Abstract
Mitochondrial disorders are deadly childhood diseases for which therapeutic remedies are an unmet need. Given that genetic suppression of the nuclear enzyme poly (adenine diphosphate-ribose) polymerase(PARP)-1 improves mitochondrial functioning, we investigated whether pharmacological inhibition of the enzyme affords protection in a mouse model of a mitochondrial disorder. We used mice lacking the Ndufs4 subunit of the respiratory complex I (Ndufs4 knockout [ KO] mice); these mice undergo progressive encephalopathy and die around postnatal day 50. Mice were treated daily with the potent PARP inhibitor N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34); neurological parameters, PARP activity, and mitochondrial homeostasis were evaluated. We found that mice receiving N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride from postnatal day 30 to postnatal day 50 show reduced neurological impairment, and increased exploratory activity and motor skills compared with vehicle-treated animals. However, drug treatment did not delay or reduce death. We found no evidence of increased PARP activity within the brain of KO mice compared with heterozygous, healthy controls. Conversely, a 10-day treatment with the PARP inhibitor significantly reduced basal poly(ADP-ribosyl)ation in different organs of the KO mice, including brain, skeletal muscle, liver, pancreas, and spleen. In keeping with the epigenetic role of PARP-1, its inhibition correlated with increased expression of mitochondrial respiratory complex subunits and organelle number. Remarkably, pharmacological targeting of PARP reduced astrogliosis in olfactory bulb and motor cortex, but did not affect neuronal loss of KO mice. In light of the advanced clinical development of PARP inhibitors, these data emphasize their relevance to treatment of mitochondrial respiratory defects.
Electronic supplementary material
The online version of this article (doi:10.1007/s13311-014-0285-y) contains supplementary material, which is available to authorized users.
Key Words: Mitochondrial diseases, complex I deficiency, Ndufs4 knockout, poly (ADP-ribose) polymerase, PARP inhibitor, mitochondrial biogenesis.
Introduction
Mitochondrial disorders are devastating, inherited diseases caused by a deficit of mitochondrial functioning. Mostly, they are caused by mutations of nuclear or mitochondrial genes coding for proteins of oxidative phosphorylation (OXPHOS) [1]. Clinical symptoms may differ among OXPHOS defects, but the most affected organs are always those with high energy expenditure, such as brain, skeletal muscle, and heart [2]. Patients with OXPHOS defects typically die within the first years of life because of severe encephalopathy [3]. Currently, there is no cure for mitochondrial disorders and symptomatic approaches only have few effects on disease severity and evolution [4].
It is widely acknowledged that a deeper understanding of the molecular mechanisms involved in neuronal death in patients affected by mitochondrial disorders can help in identifying effective therapies [5]. In this regard, animal models of OXPHOS defects are instrumental in deciphering the cascade of events that from initial deficit of mitochondrial oxidative capacity leads to neuronal demise. Transgenic mouse models of mitochondrial disorders recently became available and significantly contributed to the demonstration that the pathogenesis of OXPHOS defects is not merely due to a deficiency in the production of adenosine triphosphate (ATP) within high energy-demand tissues [6]. Indeed, several reports demonstrate that ATP and phosphocreatine levels are not reduced in patient cells or tissues of mice bearing respiratory defects [7, 8]. These findings, along with evidence that astrocyte and microglial activation takes place in the degenerating brain of mice with mitochondrial disorders [9], suggest that the pathogenesis of encephalopathy in mitochondrial patients is pleiotypic and more complex than previously envisaged. On this basis, pharmacological approaches to the OXPHOS defect must target the different pathogenetic events responsible for encephalopathy. This assumption helps us to understand why therapies designed to target specific players of mitochondrial disorders have failed, and promotes the development of innovative pleiotypic drugs.
Over the last few years we have witnessed renewed interest in the biology of the pyridine cofactor nicotinamide adenine dinucleotide (NAD). At variance with old dogmas, it is now well appreciated that the availability of NAD within subcellular compartments is a key regulator of NAD-dependent enzymes such as poly[adenine diphosphate (ADP)-ribose] polymerase (PARP)-1 [10–12]. The latter is a nuclear, DNA damage-activated enzyme that transforms NAD into long polymers of ADP-ribose (PAR) [13, 14]. Whereas massive PAR formation is causally involved in energy derangement upon genotoxic stress, ongoing synthesis of PAR recently emerged as a key event in the epigenetic regulation of gene expression [15, 16]. SIRT1 is an additional NAD-dependent enzyme able to deacetylate a large array of proteins involved in cell death and survival, including peroxisome proliferator-activated receptor gamma coactivator-1α (PGC1α) [17]. PGC1α is a master regulator of mitochondrial biogenesis and function, the activity of which is depressed by acetylation and unleashed by SIRT-1-dependent detachment of the acetyl group [18]. Several reports demonstrate that PARP-1 and SIRT-1 compete for NAD, the intracellular concentrations of which limit the two enzymatic activities [19, 20]. Consistent with this, recent work demonstrates that when PARP-1 activity is suppressed, increased NAD availability boosts SIRT-1-dependent PGC1α activation, resulting in increased mitochondrial content and oxidative metabolism [21]. The relevance of NAD availability to mitochondrial functioning is also strengthened by the ability of NAD precursors to improve both energy production and mitochondrial biogenesis [22, 23]. Although these findings point to the interplay among NAD, PARP-1, and SIRT-1 as a target to improve mitochondrial dysfunction, their relevance to mitochondrial disorders and related encephalopathy remains elusive. Remarkably, PARP-1 inhibitors have been proven to have therapeutic efficacy in different models of human disorders [24], and have recently reached the clinical arena, showing a safety profile in patients with different neoplasms [25, 26].
In this study, we took advantage of a recently developed mouse model of mitochondrial defect, the Ndufs4 KO mouse, which recapitulates the clinical phenotype of Leigh syndrome [8], to evaluate the effects of pharmacological PARP inhibition on mitochondrial function and disease progression.
Methods
Animals and Drug Treatment
Ndufs4+/– mice were bred to produce the Ndfus4–/– mice used for experiments. Mice were housed with free access to food and water, and maintained on a 12-h light/dark cycle at 22 °C . The PARP inhibitor N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34) was dissolved in saline and injected intraperitoneally. All animal manipulations were performed according to the European Community guidelines for animal care (DL 116/92, application of the European Communities Council Directive 86/609/EEC) and approved by the Committee for Animal Care and Experimental Use of the University of Florence.
Neuroscore Analysis
The neurological score was assessed as described in Table 1. Briefly, a 5-point scale was used to measure different locomotor functions/impairments, such as ataxia, hind limb clasping, balance, and limb tone. The latter was evaluated by means of a dynamometer. All the mentioned parameters were evaluated every 2 days by 2 blinded operators.
Table 1.
