Abstract
In Bacillus subtilis, the genes of the branched-chain amino acids biosynthetic pathway are organized in three genetic loci: the ilvBHC-leuABCD (ilv-leu) operon, ilvA, and ilvD. These genes, as well as ybgE, encoding a branched-chain amino acid aminotransferase, were recently demonstrated to represent direct targets of the global transcriptional regulator CodY. In the present study, the transcriptional organization and posttranscriptional regulation of these genes were analyzed. Whereas ybgE and ilvD are transcribed monocistronically, the ilvA gene forms a bicistronic operon with the downstream located ypmP gene, encoding a protein of unknown function. The ypmP gene is also directly preceded by a promoter sharing the regulatory pattern of the ilvA promoter. The ilv-leu operon revealed complex posttranscriptional regulation: three mRNA species of 8.5, 5.8, and 1.2 kb were detected. Among them, the 8.5-kb full-length primary transcript exhibits the shortest half-life (1.2 min). Endoribonucleolytic cleavage of this transcript generates the 5.8-kb mRNA, which lacks the coding sequences of the first two genes of the operon and is predicted to carry a stem-loop structure at its 5′ end. This processing product has a significantly longer half-life (3 min) than the full-length precursor. The most stable transcript (half-life, 7.6 min) is the 1.2-kb mRNA generated by the processing event and exonucleolytic degradation of the large transcripts or partial transcriptional termination. This mRNA, which encompasses exclusively the ilvC coding sequence, is predicted to carry a further stable stem-loop structure at its 3′ end. The very different steady-state amounts of mRNA resulting from their different stabilities are also reflected at the protein level: proteome studies revealed that the cellular amount of IlvC protein is 10-fold greater than that of the other proteins encoded by the ilv-leu operon. Therefore, differential segmental stability resulting from mRNA processing ensures the fine-tuning of the expression of the individual genes of the operon.
In Bacillus subtilis, three genetic loci are involved in the biosynthesis of branched-chain amino acids: the ilvBHC-leuABCD (ilv-leu) operon including genes encoding acetolactate synthase (ilvBH), ketol-acid reductoisomerase (ilvC), 2-isopropylmalate synthase (leuA), 3-isopropylmalate dehydrogenase (leuB), and 3-isopropylmalate dehydratase (leuCD); the ilvA gene encoding threonine dehydratase; and the ilvD gene encoding dihydroxy-acid dehydratase. These enzymes catalyze the formation of the α-keto acid precursors of isoleucine, valine, and leucine. In the final biosynthetic reaction, the α-keto acids are converted to the respective amino acids by transamination. Previous studies demonstrated that the ilv-leu operon is regulated in response to leucine availability by the T-box transcription antitermination system (14, 30). The common T-box-dependent regulatory mechanism of the biosynthetic ilv-leu operon and the aminoacyl-tRNA synthetase genes (15, 34) would be predicted to result in the coregulation of these genes. However, our global study of B. subtilis gene expression in response to amino acid availability (29) revealed that the ilv-leu operon was downregulated in the presence of Casamino Acids (CAA), whereas expression of the aminoacyl-tRNA synthetase genes was not affected under the same conditions. Furthermore, we found that all three transcriptional units involved in the biosynthesis of branched-chain amino acids, as well as the ybgE gene encoding a branched-chain amino acid aminotransferase, exhibited reduced expression levels in the presence of CAA. Although the genes for branched-chain amino acid biosynthesis share this common regulatory pattern, a genome-wide search revealed no T-box leader sequences upstream of ilvA, ilvD, and ybgE (6). Therefore, we were interested in identifying the regulatory mechanism responsible for the common regulation of the genes involved in branched-chain amino acid biosynthesis. Since the CodY repressor was known to mediate amino acid repression of several B. subtilis genes, we supposed that CodY may function as the common regulator of these genes. Experiments which were carried out to test whether the ilv-leu operon and the ilvA, ilvD, and ybgE genes are under the control of CodY verified this hypothesis. In the meantime, Molle et al. (33) reported on the identification of new CodY regulated genes, and the genes for branched-chain amino acid biosynthesis, including ybgE, were among these newly identified direct CodY targets.
