out by incubating labeled probe with increasing concentrations of SrrA (range, 0.3 to 2.8 μM) for 15 min at room temperature. The leftmost and rightmost lanes of each blot contained a no-protein control and an unlabeled probe competitor control, respectively. Samples were electrophoresed on a nondenaturing polyacrylamide gel and blotted onto a positively charged nylon membrane. Detection was accomplished with an antidigoxigenin antibody and CSPD chemiluminescent detection. The asterisks indicate the position of unbound probe, and the arrows indicate the positions of SrrA-bound probes. All five experimental promoter probes exhibited SrrA binding as determined by the gel shift analysis. Shifted bands were successfully competed away by an excess of unlabeled probe. (A) agr P2 promoter gel shift. Three shifted bands are indicated. (B) agr P3 promoter gel shift. Two bands are indicated. (C) tst promoter gel shift. Two shifted bands were found in the first three lanes with SrrA. The fourth lane contained two additional shifted bands. (D) spa promoter gel shift. One shifted band was found in all lanes with SrrA. Two additional shifted bands were found in the third, fourth, and fifth lanes from the left. (E) srr promoter gel shift. One shifted band is indicated. (F) rpsC gel shift. No shifted bands were observed.
Endocarditis model.
In order to determine the effects of SrrAB overexpression on the virulence of S. aureus, we employed the rabbit model of infective endocarditis. S. aureus strains RN4220, MN3050, and MN4011 were injected intravenously into American Dutch-Belted rabbits whose aortic valve leaflets had been damaged by cardiac catheterization. Cardiac vegetations were allowed to form for 4 days, and then the rabbits were sacrificed and their aortic valves were examined. Vegetations were homogenized, and S. aureus CFU were enumerated.
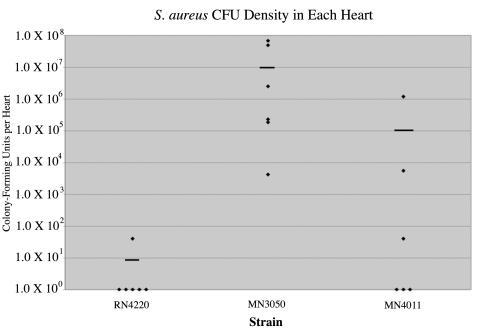
The parent organism RN4220 caused one vegetation containing 40 CFU in six hearts. MN3050, which overexpresses TSST-1, caused six vegetations in six hearts, and the average density was 2.0 × 107 CFU per heart. MN4011, which overexpresses SrrAB, caused three vegetations in six hearts, and the average cell density was 2.0 × 105 CFU per heart (Fig. 5). These data indicate that overexpression of TSST-1 in strain MN3050 significantly increased the virulence of S. aureus in this model compared to the virulence of the parent organism RN4220. Furthermore, overexpression of SrrAB in strain MN4011, which has been shown to repress TSST-1, protein A, and agr RNAIII in vitro, decreased the virulence of S. aureus 100-fold compared to the virulence of MN3050.
FIG. 5.
Overexpression of SrrAB decreases virulence. Strains RN4220, MN3050, and MN4011 were analyzed by using the rabbit model of infective endocarditis, as described in the text. Strain RN4220 caused a vegetation in one of six rabbits, and the average bacterial density was 7 CFU per heart. Strain MN3050 caused a vegetation in six of six rabbits, and the average bacterial density was 2.0 × 107 CFU per heart. Strain MN4011 caused a vegetation in three of six rabbits, and the average bacterial density was 2.0 × 105 CFU per heart. The horizontal bars indicate mean values.
Since strain RN4220 was obtained via random mutagenesis of an 8325-4 derivative, we were concerned that uncharacterized mutations in RN4220 may have confounded our results. Therefore, the endocarditis experiment was repeated by moving our overexpression vectors from RN4220 into RN6390, which is a derivative of 8325-4. Five rabbits were used in this experiment; one rabbit received RN6390, two rabbits received RN6390(pJMY10), and two rabbits received RN6390(pJMY11). Both of the rabbits that received RN6390(pJMY10) were found dead, one on day 3 and the other on day 4. The rabbit that received RN6390 was found dead on day 4. These three rabbits had large vegetations on their aortic valves, and their deaths were attributed to endocarditis. The two rabbits that received RN6390(pJMY11) were sacrificed on day 4, and each heart had one small sterile vegetation. These data are consistent with the data for the RN4220 derivatives and indicate that overexpression of SrrAB decreases the virulence of S. aureus in the rabbit model of infective endocarditis.