Neurological score evaluation
| Ataxia (grid test, 30 s) | Hind limb clasping (10 s test) | Balance (beam measurement) | Limb tone | |
|---|---|---|---|---|
| 0 | No clinical signs | No clinical signs | No clinical signs | Strength grip > 120 g |
| 1 | One foot slip during the trial period | One hind limb partially retracted for > 50 % of the trial period | Inability to turn around on the bar | 100 g < grip strength < 120 g |
| 2 | 2–4 foot slips during the trial period | One hind limb completely retracted for > 50 % of the trial period | Difficulty walking to the end of the bar without falling off | 80 g < grip strength < 100 g |
| 3 | ≥ 5 foot slips during the trial period | Both hind limbs were partially retracted for > 50 % of the trial period | The mouse can only cling to the bar and is unable to correct itself from its initial perpendicular orientation | 60 g < grip strength < 80 g |
| 4 | Foot slip without retraction during the trial period | Both hind limbs were fully retracted and touching the abdomen for > 50 % of the trial period | Postural instability as the mouse quickly falls off the bar even when placed along the long axis | 40 g < grip strength < 60 g |
| 5 | Not moving | Not moving | Not moving | Grip strength < 40 g |
The rotarod apparatus consisted of a base platform and a rotating rod with a diameter of 3 cm, with a non-slippery surface with a rod-rotating speed that was gradually accelerated from 4 rpm to 50 rpm during a 3-min test. The integrity of motor coordination was assessed according to Kuribara et al. [27] on the basis of endurance time of the animals on the rotating rod. Briefly, 1 day before the first test (i.e., postnatal day 29) the animals were placed on the rotating drum and trained twice.
The hole board apparatus consisted of an acrylic black board (31.5 cm × 31.5 cm × 20.5 cm) with 16 holes (hole diameter: 2 cm; distance between holes: 5 cm). The hole sensors were situated at a depth of 1 cm and automatically recorded the number of head-dips performed by animals. Two photo beams, crossing the plane from midpoint to midpoint of opposite sides, thus dividing the plane into 4 equal quadrants, automatically signaled the movement of the animal (counts in 5 min) on the surface of the plane (locomotor activity). Starting from postnatal day 30, mice were placed on the center of the board at 5-day intervals and allowed to move freely on the apparatus for 5 mins.
Western Blotting
Proteins for Western blotting were isolated from snap-frozen mice tissues using the NucleoSpin TriPrep method (Macherey-Nagel, Duren, Germany). After sodium dodecyl sulfate polyamide acrylic gel electrophoresis and blotting, membranes (Immobilon-P; Millipore, Bedford, MA, USA) were blocked with phosphate buffered saline (PBS) containing 0.1 % Tween-20 and 5 % skimmed milk (TPBS/5 % milk) and then probed overnight with primary antibodies (1:1000 in TPBS/5 % milk). The anti-PAR monoclonal antibody (10H) was from Alexis (Vinci, Italy). Anti-succinate dehydrogenase complex, subunit A (SDHA) and anti-β-actin antibodies were from Abcam (Cambridge, UK). Membranes were then washed with TPBS and incubated for 1 h in TPBS/5 % milk containing the corresponding peroxidase-conjugated secondary antibody (1:2000). After washing in TPBS, ECL (Amersham, UK) was used to visualize the peroxidase-coated bands. Protein oxidation detection was performed using OxyBlot Kit (Millipore Billerica, Boston, MA, USA) according to manufacturer’s instructions.
NAD Measurement
Mice were sacrificed at postnatal days 30 and 50, or after 10 days of treatment. Tissues were rapidly collected and stored at –80 °C. From each tissue, a few milligrams were processed for NAD measurement, as reported by PittellI et al. [28].
Real-Time Polymerase Chain Reaction
Genomic DNA and total RNA were extracted from mice tissues with the NucleoSpin TriPrep kit (Macherey-Nagel), and real-time polymerase chain reaction was performed as previously reported [29]. Mitochondrial content was quantified by measuring the ratio between mitochondrial ND1 and nuclear β-actin gene amplification products.
The following primers were used: for Cox1—forward 5’-TATCAATGGGAGCAGTGTTTG-3’ and reverse 5’-AGGCCCAGGAAATGTTGAG-3’; for Cox2—forward 5’-CTGAAGACGTCCTCCACTCAT-3’ and reverse 5’-TCTAGGACAATGGGCATAAAG-3’; for mt-Nd2—forward 5’-ATTATCCTCCTGGCCATCGTA-3’ and reverse 5’-AAGTCCTATGTGCAGTGGGAT-3’; for Ndufv2—forward 5’-GTGCACAATGGTGCTGGAGGAG-3’ and reverse 5’-GGTAGCCATCCATTCTGCCTTTGG-3’: for Cox15—forward 5’-GTTCTGAGATGGGCACTGGACCA-3’ and reverse 5’-GGGGCACGTGTTCCTGAATCTGT-3’: for Atp5d—forward 5’-CAGCACGGGCTGAGATCCAGAT-3’ and reverse 5’-GACAGGCACCAGGAAGCTTTAAGC-3’; for 18S—forward 5’-AAAACCAACCCGGTGAGCTCCCTC-3’ and reverse 5’-CTCAGGCTCCCTCTCCGGAATCG-3’; for mt-Nd1—forward 5’-TGCCAGCCTGACCCATAGCCATA-3’ and reverse 5’-ATTCTCCTTCTGTCAGGTCGAAGGG-3’; for β-actin—forward 5’-GCAGCCACATTCCCGCGGTGTAG-3’ and reverse 5’-CCGGTTTGGACAAAGACCCAGAGG-3’.
Mouse Primary Glial Cultures
Primary cultures of glial cells were prepared from P1 mice as previously described [30]. Briefly, cortices were isolated in cold PBS and then incubated for 30 mins at 37 °C in PBS containing 0.25 % trypsin and 0.05 % DNase. After blocking enzymatic digestion with the addition of 10 % heat-inactivated fetal bovine serum, cortices were mechanically disrupted by pipetting. Cells obtained from each cortex were washed, resuspended in Dulbecco’s modified Eagle medium plus 10 % fetal bovine serum (GIBCO, Life Technologies, Rockville, MD, USA) and plated separately. Glial cells from Ndufs4 knockout (KO) mice were identified by genotyping and used for mitochondrial membrane potential evaluation at 7 days in vitro (DIV).
Evaluation of Mitochondrial Membrane Potential
Mitochondrial membrane potential was evaluated by means of flow cytometry [29]. Glial cells from Ndufs4 KO mice were treated with vehicle or with the 2 PARP inhibitors, PJ34 (20 μM) or Olaparib (100 nM), for 72 h. Cells were then detached, incubated with tetramethylrhodamine ethyl ester (TMRE) 2.5 nM, and analyzed with a Coulter EPICS XL flow cytometer (Beckman Coulter, Fullerton, CA, USA) equipped with the EXPO32 Flow Cytometry ADC software (Beckman Coulter).
Transmission Electron Microscopy
Tissues were fixed in 4 % glutaraldehyde, postfixed in 1 % osmium tetroxide, and embedded in Epon 812. Ultrathin sections were stained with uranyl acetate and alkaline bismuth subnitrate and examined under a JEM 1010 electron microscope (Jeol, Tokyo, Japan) at 80 kV.
Micrographs were taken throughout the whole motor cortex, skeletal muscle, and liver at final magnifications of 12,000× and 50,000× using a MegaView III digital camera and interfacing software (SIS-Soft Imaging System, Munster, Germany). The first ones were used for determination of the amount of mitochondria, and the latter ones for analysis of mitochondria and internal cristae volumes. Briefly, to analyze the number of mitochondria, 5 cytoplasmic fields (test area per field 97.8 μm2) for each section were chosen at random and only mitochondria unequivocally present within neuronal structures were counted/analyzed. Areas of mitochondria and areas of cristae were measured using iTEM image analysis software (SIS).