Many early-stationary-phase genes of B. subtilis are repressed by the global regulator CodY. Ratnayake-Lecamwasam et al. (35) demonstrated that CodY represents a GTP-sensing protein that functions as a repressor under high-GTP conditions, i.e., during exponential growth in nutrient-rich medium. When B. subtilis cells encounter nutrient-limiting conditions, the cellular GTP pool drops, due either to the depletion of precursors of guanine nucleotides or to the activation of the stringent response (26). Since CodY-regulated genes are repressed during growth in media containing amino acid mixtures (2), the cellular GTP pool might be affected by amino acid availability via the stringent response due to partial amino acid limitation in minimal medium. However, recent in vitro experiments carried out in the Sonenshein laboratory demonstrated that branched-chain amino acids interact directly with CodY, thereby stimulating the binding of CodY to many of its targets (33).
In the present work, we analyzed the transcriptional organization of the genes encoding the branched-chain amino acid biosynthetic enzymes, identifying ilvD and ybgE as monocistronic transcriptional units and cotranscription of ilvA with the ypmP gene. The heptacistronic ilvBHC-leuABCD operon was demonstrated to exhibit posttranscriptional regulation by mRNA processing and differential segmental stabilities of the processing products, which results in different amounts of the encoded proteins.
Relatively little is known about mRNA degradation, mRNA processing, and mechanisms of posttranscriptional regulation in B. subtilis. In gram-negative bacteria, the extensively analyzed pufQBALMX operon of Rhodobacter capsulatus represents a model system for studying gene regulation by differential segmental mRNA stability of polycistronic operons (3, 16, 17, 23, 24). The pufBA genes encode the proteins of the light-harvesting I complex, while the pufLM genes encode the pigment-binding proteins of the reaction center. There is a 10- to 15-fold excess of light-harvesting I complexes over reaction center complexes in R. capsulatus cells grown under low-oxygen tension, which at least partially results from differential stabilities of the individual puf mRNA segments (23). As exemplified in the puf operon, the endoribonuclease RNase E represents a key factor in the modulation of RNA degradation and processing in gram-negative bacteria (8). In Escherichia coli as well as R. capsulatus, RNase E, the exoribonuclease PNPase, and the RNA helicase RhlB, which facilitates the unwinding of structured RNA, are organized in a large complex called the degradosome (8, 22). According to current models, degradation of the majority of the mRNAs is initiated by the binding of RNase E to the 5′ end. Subsequently, the degradosome proceeds in the 5′-3′ direction, thereby “scanning” the bound RNA for AU-rich, single-stranded RNA regions representing RNase E recognition sites. The resulting cleavage fragments are rapidly degraded by the exoribonucleases PNPase and RNase II. Both enzymes are impeded by secondary structures frequently located at the 3′ ends of primary transcripts, functioning both as transcriptional terminators and as 3′-RNA stabilizers. Therefore, the decay of structured 3′ ends is postulated to require successive rounds of addition and degradation of poly(A) tails which function as “on-ramps” for the exonucleases, particularly the PNPase. This RNA polyadenylation is performed by the poly(A) polymerase (PAPI) (8).
Although an RNase E-encoding gene cannot be derived from the genome sequence of B. subtilis, an RNase E-like enzymatic activity has been found (9). Whereas PNPase is present in B. subtilis, a gene which encodes the orthologue of PAPI is absent, although polyadenylation of mRNA has been observed (10). Obviously, the enzymatic apparatus responsible for mRNA degradation and processing differs markedly between E. coli and B. subtilis. However, the structural elements influencing mRNA stability are similar: the accessibility of 5′ and 3′ ends is crucial. Protection of the 5′ end by a stem-loop structure and frequently or strongly binding ribosomes confers stability to a particular mRNA in both species, as well as stable stem-loop structures at the 3′ end (10).
Up to now, two polycistronic operons exhibiting differential segmental mRNA stability were described in B. subtilis, the heptacistronic dnaK operon (20, 21) and the hexacistronic gapA operon (27, 31). In both cases, the processing of primary transcripts generates products with significantly different half-lives, resulting in different amounts of the encoded proteins. However, the identity of the endoribonucleases performing the mRNA-processing reactions is still unknown.