DISCUSSION
Many studies have demonstrated the importance of environmental signaling in S. aureus (22). In particular, oxygen has been shown to play a role in the regulation of TSST-1 (15, 29, 38). Previous work in our laboratory led to the description of SrrAB, a novel two-component system in S. aureus that regulates agr RNAIII, protein A, SrrAB, and TSST-1 levels and appears to respond to oxygen availability. Here, we investigated the subcellular localization of SrrA and SrrB, obtained insight into the mechanism by which SrrA regulates its target genes, and obtained evidence that SrrAB can modify the virulence of an organism.
Our results demonstrate that SrrAB is properly localized to function as a two-component system. The response regulator, SrrA, localizes to the cytoplasmic fraction. SrrB, the transmembrane histidine kinase, localizes to the membrane fraction. Immunostaining experiments with anti-SrrB* also demonstrated the membrane localization of SrrB. In addition, this study also provided evidence of the orientation of SrrB in the membrane. As the anti-SrrB* antibody recognizes only the portion of SrrB that is predicted to be extracellular, staining of the surface of S. aureus MN8 with this antibody indicated that this portion of the molecule (amino acids 36 to 170) is indeed extracellular. This information was incorporated into our model of SrrAB localization and function (Fig. 6).
FIG. 6.
SrrAB model. SrrB is present in the membrane and is oriented as indicated. Upon signaling, SrrB is autophosphorylated and transfers the phosphate to SrrA, which is located in the cytoplasm. SrrA binds to the spa, srr, tst, and agr promoters. Multiple SrrA molecules may bind to the agr, tst, and spa promoters. SrrA-promoter binding is probably responsible for the ability of SrrAB to alter the expression of protein A, SrrAB, TSST-1, and agr RNAIII.
Primer extension analysis of the srrAB promoter showed that the transcriptional start site of srrAB is located at a G residue 24 nucleotides upstream of the SrrA start codon. No primer extension product was observed for srrB, indicating that srrB must be cotranscribed with srrA. This finding is consistent with a previously reported (37) Northern blot analysis of srrAB transcripts. In this assay, srrB was detected only as a 2.5-kb band, which corresponded to a full-length srrAB mRNA. In contrast, a small amount of the full-length 2.5-kb srrA transcript was detected, as was a much larger amount of a 0.7-kb transcript that represented srrA mRNA alone. An inverted repeat present at the end of the SrrA coding sequence and at the beginning of the SrrB coding sequence may form a stem-loop structure that inhibits the elaboration of a full-length srrAB transcript (37). This structure may contribute to the relatively small amount of full-length transcript compared to the amount of srrA transcript alone. It is not surprising that much larger quantities of srrA than of srrB are transcribed, as the quantities of response regulators are typically much larger than the quantities of the corresponding transmembrane histidine kinases (17).
Previous work on SrrAB indicated that SrrAB is able to regulate virulence factors and regulatory elements, such as agr RNAIII, protein A, and TSST-1. In addition, we observed increased transcription of a truncated chromosomal copy of srrAB when srrAB was expressed in trans on a multicopy plasmid, indicating that SrrAB may be able to upregulate its own expression. In order to determine if these effects on agr, protein A, TSST-1, and SrrAB expression are mediated directly by SrrA or through another virulence regulator, gel shift assays were performed. The agr, spa, srr, and tst promoters all exhibited binding to SrrA, which indicated that SrrAB is able to exert its effects through agr-dependent mechanisms as well as agr-independent mechanisms. The agr P2, agr P3, tst, and spa promoters all exhibited multiple shifted bands, suggesting that these promoters may bind SrrA as a dimer or at multiple sites. Since agr RNAII and RNAIII are divergently transcribed and P2 and P3 are found in the same 236-bp region, the agr gel shift assays demonstrated that the agr intergenic region is probably capable of binding multiple SrrA molecules. The identification of multiple shifted bands in these gel shift assays was not surprising, as gel shift studies with OmpR and SarA revealed multiple shifted bands at target promoters (14, 27). An identical gel shift reaction with 100 bp of the S. aureus ribosomal protein rpsC did not produce a shifted band, indicating that the SrrA binding to agr, spa, srr, and tst promoters is not due to nonspecific association with staphylococcal DNA. Our gel shift assays demonstrated that DNA binding occurred at relatively high concentrations of SrrA compared to the results of other gel shift studies, which we believe was due to the instability of purified SrrA. Purified SrrA is unstable at 4°C and room temperature. The SrrA used in the gel shift assays was purified from E. coli and frozen in single-use aliquots immediately upon purification. In light of this information, it seems likely that not all of the purified SrrA used in these assays was active. In addition, phosphorylation of SrrA likely modifies the ability of SrrA to bind DNA. It has been shown that phosphorylation of OmpR greatly increases its affinity for its target DNA sequences (12, 14). Unfortunately, the instability of purified SrrA makes in vitro phosphorylation difficult to accomplish.