Immunohistochemistry
Immunohistochemistry was performed as previously described [31], according to standard procedure. Briefly, snap-frozen brain was embedded in embedding matrix (CellPath Ltd., UK) (OCT) and cut with a cryostat (Leica, Solms, Germany). Brain section (14 μm) were fixed with 4 % paraformaldehyde and incubated in PBS with 0.3 % Triton X-100 (Sigma, St. Louis, MO, USA) and 2 % of bovine albumin. Sections were double-stained with anti-Neuronal Nuclei (NeuN) monoclonal antibody (mouse monoclonal, 1:100; Chemicon International, Temecula, CA, USA) and anti-glial fibrillary acidic protein (GFAP; monoclonal, clone G-A-5, 1:200; Sigma). To-pro3 (Molecular Probes, Eugene, OR, USA) was used as nuclear counterstain. Quantification of fluorescence was performed using Metamorph/Metafluor software. Values correspond to the mean ± SEM of 5 different microscopic fields per 3 different mouse brain sections per brain (4 brain per group).
Data Analysis
Data were analyzed using WinLTP 1.11 reanalysis program and GraphPad Prism (version 4.0; GraphPad, San Diego, CA, USA). All numerical data are expressed as mean ± SEM. Statistical significance of differences between results was evaluated by performing analysis of variance followed by Tukey's w test for multiple comparisons.
Results
Inhibition of PARP Improves Neuroscore and Delays Disease Development of Ndufs4 KO Mice
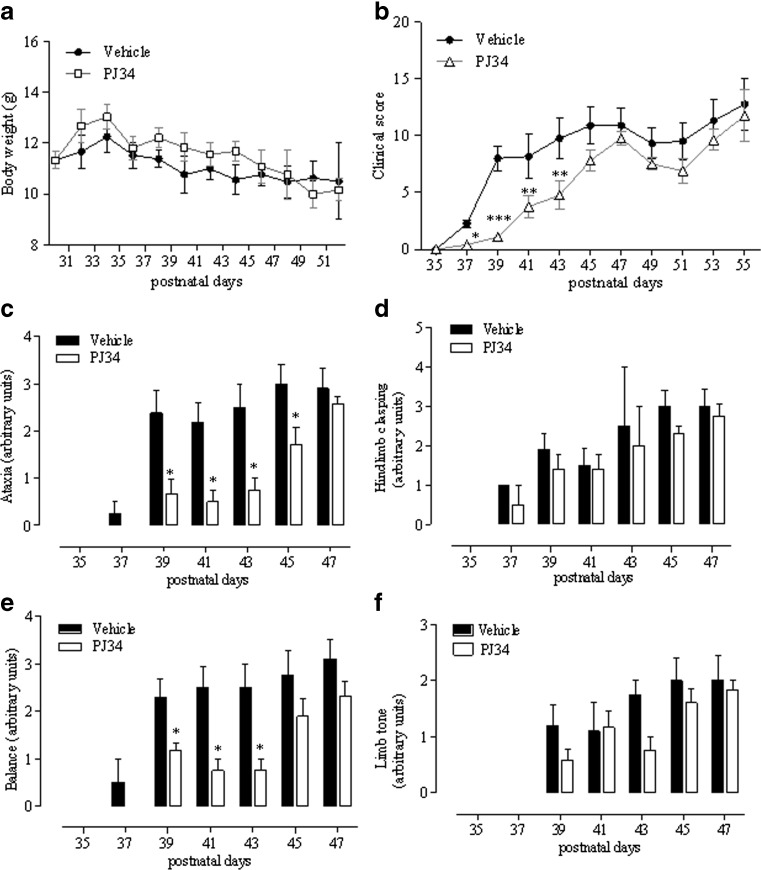
To unravel the pathogenetic role of PARP-1 in the development of mitochondrial encephalopathy and to understand the therapeutic potential of its inhibition in patients with OXPHOS defects, we evaluated the effect of pharmacological PARP suppression on disease development in KO mice. We treated animals with daily intraperitoneal injections of PJ34 (20 mg/kg body weight), a water-soluble, potent PARP inhibitor [24]. We found that the number of pups per litter was low (4–5), even though the KO mice in the offspring were at the expected Mendelian ratio. To adopt a clinically relevant treatment protocol, we start injecting mice at day 30 when hair loss, the first sign of disease development, is almost complete [8]. As shown in Fig. 1A, treatment did not alter mouse weight compared with vehicle-injected animals, although a tendency to higher values in the PJ34-treated group was evident. Evolution of encephalopathy was assessed by evaluator-blind analysis of neurological impairment [8]. We found that significant worsening of clinical score occurred at day 37 and motor impairment inexorably increased up to postnatal day 53–55, when mice died. In mice receiving PJ34, the clinical score was significantly delayed from postnatal day 37 to postnatal day 43 (Fig. 1B). At later time points, mice treated with the PARP inhibitor had a neuroscore that did not differ from that of vehicle-injected animals, although, again, a tendency to slight reduction was obtained (Fig. 1B).
Fig. 1.
Effects of N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34) on symptom development of Ndufs4 knockout mice. PJ34 (20 mg/kg) was injected intraperitoneally daily from postnatal day 30 and the effects on (A) weight and (B) clinical score evaluated every other day. The drug’s effect on the evolution of (C) ataxia, (D) hindlimb clasping, (E) balance, and (F) limb tone is also shown. Each point/columns represent the mean ± SEM of 6 (vehicle) and 8 (PJ34) animals per group. *p < 0.05 vs vehicle, analysis of variance plus Tukey’s post hoc test
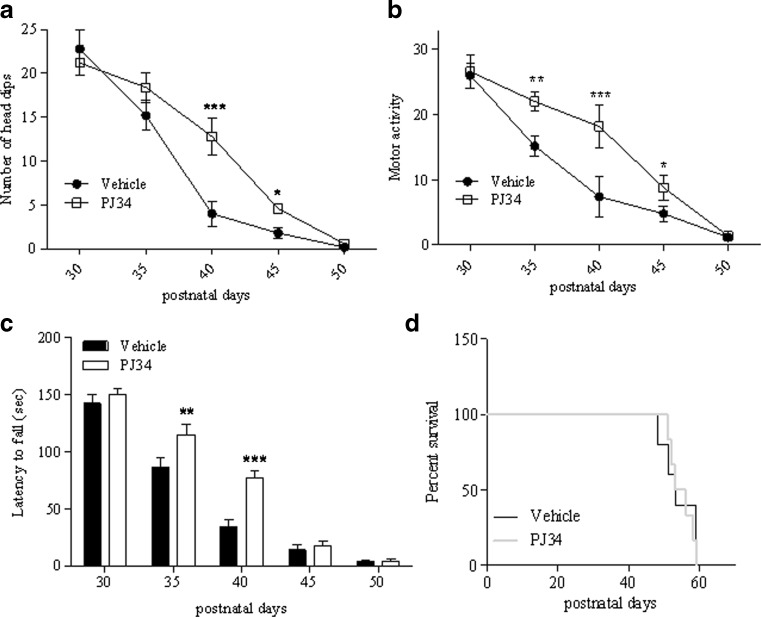
Detailed analysis of specific symptoms indicates that treatment reduced the severity of ataxia and improved balance, having no effects on hind limb clasping and limb tone (Fig. 1C–F). Of note, analysis of exploratory and motor activity also revealed that treatment with the PARP inhibitor improved both parameters during postnatal days 40–45 and 35–45, respectively (Fig. 2A, B). When motor skill was evaluated by means of rota-rod assay, we found that KO mice receiving PJ34 showed significantly prolonged latency to fall at P35-40 compared with vehicle-injected animals (Fig. 2C). However, PJ34 only delayed worsening of motor performances, given that at later time points (day 50) the therapeutic effects disappeared. In keeping with this, drug treatment did not prolong survival of the KO mice (Fig. 2D).
Fig. 2.