The ilv-leu operon is the third example of a polycistronic operon in B. subtilis exhibiting modulation of the amounts of its encoded proteins by mRNA processing and differential segmental mRNA stability. Besides regulation of the whole operon via CodY repression and T-box-mediated antitermination, this additional level of regulation fine-tunes the expression of its individual genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. subtilis strains used in this study were B. subtilis 168 (trpC2) (1), PS29 (trpC2 unkU::spec), and its isogenic ΔcodY derivative PS37 (37). All B. subtilis strains were grown in minimal medium (38) supplemented with glucose (0.5%) and tryptophan (16 mg/liter). If indicated, CAA (Sigma) were added to a final concentration of 0.2%. Cells were harvested in the exponential growth phase after reaching an optical density at 500 nm (OD500) of 0.5.
The ilvA::cat mutant strain UM101 was constructed as follows. The complete ilvA coding region was amplified from B. subtilis 168 chromosomal DNA using Pfu polymerase (Stratagene) and primers ILVACLONE-5′ (5′-GGCCATGGATCCATGAAACCGTTGCTTAAAGAAAAC-3′) and ILVACLONE-3′ (5′-GGCCATGGATCCAGATTAGCAAATGGAACAAGTCCT-3′). The PCR product carried BamHI restriction sites at both ends (underlined in the primer sequences). After hydrolysis with BamHI, this fragment was inserted in BamHI-linearized pSKII(+) (Stratagene), resulting in the plasmid pilvA. Subsequently, pilvA was hydrolyzed using Eco47III. Using this enzyme, a 464-bp fragment was removed from the ilvA coding sequence, leaving 426 bp of the 5′ region and 376 bp of the 3′ region of the gene. After removal of the central 464-bp fragment, the Eco47III-hydrolyzed pilvA was ligated with a 950-bp EcoRV fragment (released from pSK::cat [39]) carrying a chloramphenicol resistance gene with its own promoter and two transcriptional terminators, one at each end of the resistance cassette. One of the resulting plasmids, in which transcription of the cat and ilvA genes occurs in the same direction, was named pilvA::cat. The pilvA::cat plasmid was linearized with ScaI and used for transformation of B. subtilis 168, with subsequent selection on chloramphenicol-containing agar plates. One of the transformants was checked by PCR for the correct integration of the cat cassette in the ilvA locus and designated UM101.
The ilvB::cat mutant strain UM102 was constructed as follows. The complete ilvB coding region was amplified from B. subtilis 168 chromosomal DNA using Pfu polymerase and primers ILVBCLONE-5′ (5′-GGCCATTCTAGAATGGGGACTAATGTACAGGTGGAT-3′) and ILVBCLONE-3′ (5′-GGCCATCTCGAGAGGTTTCACCCCCACCATTTCATG-3′). The PCR product carried a XbaI restriction site at its 5′ end an a XhoI restriction site at its 3′ end (underlined in the primer sequences). After hydrolysis with XbaI and XhoI, this fragment was inserted into XbaI-XhoI-digested pBlue SalI SmaIΔ (20), resulting in plasmid pilvB. Subsequently, pilvB was linearized with SmaI. Since this plasmid contains one single SmaI restriction site within ilvB, the hydrolysis cleaved the ilvB coding sequence in a 5′-terminal 581-bp fragment and a 3′-terminal 1,141-bp fragment. The SmaI-linearized pilvB was ligated with the same chloramphenicol resistance cassette used for the construction of UM101. One of the resulting plasmids in which transcription of the cat and ilvB genes occurs in the same direction was chosen and named pilvB::cat. B. subtilis 168 was transformed with the ScaI-linearized pilvB::cat plasmid, with subsequent selection on chloramphenicol containing agar plates. One of the transformants was checked by PCR for correct integration of the cat cassette in the ilvA locus and designated UM102.
The B. subtilis 168 strains carrying translational fusions of lacZ to the upstream regions of the ilvA, ilvB, ilvD, and ybgE genes were kindly provided by Jörg Stülke. The fusions were introduced into the codY null mutant strain PS37 and its isogenic wild-type strain, PS29, by transformation with chromosomal DNA and subsequent selection on agar plates containing chloramphenicol and spectinomycin. In the case of the PS37 derivative strains, the codY deletion was verified by PCR. In all cloning experiments, E. coli DH10B (Invitrogen) was used as the host strain, which was routinely grown in Luria-Bertani medium containing ampicillin (200 μg/ml) for all plasmid-bearing strains. Chloramphenicol and spectinomycin were added at 5 and 100 μg/ml, respectively.