Our findings indicate that the ability of SrrAB to influence expression of agr RNAIII, protein A, TSST-1, and SrrAB is likely due to its ability to bind the agr, spa, tst, and srr promoters. Therefore, SrrAB may modify virulence factor expression through agr-dependent and agr-independent mechanisms.
It remains to be determined exactly where the SrrA binding site is in each promoter region. Although alignments of the promoters obtained with the ClustalW multiple-sequence-alignment program (http://www.ebi.ac.uk/clustalw/) have revealed several regions of homology for the five promoters, no clear area of high homology has emerged. It is likely that the consensus sequence necessary for SrrA-promoter binding will emerge only after the SrrA binding activity has been localized to a smaller area of the promoter. DNase I footprinting studies currently under way in our laboratory should confirm our results and should provide insight into the consensus sequence necessary for SrrA binding.
The identification of a consensus sequence and its location relative to the transcriptional start site should aid in the development of a model of SrrA-mediated transcriptional regulation. This information should prove to be quite interesting, since it appears that SrrA is able to repress transcription of the agr, spa, and tst loci while activating transcription at the srr locus. Perhaps the number of SrrA binding sites, their relative affinities for SrrA, or their locations relative to the transcriptional start site are important in controlling the activation or repression of a particular locus. This strategy of activating or repressing transcription based on the location or the relative affinity of a response regulator for a target promoter sequence has been proposed as a mechanism for OmpR-mediated regulation of ompF and ompC transcription (12).
Our initial data regarding SrrAB-mediated repression of virulence factors in vitro is supported by the in vivo data presented here. Our endocarditis studies with RN4220 derivatives demonstrated that overexpression of SrrAB decreases the virulence of a strain 100-fold compared to the virulence of an identical strain that does not overexpress SrrAB. It is likely that the ability of SrrAB to repress virulence factors such as agr RNAIII, protein A, and TSST-1 is responsible for the observed decrease in virulence. Although the RN6390 derivatives appeared to be more virulent overall, the trend in virulence was consistent for RN4220 derivatives and RN6390 derivatives. In both groups, the most virulent strain overexpressed TSST-1, while overexpression of SrrAB decreased the virulence of the organism. These data indicate that overexpression of SrrAB decreases the virulence of S. aureus in the rabbit model of infective endocarditis.
The studies presented here allowed us to develop a working model (Fig. 6) of the role that SrrAB plays in S. aureus pathogenesis. SrrB is located in the membrane, where its transmembrane or extracellular domains are able to detect an environmental signal. Although SrrAB seems to respond to oxygen levels, it is unlikely that oxygen itself directly signals SrrB. Other factors that reflect the oxygen level or redox state of the environment, such as pH or components of the respiratory system, may act as the signal for the SrrAB system. Once SrrB has received the signal, it is autophosphorylated at a conserved histidine residue. Association with SrrA, located in the cytoplasm, allows the transfer of a phosphate group to a conserved aspartate residue on SrrA. Phosphorylation of SrrA alters the DNA-binding activity of this response regulator. Binding of SrrA to the promoter regions of the agr, spa, srr, and tst loci alters the transcription of these loci. Alteration of the expression of these virulence factors in response to oxygen modifies the virulence of the organism.
In this study we characterized SrrAB, a novel two-component regulatory system in S. aureus. We demonstrated that it is properly localized and oriented in the membrane to function as a two-component system. Here we describe the srrAB promoter and the srrA and srrAB transcripts. We demonstrated that SrrA likely exerts its effects on agr RNAIII, protein A, SrrAB, and TSST-1 expression by binding to the agr P2, agr P3, spa, srr, and tst promoters. In addition, overexpression of SrrAB decreases virulence in a rabbit model of endocarditis.
Acknowledgments
We gratefully acknowledge the Minnesota Medical Foundation for financial support. A.A.P. was supported by an NIAID predoctoral fellowship (training grant T32 AI 07421). J.M.Y. was supported by a Howard Hughes Predoctoral Fellowship in the Biological Sciences.
We thank Gregory Bohach for providing S. aureus strains DU5875 and RN6390, Diane Maher for assistance with immunostaining studies, and Tim Leonard for assistance with preparation of the figures.