Effects of N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34) on motor activity and survival of Ndufs4 knockout mice. PJ34 (20 mg/kg) was injected intraperitoneally daily from postnatal day 30, and the effects on (A) exploratory and (B) motor activity, as well as on (C) motor skill evaluated at the indicated time points. (D) Survival curves of vehicle and PJ34-injected mice. In (A–C) each point/column represents the mean ± SEM of 6 (vehicle) and 8 (PJ34) animals per group. *p < 0.05, **p < 0.01, ***p < 0.001 vs vehicle, analysis of variance plus Tukey’s post hoc test
Oxidative Stress, PARP Activity, and NAD Levels in Ndufs4 KO Mice
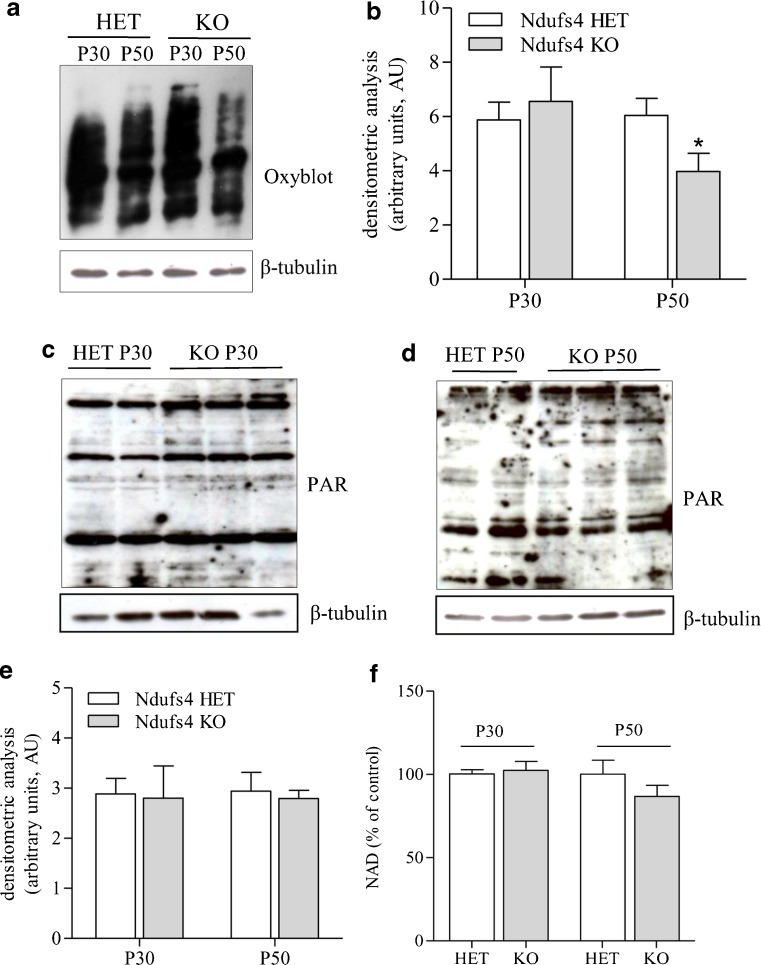
OXPHOS defects are typically characterized by derangement of electron transfer through the respiratory chain, a condition leading to the formation of reactive oxygen species and oxidative stress. The latter is thought to play a key pathogenetic role in encephalopathy of patients with mitochondrial disorders [32]. Given that PARP-1 is hyperactivated in condition oxidative stress and causes massive energy consumption [33], we reasoned that PARP-1 activation-dependent ATP depletion could further compromise the precarious energy homeostasis in the brains of KO mice. Therefore, we evaluated whether oxidative stress occurs within the motor cortex of these animals at different stages of disease development. As a marker of oxidative stress in vivo, we analyzed protein carbonylation by means of Oxyblot in KO and heterozygous mice. The latter are healthy, indistinguishable from wild-type mice, and have previously been used as controls [8]. Although prior work demonstrates increased protein carbonylation in the olfactory bulb of KO mice [9], we found that this marker of oxidative stress did not differ between KO and heterozygous mice at postnatal day 30, whereas it was reduced in KO animals at postnatal day 50 (Fig. 3A, B).
Fig. 3.
Protein carbonylation, poly(ADP-ribose) (PAR) and nicotinamide adenine dinucleotide (NAD) content in the motor cortex of heterozygous (HET) and Ndufs4-null mice. (A) Oxyblot analysis of protein carbonylation in the motor cortex of heterozygous (HET) and knockout (KO) mice at postnatal days 30 (P30) and 50 (P50). (B) Densitometric analysis of oxyblots. Western blotting evaluation of PAR content in the motor cortex of HET and KO mice at (C) P30 and (D) P50. (E) Densitometric analysis of Western blots of PAR. (F) NAD contents in the motor cortex of HET and KO mice at P30 and P50. Basal NAD content was 0.73 ± 0.12 μmol/g tissue. In (A), (C), and (D), each blot is representative of 6 animals per group. In (B), (E), and (F), each column represents the mean ± SEM of 6 animals per group
Western blot analysis of poly(ADP-ribosyl)ated proteins is typically used as an index of PARP activity. Therefore, we evaluated basal poly(ADP-ribosyl)ation in the motor cortex of heterozygous and KO mice. In keeping with the lack of oxidative stress, levels of poly(ADP-ribosyl)ated proteins did not differ between the 2 mouse strains at postnatal day 30 and postnatal day 50 (Fig. 3C–E). A reduction in NAD content typically occurs in tissues undergoing PARP-1 hyperactivity [33].Hence, as an additional index of PARP activity, we quantified the NAD content in the motor cortex of heterozygous and KO mice. Again, we were unable to find any difference in the content of NAD in the cortices of the two mouse strains at both p30 and p50 (Fig. 3F).
Inhibition of PARP Increases the Expression of Respiratory Complex Subunits and Promotes Mitochondrial Biogenesis in Ndufs4 KO Mice
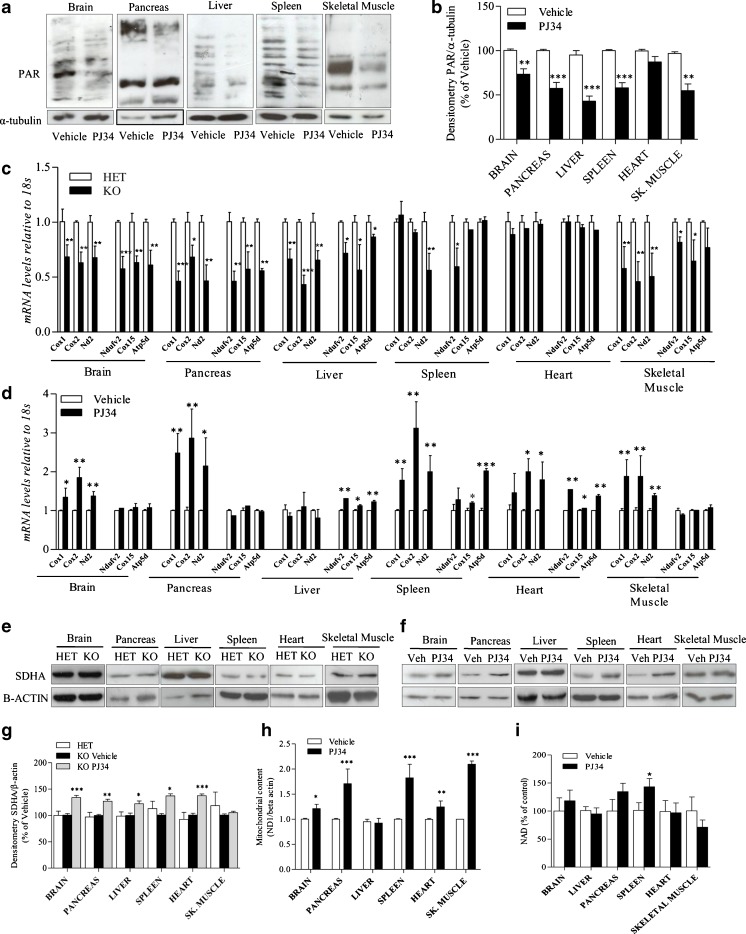
To obtain evidence that PJ34 was, indeed, inhibiting PARP in KO mice, we analyzed PAR content in their tissues after10 days of treatment (i.e., postnatal day 40). In keeping with the pharmacodynamic effect of the drug, we found a reduced PAR content in brain, pancreas, liver, spleen, and skeletal muscle of animals challenged with PJ34 compared with vehicle-injected mice (Fig. 4A, B).