Assay of β-galactosidase activity, RNA preparation, and Northern analysis.
The β-galactosidase assay was carried out as described by Miller (32). The preparation of total RNA and the Northern hybridization procedures were carried out as described previously (13). Chemiluminescence was detected with the Lumi-Imager (Roche Diagnostics), and chemiluminographs were quantified using the Lumi-Analyse software package (Roche Diagnostics). Transcript sizes were determined by comparison with an RNA size marker (Invitrogen). The positions of the molecular size markers are depicted on the left in all Northern blot images.
The digoxigenin-labeled RNA probes were synthesized by in vitro transcription with T7 RNA polymerase and specific PCR products as templates. Synthesis of the templates by PCR was performed using the following oligonucleotide pairs: for the ilvA probe; ILVA-5′ (5′-ATGAAACCGTTGCTTAAAGA-3′) and ILVA-3′ (5′-CTAATACGACTCACTATAGGGAGATGTACCTACCCCAGAGAGAA-3′); for ilvB, ILVB-5′ (5′-ATGGGGACTAATGTACAGGT-3′) and ILVB-3′ (5′-CTAATACGACTCACTATAGGGAGATGTTGTCGGCTGGTATCCCG-3′); for ilvC, ILVC-5′ (5′-TATAACGGTGATATCAAAGA-3′) and ILVC-3′ (5′-CTAATACGACTCACTATAGGGAGAGCCGCAAAGAACTGCTTGCT-3′); for ilvD, ILVD-5′ (5′-GAGGATCACCATGGCAGAAT-3′) and ILVD-3′ (5′-CTAATACGACTCACTATAGGGAGAAGACAGTTCATTGAGTTCGC-3′); for ilvH, ILVH-5′ (5′-ACATTGACTGTGGTGAACCG-3′) and ILVH-3′ (5′-CTAATACGACTCACTATAGGGAGAGCTGGTTCCCCTCGCAAAAG-3′); for leuA, LEUA-5′ (5′-GGTCTCCGTTGCGCAAAATT-3′) and LEUA-3′ (5′-CTAATACGACTCACTATAGGGAGAGACTGCCATTCCCAAATCATC-3′); for leuC, LEUC-5′ (5′-ATGATGCCTCGAACAATCAT-3′) and LEUC-3′ (5′-CTAATACGACTCACTATAGGGAGAGACGTAGCCTGTGCCGAATT-3′); for leuD, LEUD-5′ (5′-ATGGAACCTTTGAAATCACATAC-3′) and LEUD-3′ (5′-CTAATACGACTCACTATAGGGAGAGGCTTGAAGCCAAGAGCTTC-3′); for ybgE, YBGE-5′ (5′-CTTCCTTGGGGTTTGGACAA-3′) and YBGE-3′ (5′-CTAATACGACTCACTATAGGGAGATGAAGACTGGCGGCATAGTT-3′); and for ypmP, YPMP-5′ (5′-TTGTTACTCAAAAGCCTAGA-3′) and YPMP-3′ (5′-CTAATACGACTCACTATAGGGAGACGCGTTGTGCTTTAACACCT-3′). The underlined sequences indicate the T7 promoter region.
Primer extension analysis.
Primer extension was carried out as described previously (40) using the 32P-labeled primer ILVC-PEX (5′-TTCAGGGCATGTGCGTGGCC-3′). The DNA-sequencing reactions were carried out with the same primer and an 1,567-bp ilvH-ilvC DNA template which was generated by PCR using the primer pair ILVH-5′B (5′-CTAATACGACTCACTATAGGGAGATTGAAAAGAATTATCACATT-3′) and ILVC-3′B (5′-ATTTTGCGCAACGGAGACCA-3′).
Preparation of protein extracts and 2-D protein gel electrophoresis.