REFERENCES
- 1.Blomster-Hautamaa, D. A., and P. M. Schlievert. 1988. Preparation of toxic shock syndrome toxin-1. Methods Enzymol. 165:37-43. [DOI] [PubMed] [Google Scholar]
- 2.Chan, P. F., and S. J. Foster. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 144:2469-2479. [DOI] [PubMed] [Google Scholar]
- 3.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chien, Y., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273:2645-2652. [DOI] [PubMed] [Google Scholar]
- 6.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 7.Giraudo, A. T., A. Calzolari, A. A. Cataldi, C. Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15-22. [DOI] [PubMed] [Google Scholar]
- 8.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 9.Giraudo, A. T., C. G. Raspanti, A. Calzolari, and R. Nagel. 1994. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can. J. Microbiol. 40:677-681. [DOI] [PubMed] [Google Scholar]
- 10.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 12.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 13.Heinrichs, J. H., M. G. Bayer, and A. L. Cheung. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, K. J., C. Y. Lan, and M. M. Igo. 1997. Phosphorylation stimulates the cooperative DNA-binding properties of the transcription factor OmpR. Proc. Natl. Acad. Sci. 94:2828-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kass, E. H., M. I. Kendrick, Y. C. Tsai, and J. Parsonnet. 1987. Interaction of magnesium ion, oxygen tension, and temperature in the production of toxic-shock-syndrome toxin-1 by Staphylococcus aureus. J. Infect. Dis. 155:812-815. [DOI] [PubMed] [Google Scholar]
- 16.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 17.Magasanik, B. 1995. Historical perspective, p. 1-5. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 18.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 19.McCormick, J. K., H. Hirt, C. M. Waters, T. J. Tripp, G. M. Dunny, and P. M. Schlievert. 2001. Antibodies to a surface-exposed, N-terminal domain of aggregation substance are not protective in the rabbit model of Enterococcus faecalis infective endocarditis. Infect. Immun. 69:3305-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1995. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol. Microbiol. 16:895-907. [DOI] [PubMed] [Google Scholar]
- 21.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52:165-190. [DOI] [PubMed] [Google Scholar]
- 22.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 23.Novick, R. P. 1990. The Staphylococcus as a molecular genetic system, p. 1-40. In Molecular biology of the staphylococci. VCH Publishers, Inc., New York, N.Y.
- 24.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 26.Qazi, S. N., E. Counil, J. Morrissey, C. E. Rees, A. Cockayne, K. Winzer, W. C. Chan, P. Williams, and P. J. Hill. 2001. agr expression precedes escape of internalized Staphylococcus aureus from the host endosome. Infect. Immun. 69:7074-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rechtin, T. M., A. F. Gillaspy, M. A. Schumacher, R. G. Brennan, M. S. Smeltzer, and B. K. Hurlburt. 1999. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol. Microbiol. 33:307-316. [DOI] [PubMed] [Google Scholar]
- 28.Recsei, P., B. Kreiswirth, M. O'Reilly, P. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 202:58-61. [DOI] [PubMed] [Google Scholar]
- 29.Ross, R. A., and A. B. Onderdonk. 2000. Production of toxic shock syndrome toxin 1 by Staphylococcus aureus requires both oxygen and carbon dioxide. Infect. Immun. 68:5205-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schlievert, P. M., P. J. Gahr, A. P. Assimacopoulos, M. M. Dinges, J. A. Stoehr, J. W. Harmala, H. Hirt, and G. M. Dunny. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Throup, J. P., F. Zappacosta, R. D. Lunsford, R. S. Annan, S. A. Carr, J. T. Lonsdale, A. P. Bryant, D. McDevitt, M. Rosenberg, and M. K. Burnham. 2001. The srhSR gene pair from Staphylococcus aureus: genomic and proteomic approaches to the identification and characterization of gene function. Biochemistry 40:10392-10401. [DOI] [PubMed] [Google Scholar]
- 33.Wesson, C. A., L. E. Liou, K. M. Todd, G. A. Bohach, W. R. Trumble, and K. W. Bayles. 1998. Staphylococcus aureus Agr and Sar global regulators influence internalization and induction of apoptosis. Infect. Immun. 66:5238-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 35.Wong, A. C., and M. S. Bergdoll. 1990. Effect of environmental conditions on production of toxic shock syndrome toxin 1 by Staphylococcus aureus. Infect. Immun. 58:1026-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarwood, J. M., and P. M. Schlievert. 2000. Oxygen and carbon dioxide regulation of toxic shock syndrome toxin 1 production by Staphylococcus aureus MN8. J. Clin. Microbiol. 38:1797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]