Fig. 4.
Effect of N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34) on tissue poly(ADP-ribose) (PAR) content, respiratory complex subunits expression and mitochondrial DNA (mtDNA) content in Ndufs4 knockout (KO) mice. (A) The effects of a 10-day treatment (postnatal days 30–40) with PJ34 (daily intraperitoneal injections of 20 mg/kg) on tissue PAR content is shown. (B) Densitometric analysis of the effects of PJ34 on tissue PAR content of Ndufs4 KO mice. (C) mRNA levels of several mitochondrial [cyclooxygenase (COX)1, COX2, NADH dehydrogenase 2 (ND2)] and nuclear (NADH dehydrogenase (ubiquinone) flavoprotein 2 (NDUFV2), COX15, and ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit (ATP5D)) respiratory complex subunits in different organs of Ndufs4 heterozygous (HET) and KO mice. (D) The effects of PJ34 on transcripts levels of the respiratory complex subunits in KO mice are also shown. Succinate dehydrogenase complex, subunit A (SDHA) expression levels in different organs of (E) heterozygous and (F) KO mice treated or not with PJ34 is shown by Western blotting and (G) Densitometric analysis. (H) Effects of PJ34 on mitochondrial content (expressed as ND1/beta actin gene ratio) or (I) nicotinamide adenine dinucleotide (NAD) levels in different organs of Ndufs4 KO mice. Basal NAD content was 0.73 ± 0.12 μmol/g tissue, 0.64 ± 17 μmol/g tissue, 35 ± 0.08 μmol/g tissue, 0.1 ± 0.005 μmol/g tissue, 0.67 ± 0.21 μmol/g tissue, 0.59 ± 0.16 μmol/g tissue in the brain, pancreas, liver, spleen, heart, and skeletal muscle (sk. muscle), respectively. (A, E, F) A blot representative of 4 mice per group is shown. (B, C, D, G, H, I), columns represent the mean ± SEM of 4 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001 vs vehicle, analysis of variance plus Tukey’s post hoc test
We next wondered whether the expression of different respiratory complex subunits is altered in KO compared with heterozygous mice. Interestingly, we found a significant reduction of transcripts for mitochondrial- and nuclear-encoded respiratory subunits, such as cyclooxygenase (COX)1, COX2, NADH dehydrogenase 2 (ND2), COX15, NADH dehydrogenase (ubiquinone) flavoprotein 2 (NDUFV2), and ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit (ATP5D), in different mouse organs, with the exception of the heart (Fig. 4C). It has previously been reported that PARP-1-dependent NAD consumption limits PGC1α transcriptional activity and overall mitochondrial efficiency [21]. Therefore we evaluated whether treatment with PJ34 promotes transcription of mitochondrial- and nuclear-encoded respiratory complex subunits. Notably, we found that the PARP1 inhibitor increased the transcript levels of the different respiratory subunits in an organ-specific manner. Specifically, the mRNA levels of mitochondrial genes Cox1, Cox2, and mt-Nd2 increased in all the organs tested (brain, pancreas, spleen, heart, and skeletal muscle) with the exception of liver. Conversely, transcripts of the nuclear genes Ndufv2, Cox5, and Atp5d were only augmented in liver, spleen, and heart (Fig. 4D). We also evaluated expression of the SDHA subunit of succinate dehydrogenase, and found that it was not affected in KO mice compared with heterozygous ones, whereas it increased in the organs of PJ34-treated mice, with the exception of skeletal muscle (Fig. 4E–G).
The increased mitochondrial content reported in PARP-1 KO mice prompted us to evaluate whether the same phenotype could be recapitulated by pharmacological PARP inhibition [21]. As a prototypical index of mitochondrial content we quantitated the mitochondrial DNA (mtDNA) gene mt-Nd1 in the different organs of KO mice treated or not with PJ34. As shown in Fig. 4H, a 10-day treatment with the PARP inhibitor increased the content of mtDNA in all the organs tested except the liver. Notably, with the exception of the spleen, the NAD content in the mouse organs was not increased by the PARP inhibitor (Fig. 4I).
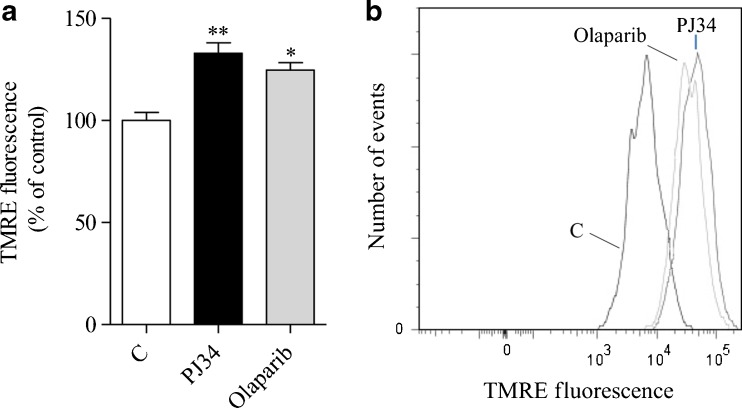
To corroborate the evidence that PARP inhibitors improve mitochondrial function and that the effects of PJ34 are due to PARP inhibition, we next evaluated the impact of PJ34 and a structurally unrelated, very potent PARP inhibitor such as Olaparib, on mitochondrial membrane potential of cultured glial cells from Ndufs4 KO mice. As shown in Fig. 5, we found that both compounds increased the mitochondrial membrane potential by approximately 25 % upon 72 h of treatment, at concentrations consistent with their relative IC50 on PARP-1 [34].
Fig. 5.