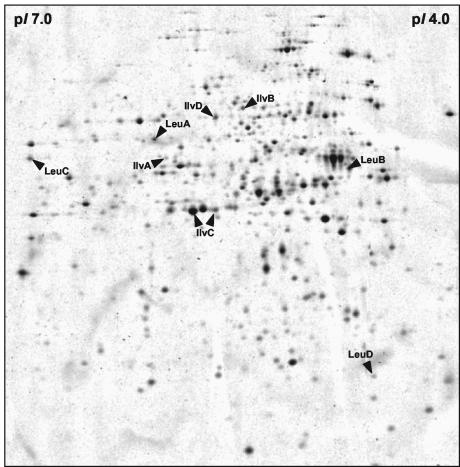
B. subtilis strain 168 growing in minimal medium at 37°C was harvested in the exponential growth phase after reaching an OD500 of 0.5. Protein extracts were prepared as described previously (5). IPG strips in the pH range from 4 to 7 (Amersham Biosciences) were loaded with 100 μg of crude protein extract. Isoelectric focusing using the Multiphor II unit (Amersham Biosciences) and sodium dodecyl sulfate polyacrylamide gel electrophoresis using the Investigator two-dimensional (2-D) electrophoresis system (Genomic Solutions) were performed as described previously (5). The resulting 2-D gels were fixed with 50% (vol/vol) methanol-7% (vol/vol) acetic acid and stained with SYPRO Ruby protein gel stain (Molecular Probes). Fluorescence was detected using a Storm860 imager (Molecular Dynamics). The 2-D gel image analysis and quantification of the protein amounts were performed with the Delta 2-D software (Decodon). Protein extracts from three independent cultures were used for quantification, and each sample was resolved twice. The identification of proteins was performed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry as described previously (5).
RESULTS
Analysis of regulation and transcriptional organization of the genes encoding the branched-chain amino acid biosynthetic enzymes.
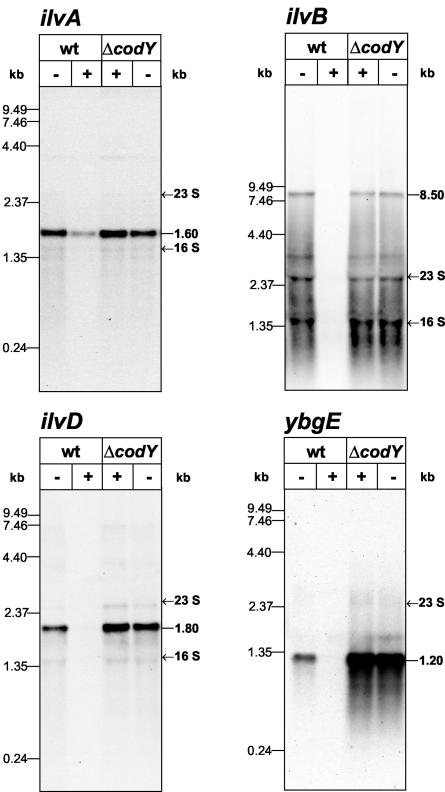
The results of our global gene expression analysis of B. subtilis grown in minimal medium with and without CAA (29) prompted us to conclude that CodY-dependent regulation of the genes for the biosynthesis of branched-chain amino acids (ilv-leu operon, ilvA, ilvD, and ybgE) was occurring. To verify this hypothesis and to obtain information about the transcriptional organization of these genes, Northern analyses using probes with specificity for ilvA, ilvB, ilvD, and ybgE were carried out. To this end, RNA was prepared from B. subtilis PS29 and the isogenic ΔcodY mutant PS37 grown in minimal medium in the presence or absence of CAA until the OD500 reached 0.5. The results are given in Fig. 1. The ilvA-specific probe detected a single mRNA of around 1.6 kb, indicating contranscription of ilvA and the downstream gene ypmP. The ilvB Northern blot demonstrated a large (8.5-kb) mRNA predicted to encompass the coding regions of ilvB, ilvH, ilvC, leuA, leuB, leuC, and leuD. The ilvD and ybgE genes were found to be monocistronically transcribed as mRNAs of 1.8 and 1.2 kb, respectively.
FIG. 1.
Northern analysis of ilvA, ilvB, ilvD, and ybgE in PS29 (wt) and PS37 (ΔcodY). RNA was prepared from cells growing exponentially in minimal medium in the presence (+) or absence (−) of CAA (0.2%). Electrophoretic separation of the RNA (5 μg per lane) was performed using 1.2% (ilvA, ilvD, and ybgE) or 0.6% (ilvB) agarose gels.