Effects of poly(adenosine diphosphate-ribose) polymerase (PARP) inhibitors on mitochondrial membrane potential in Ndufs4 knockout (KO) cultured glial cells. The effect of a 72-h treatment with N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34) (20 μM) or Olaparib (100 nM) on mitochondrial membrane potential [measured by means of potentiometric, fluorescent dye tetramethylrhodamine ethyl ester (TMRE)] of cultured glial cells from Ndufs4 KO mice is shown as (A) the mean ± SEM of 2 experiments conducted in triplicate and (B) a representative cytofluorimetric plot. *p < 0.05, **p < 0.01, vs control, analysis of variance plus Tukey’s post hoc test
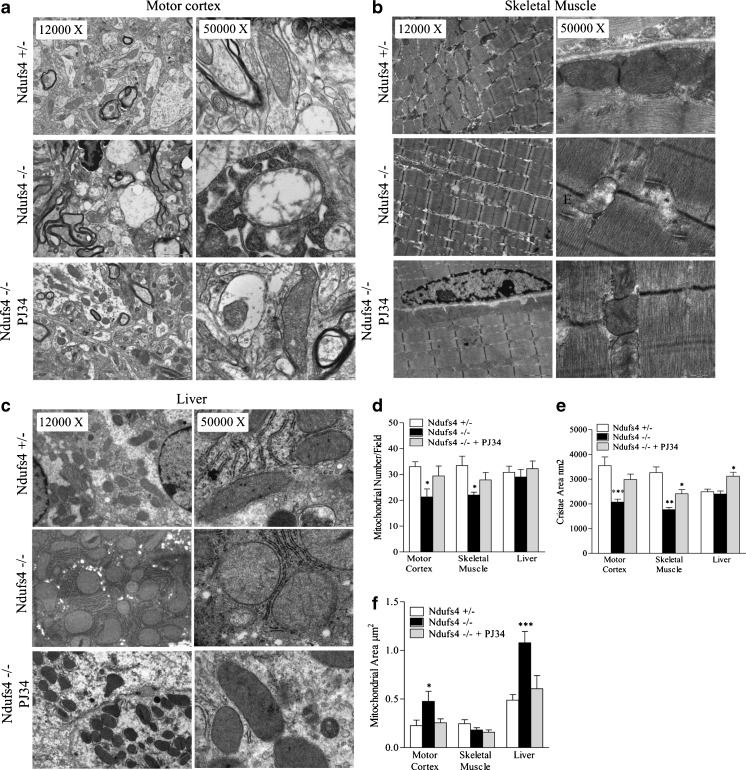
These findings taken together with knowledge that transcriptional networks leading to increased oxidative capacity also regulate mitochondrial biogenesis [35], prompted us to evaluate whether mitochondrial number and morphology of KO mice was affected by PARP inhibition. Electron microscopy revealed that mitochondrial number and cristae area were reduced in motor cortex and skeletal muscle but not in liver of KO mice compared with heterozygous animals at postnatal day 40 (Fig. 6). We also found that the mitochondrial area increased in motor cortex and liver but not in skeletal muscle of KO mice (Fig. 6). Remarkably, a reduction in mitochondrial number, as well as changes in organelle morphology, were prevented in KO mice treated with PJ34 from postnatal day 30 to postnatal day 40 (Fig. 6). Also, the area of mitochondrial cristae in the liver was increased by drug treatment even if it was not reduced in KO mice (Fig. 6F).
Fig. 6.
Mitochondrial number and morphology of Ndufs4 heterozygous and knockout mice treated or not with N-(6-oxo-5,6-dihydrophenanthridin-2-yl)-(N,N-dimethylamino)acetamide hydrochloride (PJ34). Mitochondrial morphology and number in shown in representative electron microscopy images at 2 different magnifications for (A) motor cortex, (B) skeletal muscle, and (C) liver. Data summarizing the effects of Ndufs4 deletion in the presence or absence of PJ34 on (D) mitochondrial number, (E) cristae area, and (F) mitochondrial area in the different tissues is shown. Each column is the mean ± SEM of 5 microscopic fields per 5 (+/–), 3 (–/–), and 4 (–/– treated with PJ34) animals per group. *p < 0.05, **p < 0.01, ***p < 0.001 vs Ndufs4+/– mice, analysis of variance plus Tukey’s post hoc test
Effects of PARP Inhibition on Astrogliosis and Neuronal Loss in Ndufs4 KO Mice
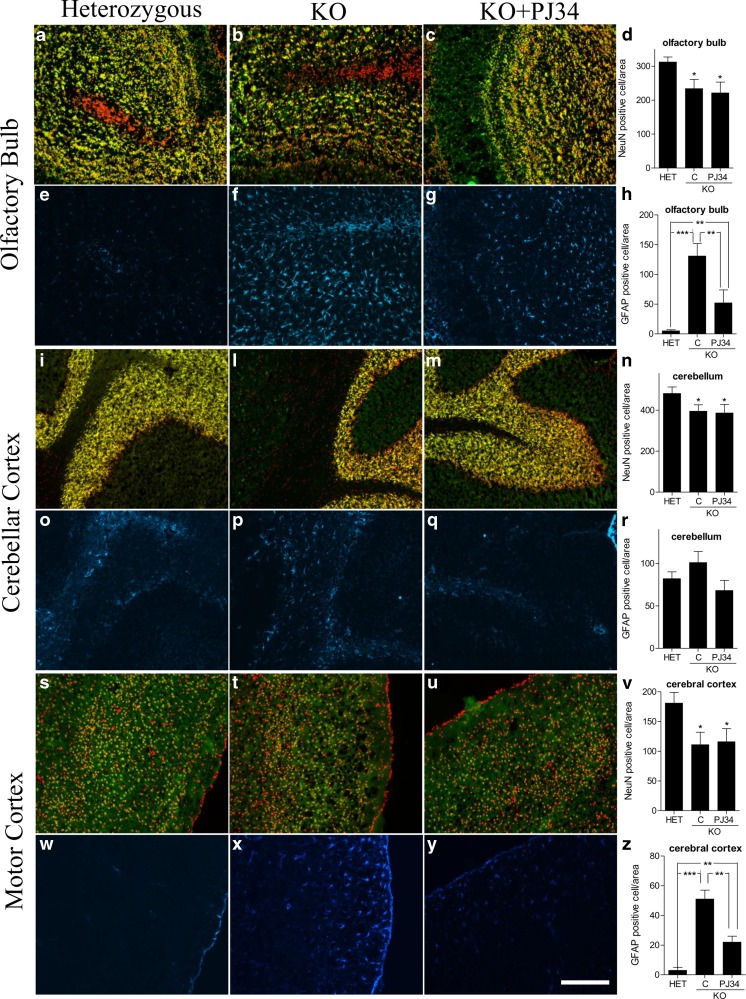
Improved neurological score by PJ34, along with the notion that neurodegeneration takes place in the olfactory bulb and cerebellum of Ndufs4 mice [9], prompted us to evaluate the impact of PJ34 on neuronal loss and astrogliosis in these mice. We found that a robust increase of GFAP-positive cell number (a prototypical marker of astrogliosis) occurred at the level of the olfactory bulb and motor cortex of Ndufs4 mice at p40, but not in the cerebellum. Of note, treatment with the PARP inhibitor significantly reduced GFAP expression in these brain regions. However, neuronal loss occurring at p40 in olfactory bulb, cerebellum and motor cortex was not affected by drug treatment (Fig. 7).
Fig. 7.
Neuronal loss and astrogliosis in different brain regions of Ndufs4 heterozygous (HET) and knockout (KO) mice treated or not with PJ34. Neuronal loss and astrogliosis have been evaluated in (A–H) olfactory bulb, (I–R) cerebellar, and (S–Z) motor cortex. Neuronal loss has been evaluated according to Chiarugi et al. [9] by staining neurons with NeuN (green) and nuclei with To-pro3 (red). Co-localization of both labels is shown in yellow. Astrocyte activation has been evaluated by means of glial fibrillary acidic protein (GFAP) staining (blue). Images representative of 4 brains per group are shown. (D, H, N, R, V, Z) Each column is the mean ± SEM of 5 different microscopic fields per 3 different mouse brain sections per brain. *p < 0.05, **p < 0.01, ***p < 0.001 vs Ndufs4+/– mice, analysis of variance plus Tukey’s post hoc test. Bar = 500 μm. C = Vehicle treated mice
Discussion
We report that a pharmacological inhibitor of PARP delays the development of encephalomyopathy in a mouse model of mitochondrial disorder. We also show that PARP inhibition prompts a transcriptional program leading to increased expression of respiratory complex subunits and mitochondrial biogenesis. In light of the urgent need for drugs able to improve symptoms in patients with OXPHOS defects [5, 32], along with the apparent safety profile shown by PARP1 inhibitors in clinical trials [26], the present study might have realistic clinical implications.
Several transgenic mouse models of OXPHOS defects have recently been developed; among them, those related to genetic mutations of respiratory complex I subunits appear to reproduce closely the symptomatology of patients [6]. The KO mice used in our study lack exon 2 of Ndufs4 so that the corresponding 18-kDa protein is absent because of frameshift. This mouse develops a phenotype resembling Leigh syndrome and dies by fatal encephalomyopathy within approximately 50 days [8]. Notably, mice bearing the same Ndufs4 mutation selectively in neural cells display a phenotype almost identical to those with systemic mutation [9]. This finding indicates that the therapeutic effects exerted by the PARP inhibitor should be ascribed to its ability to reduce neurodegeneration during the development of mitochondrial encephalopathy. This assumption is in keeping with the large body of evidence that PARP inhibitors, including PJ34, have remarkable neuroprotective effects in different models of neuronal death in vitro and in vivo [24]. Of note, we show that tissue PAR content is reduced in KO mice upon PJ34 administration, which is in keeping with the notion that PARP-1 contributes to the majority of PAR formations [13, 14]. However, given that the drug is not strictly PARP-1 selective [36], we cannot rule out the possibility that inhibition of additional PARPs, including PARP-2 [37], may have contributed to the pharmacodynamic effects of PJ34.
In principle, PARP inhibition might exert its therapeutic effect in KO mice by different mechanisms. For instance, necrotic neuronal death occurs in the brain of KO mice [9], and numerous reports demonstrate the ability of PARP inhibitors to protect from this form of neuronal demise [33]. However, our findings showing lack of oxidative stress, PARP activation, and NAD depletion in the motor brain cortex of KO mice at different stages of encephalopathy suggest that PARP1 is not causative in necrotic neuronal death in this model of mitochondrial disorder. Although data are consistent with prior work showing no increase of ROS in fibroblasts from a patient with a nonsense mutation in Ndufs4 [38], recent findings in Ndufs4 KO mice show the occurrence of oxidative stress in the olfactory bulb during disease progression [9]. In this regard, even though our electron microscopy analysis and immunohistochemistry reveal mitochondrial morphological abnormalities, astrogliosis and neuronal loss in the motor cortex, the olfactory bulb is the first and most compromised brain structure in KO mice [9]. Therefore, we speculate that mechanisms underlying neurodegeneration in KO mice are brain region-specific. The decrease of protein carbonylation in KO mice compared with heterozygous mice at P50 could be ascribed to the moribund conditions of the animals and the related breathing defect resulting in reduced blood perfusion and oxygenation [39]
PARP-1 is a key player of apoptosis inducing factor-dependent apoptosis during neurodegeneration [40]. However, given that the extrinsic (i.e., mitochondrial independent) apoptotic pathway is triggered in the brain of KO mice [9], it is unlikely that prevention of AIF release and apoptosis is a major mechanism responsible for the PJ34 effect. Interestingly, in keeping with evidence that astrocyte and microglia activation occurs in the degenerating brain regions of Ndufs4 KO mice [9], we show that GFAP immunoreactivity is increased in olfactory bulb and motor cortex. Although the pathogenetic relevance of this inflammatory event still needs to be clarified, it is tempting to speculate that the ability of PARP inhibitors to suppress astroglia activation contributed to reduce the severity of encephalopathy and related symptoms [41].
In addition to the possibility that PARP inhibition counteracts neurodegeneration by blocking neurotoxic events in the KO mice, pharmacological suppression of PARP could also prompt neuroprotective mechanisms. In this regard, a key pathway of relevance to neuroprotection in these animals might be that prompted by PGC1α. Indeed, both genetic or pharmacological suppression of PARP-1 promotes SIRT-1-dependent PGC1α activation which leads to increased oxidative capacity and mitochondrial content [21]. Accordingly, we found that PJ34 induced the expression of respiratory complex subunits and mitochondrial biogenesis. This finding, along with evidence that mRNAs for respiratory complex subunits are reduced in KO compared with heterozygous mice, is of particular importance because it suggests that the therapeutic effects of PARP inhibition may be due to a restoration of homeostatic transcript levels. Notably, KO mice receiving the PARP inhibitor showed increased mRNA abundance of both nuclear- and mitochondrial-encoded respiratory complex subunits. We reason that this occurred because, in addition to the activation of the PGC1α-dependent transcriptional program, PARP inhibition also alters nuclear transcription directly. Indeed, it is well appreciated that PARP-1 activity epigenetically regulates transcription of numerous genes by direct interaction with both gene promoters and basal transcriptional machinery [15]. PARP1 can also regulate the activity of several transcription factors, including YY1 or NRF-1 [42, 43], which are of relevance to mitochondrial functioning. Interestingly, nuclear respiratory factor (NRF)-1, a key regulator of nuclear genes involved in mitochondrial respiration and mtDNA duplication, is negatively regulated by PARP-1 activity [43]. Therefore, inhibition of PARP-1 by PJ34 might have unleashed NRF-1, thereby potentiating PGC1α-dependent mitochondrial biogenesis. Evidence that NAD content increased only in the spleen of KO mice treated with PJ34 is in line with the hypothesis that mechanisms in addition to SIRT1-dependent PGC1α activation contribute to mitochondrial biogenesis. The selective NAD increase in the spleen is also in keeping with our recent study that showed a high NAD turnover in this mouse organ [28].
At present we do not know why PJ34 affected mitochondrial number and morphology in some organs but not in others. Possibly, this is owing to tissue-specific mechanisms of epigenetic regulation, as well as to different impairment of tissue homeostasis during disease development. Accordingly, we previously reported that PJ34 impairs mitochondrial DNA transcription in cultured human tumor cells [44]. We speculate that the reason(s) of this apparent inconsistency can be ascribed to differences in experimental settings, that is in vivo versus in vitro and/or acute versus chronic exposure to PJ34.
Unfortunately, in spite of the ability of PJ34 to reduce neurological impairment after a few days of treatment, neither neuronal loss nor death of mice was reduced or delayed. Although this KO mouse model is extremely severe, showing a shift from healthy condition to fatal breathing dysfunction in only 20 days [39], recent work demonstrates that rapamycin increases median survival of male Ndufs4 KO mice from 50 to 114 days [45]. In light of this, we speculate that inhibition of PARP prompts a cascade of events, such as mitochondrial biogenesis or increased oxidative capacity, that is of symptomatic relevance, but eventually unable to counteract specific mechanisms responsible for neurodegeneration and disease development. Still, symptom improvement obtained with PJ34 is of pathogenetic and therapeutic significance, and might be potentiated by different means such as use of ultra-potent PARP inhibitors [24] and co-treatment with symptomatic drugs already used in mitochondrial patients. In keeping with this hypothesis, very recent studies report improvement of mitochondrial functioning and muscle fitness in mice challenged with PARP inhibitors [46, 47].