As shown in Fig. 1, significantly smaller amounts of ilvA, ilvB, ilvD, and ybgE mRNA were observed in the wild type in presence of CAA whereas similar amounts of mRNA were detected under both growth conditions in the codY mutant. In agreement with the study by Molle et al. (33), these results demonstrated CodY-dependent regulation of the branched-chain amino acid biosynthesis genes. However, the extent of CodY repression was different: whereas no ilvB, ilvD, and ybgE specific transcripts were detected in the wild type in the presence of CAA, a faint band was visible for ilvA, indicating weaker CodY repression. Furthermore, the ilvA, ilvB, and ilvD specific mRNAs were present in comparable amounts in the wild-type strain in the absence of CAA and in the codY mutant. In contrast, the amount of ybgE mRNA was significantly larger in the mutant strain. This indicated that in wild-type cells, CodY strongly represses the ybgE gene even in minimal medium without CAA. These results were confirmed by β-galactosidase activity assays of wild-type and ΔcodY strains carrying chromosomal lacZ fusions to the ilvA, ilvB, ilvD, and ybgE promoter regions with samples taken under the same growth conditions as for the Northern blot analyses (data not shown).
Analysis of the transcriptional units carrying the genes encoding the branched-chain amino acids biosynthesis enzymes after GTP depletion.
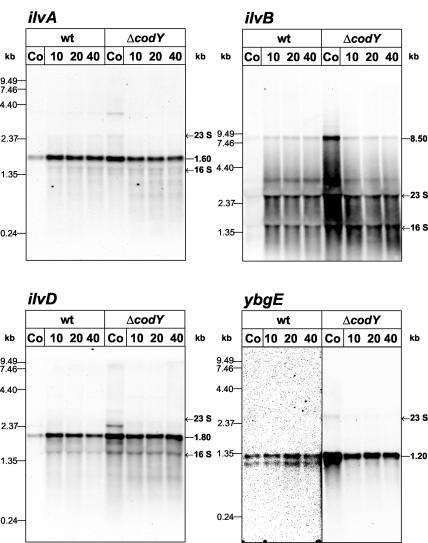
It was previously shown that the CodY repressor is active under conditions of high cellular GTP concentration (35). Consequently, GTP depletion causes derepression of CodY-regulated genes. Ratnayake-Lecamwasam et al. (35) demonstrated induction of the CodY-repressed dpp operon by treatment of cells with decoyinine, a GMP synthetase inhibitor. In the present study, mycophenolic acid, which inhibits the IMP dehydrogenase and thus the synthesis of guanine nucleotides, was used to decrease the cellular GTP pool. Cultures of B. subtilis PS29 and the isogenic ΔcodY mutant PS37 were grown in minimal medium with CAA. When the OD500 had reached 0.5, mycophenolic acid was added to a final concentration of 100 μM. Samples for RNA preparation were removed before and 10, 20, and 40 min after addition of mycophenolic acid. The RNA preparations were used in Northern hybridizations with ilvA-, ilvB-, ilvD-, and ybgE-specific probes (Fig. 2).
FIG. 2.
Northern analysis of ilvA, ilvB, ilvD, and ybgE under conditions of GTP depletion. PS29 (wt) and PS37 (ΔcodY) were grown in minimal medium in the presence of CAA (0.2%). After the culture reached an OD500 of 0.5, mycophenolic acid (Sigma) was added to a final concentration of 100 μM. RNA was prepared from cells harvested before (Co [control]) and at the indicated times (10, 20, and 40 min) after the addition of mycophenolic acid. Electrophoretic separation of the RNA (5 μg per lane) was performed using 1.2% (ilvA, ilvD, and ybgE) or 0.6% (ilvB) agarose gels. Note that in the case of the ybgE luminograph, different exposure times for the wild-type and ΔcodY strains were used. Whereas a long exposure time was necessary to detect the wild-type bands, the very strong signals in the ΔcodY strain required a short exposure time to avoid overexposition.