Electronic supplementary material
(PDF 1224 kb)
Acknowledgments
This work was supported by grants from Regione Toscana Health Projects 2009 (recipient A.C.) and 2012 (recipient A. L.), Association of Amyotrophic Lateral Sclerosis (ARISLA), and Ente Cassa di Risparmio di Firenze. The authors gratefully acknowledge R.D. Palmiter for the kind gift of Ndufs4 KO mice and helpful comments.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
References
- 1.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sofou K. Mitochondrial disease: a challenge for the caregiver, the family, and society. J Child Neurol. 2013;28:663–667. doi: 10.1177/0883073813481622. [DOI] [PubMed] [Google Scholar]
- 4.Pfeffer G, Majamaa K, Turnbull DM, Thorburn D, Chinnery PF. Treatment for mitochondrial disorders. Cochrane Database Syst Rev. 2012;4 doi: 10.1002/14651858.CD004426.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koene S, Willems PH, Roestenberg P, Koopman WJ, Smeitink JA. Mouse models for nuclear DNA-encoded mitochondrial complex I deficiency. J Inherit Metab Dis. 2011;34:293–307. doi: 10.1007/s10545-009-9005-x. [DOI] [PubMed] [Google Scholar]
- 7.Valsecchi F, Koopman WJ, Manjeri GR, et al. Complex I disorders: causes, mechanisms, and development of treatment strategies at the cellular level. Dev Disabil Res Rev. 2010;16:175–182. doi: 10.1002/ddrr.107. [DOI] [PubMed] [Google Scholar]
- 8.Kruse SE, Watt WC, Marcinek DJ, et al. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell Metab. 2008;7:312–320. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quintana A, Kruse SE, Kapur RP, Sanz E, Palmiter RD. Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc Natl Acad Sci U S A. 2010;107:10996–11001. doi: 10.1073/pnas.1006214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiarugi A, Dolle C, Felici R, Ziegler M. The NAD metabolome – a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12:741–752. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- 11.Canto C, Auwerx J. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:291–298. doi: 10.1101/sqb.2012.76.010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houtkooper RH, Auwerx J. Exploring the therapeutic space around NAD+ J Cell Biol. 2012;199:205–209. doi: 10.1083/jcb.201207019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Amours D, Desnoyers S, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342:249–268. doi: 10.1042/0264-6021:3420249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 15.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/S0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 17.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T, Berrocal JG, Frizzell KM, et al. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009;284:20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. J Biol Chem. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 21.Bai P, Canto C, Oudart H, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pittelli M, Felici R, Pitozzi V, et al. Pharmacological effects of exogenous NAD on mitochondrial bioenergetics, DNA repair, and apoptosis. Mol Pharmacol. 2011;80:1136–1146. doi: 10.1124/mol.111.073916. [DOI] [PubMed] [Google Scholar]
- 23.Canto C, Houtkooper RH, Pirinen E, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagtap P, Szabo C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 25.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papeo G, Forte B, Orsini P, et al. Poly(ADP-ribose) polymerase inhibition in cancer therapy: are we close to maturity? Expert Opin Ther Pat. 2009;19:1377–1400. doi: 10.1517/13543770903215883. [DOI] [PubMed] [Google Scholar]
- 27.Kuribara H, Higuchi Y, Tadokoro S. Effects of central depressants on rota-rod and traction performances in mice. Jpn J Pharmacol. 1977;27:117–126. doi: 10.1254/jjp.27.117. [DOI] [PubMed] [Google Scholar]
- 28.Pittelli M, Cavone L, Lapucci A, et al. Nicotinamide phosphoribosyltransferase (NAMPT) activity is essential for survival of resting lymphocytes. Immunol Cell Biol. 2014;92:191–199. doi: 10.1038/icb.2013.85. [DOI] [PubMed] [Google Scholar]
- 29.Felici R, Lapucci A, Ramazzotti M, Chiarugi A. Insight into molecular and functional properties of NMNAT3 reveals new hints of NAD homeostasis within human mitochondria. PLoS One. 2013;8:e76938. doi: 10.1371/journal.pone.0076938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faraco G, Pittelli M, Cavone L, et al. Histone deacetylase (HDAC) inhibitors reduce the glial inflammatory response in vitro and in vivo. Neurobiol Dis. 2009;36:269–279. doi: 10.1016/j.nbd.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Faraco G, Pancani T, Formentini L, et al. Pharmacological inhibition of histone deacetylases by suberoylanilide hydroxamic Acid specifically alters gene expression and reduces ischemic injury in the mouse brain. Mol Pharmacol. 2006;70:1876–1884. doi: 10.1124/mol.106.027912. [DOI] [PubMed] [Google Scholar]
- 32.Dimauro S, Rustin P. A critical approach to the therapy of mitochondrial respiratory chain and oxidative phosphorylation diseases. Biochim Biophys Acta. 2009;1792:1159–1167. doi: 10.1016/j.bbadis.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 33.Chiarugi A. PARP-1: killer or conspirator? The suicide hypothesis revisited. Trends Pharmacol Sci. 2002;23:122–129. doi: 10.1016/S0165-6147(00)01902-7. [DOI] [PubMed] [Google Scholar]
- 34.Wahlberg E, Karlberg T, Kouznetsova E, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 35.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 36.Pellicciari R, Camaioni E, Costantino G, et al. On the way to selective PARP-2 inhibitors. Design, synthesis, and preliminary evaluation of a series of isoquinolinone derivatives. Chem Med Chem. 2008;3:914–923. doi: 10.1002/cmdc.200800010. [DOI] [PubMed] [Google Scholar]
- 37.Bai P, Canto C, Brunyanszki A, et al. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011;13:450–460. doi: 10.1016/j.cmet.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iuso A, Scacco S, Piccoli C, et al. Dysfunctions of cellular oxidative metabolism in patients with mutations in the NDUFS1 and NDUFS4 genes of complex I. J Biol Chem. 2006;281:10374–10380. doi: 10.1074/jbc.M513387200. [DOI] [PubMed] [Google Scholar]
- 39.Quintana A, Zanella S, Koch H, et al. Fatal breathing dysfunction in a mouse model of Leigh syndrome. J Clin Invest. 2012;122:2359–2368. doi: 10.1172/JCI62923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiarugi A, Moskowitz MA. Poly(ADP-ribose) polymerase-1 activity promotes NF-kappaB-driven transcription and microglial activation: implication for neurodegenerative disorders. J Neurochem. 2003;85:306–317. doi: 10.1046/j.1471-4159.2003.01684.x. [DOI] [PubMed] [Google Scholar]
- 42.Oei SL, Shi Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem Biophys Res Commun. 2001;285:27–31. doi: 10.1006/bbrc.2001.5115. [DOI] [PubMed] [Google Scholar]
- 43.Hossain MB, Ji P, Anish R, Jacobson RH, Takada S. Poly(ADP-ribose) polymerase 1 interacts with nuclear respiratory factor 1 (NRF-1) and plays a role in NRF-1 transcriptional regulation. J Biol Chem. 2009;284:8621–8632. doi: 10.1074/jbc.M807198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapucci A, Pittelli M, Rapizzi E, et al. Poly(ADP-ribose) polymerase-1 is a nuclear epigenetic regulator of mitochondrial DNA repair and transcription. Mol Pharmacol. 2011;79:932–940. doi: 10.1124/mol.110.070110. [DOI] [PubMed] [Google Scholar]
- 45.Johnson SC, Yanos ME, Kayser EB, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerutti R, Pirinen E, Lamperti C, et al. NAD-dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 2014;19:1042–1049. doi: 10.1016/j.cmet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pirinen E, Canto C, Jo YS, et al. Pharmacological inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab. 2014;9:1034–1041. doi: 10.1016/j.cmet.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1224 kb)