The ilvA-, ilvB-, and ilvD-specific mRNAs were significantly upregulated in the wild type by 10 min after addition of mycophenolic acid. However, the derepressed wild-type mRNA levels did not reach that of the codY mutant before mycophenolic acid treatment, although the amount of ilvA mRNA was very similar to that of the mutant. In the codY mutant strain, neither the ilvA- nor the ilvB- and ilvD-specific mRNAs were induced on mycophenolic acid addition. In contrast, the amounts of mRNA were significantly reduced, most obviously in the case of ilvB. Whereas CodY-dependent derepression of ilvA, ilvB, and ilvD could be confirmed following GTP depletion, the monocistronic ybgE exhibited a different pattern of expression. Although ybgE was clearly CodY dependent, it remained strongly repressed after mycophenolic acid treatment and was not induced by GTP depletion. The expression of chromosomal fusions between lacZ and the promoter regions of ilvA, ilvB, ilvD, and ybgE, measured before and 40 min after the addition of mycophenolic acid, confirmed the results of the Northern blot analyses (data not shown).
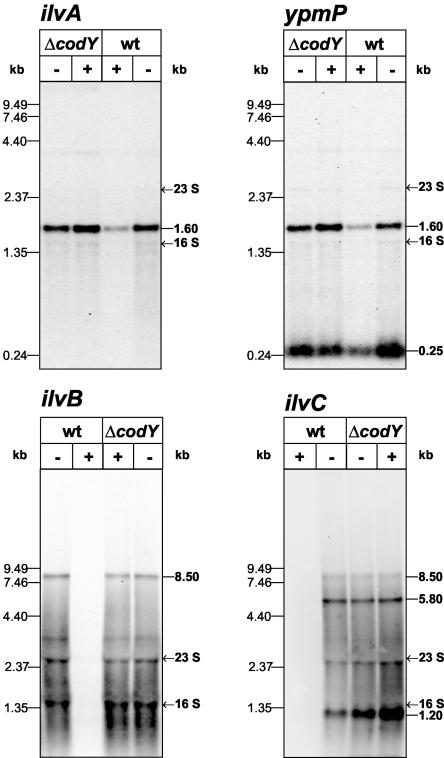
Northern blot analysis of ypmP and ilvC.
The results of the Northern hybridizations demonstrated single monocistronic ilvD and ybgE mRNAs, whereas a bicistronic ilvA-ypmP mRNA and a heptacistronic ilv-leu mRNA could be predicted. To complete the analysis of the two polycistronic operons, RNA prepared from the wild type (PS29) and ΔcodY mutant (PS37), after growth in minimal medium in the presence or absence of CAA, was used in Northern hybridizations with ypmP- and ilvC-specific probes (Fig. 3). In addition to the 1.6-kb ilvA-ypmP mRNA, which was already demonstrated using the ilvA probe, the ypmP probe detected an additional mRNA of 0.25 kb, which exhibited the expression pattern of ilvA. This monocistronic ypmP mRNA was present in larger amounts compared to the bicistronic transcript. For ilvC, two further mRNAs were detected in addition to the 8.5-kb mRNA, one of around 5.8 kb and one of 1.2 kb. The 5.8-kb mRNA was present in larger amounts than the 8.5-kb mRNA, and the 1.2-kb mRNA was the most abundant transcript. All three mRNA species detected with the ilvC probe had the same expression pattern. The newly identified mRNA species could be generated by endoribonucleolytic processing of the 1.6-kb ilvA-ypmP mRNA and the 8.5-kb ilv-leu mRNA or by the activities of additional internal promoters. To differentiate between these possibilities, several experiments were carried out.
FIG. 3.
Northern analysis of ilvA, ypmP, ilvB, and ilvC in PS29 (wt) and PS37 (ΔcodY). RNA was prepared from cells growing exponentially in minimal medium in the presence (+) or absence (−) of CAA (0.2%). Electrophoretic separation of the RNA (5 μg per lane) was performed using 1.2% (ilvA and ypmP) or 0.6% (ilvB and ilvC) agarose gels.
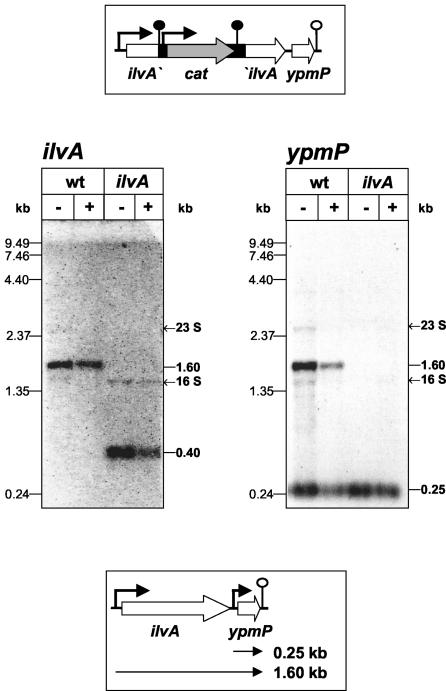
Detection of an internal promoter upstream of ypmP.
The results obtained with strain PS29 and the isogenic ΔcodY mutant PS37 concerning the regulation of the four transcriptional units as well as all detected mRNA species were reproducible in the B. subtilis 168 background. Since this strain is routinely used in our laboratory, all further experiments were carried out with B. subtilis 168. To analyze the origin of the 0.25-kb ypmP mRNA, a mutant was constructed to separate ypmP transcription from the ilvA promoter. Therefore, part of the ilvA coding region was replaced by a chloramphenicol resistance cassette carrying its own promoter and two transcriptional terminators, as described in Materials and Methods (Fig. 4). If the 0.25-kb ypmP mRNA was generated by endoribonucleolytic cleavage of the 1.6-kb ilvA-ypmP mRNA, it should be absent when transcription from the promoter upstream of ilvA is blocked. However, if an internal promoter upstream of ypmP was responsible for the synthesis of the 0.25-kb mRNA, this should be present even when the ypmP gene and the ilvA promoter were uncoupled. RNA was prepared from the ilvA mutant and from the B. subtilis 168 wild type growing exponentially in minimal medium supplemented with 1 mM isoleucine or CAA. Growth of the ilvA mutant requires isoleucine due to the loss of the threonine dehydratase activity. These RNA preparations were used in Northern experiments using ilvA- and ypmP-specific probes (Fig. 4).
FIG. 4.
Northern analysis of the ilvA::cat mutant strain UM101. RNA was prepared from UM101 (ilvA) and B. subtilis 168 (wt) growing exponentially in minimal medium supplemented with 1 mM isoleucine in the presence (+) or absence (−) of CAA (0.2%). RNA electrophoresis (5 μg per lane) was performed using 1.2% agarose gels. Hybridization was carried out using ilvA- and ypmP-specific probes. The top drawing shows the chromosomal organization of the ilvA-ypmP locus in UM101. The detected transcriptional organization of the wild-type locus is depicted in the bottom drawing.
Both probes detected the 1.6-kb ilvA-ypmP mRNA in the wild-type strain but not in the ilvA mutant, demonstrating the uncoupling of ypmP transcription from the ilvA promoter. In the ilvA mutant, the ilvA probe additionally hybridized to an mRNA of 0.4 kb that showed the same regulatory pattern as the 1.6-kb mRNA of the wild type. This transcript most probably results from transcription initiation at the ilvA promoter and termination at the first terminator within the chloramphenicol resistance cassette. However, the 0.25-kb ypmP mRNA was still present in the mutant strain, clearly indicating the presence of an additional promoter immediately upstream of ypmP, which is likely to be located in the relatively large intercistronic region of 89 bp between ilvA and ypmP (Fig. 4). Thus, generation of the 0.25-kb mRNA by mRNA processing could be excluded. Since this transcript still exhibited CAA-dependent regulation, the ypmP promoter might also be under CodY control.
Detailed Northern analysis of the ilv-leu operon.
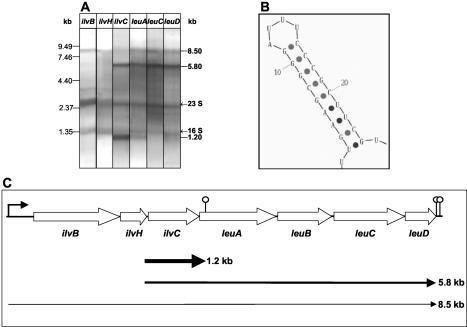
To unravel the transcriptional organization of the ilv-leu operon, a comprehensive Northern analysis was carried out. Wild-type cells grown in minimal medium were harvested during exponential growth (OD500 of 0.5) for RNA preparation. This RNA was hybridized to probes with specificity for ilvB, ilvH, ilvC, leuA, leuC, and leuD (Fig. 5A). The ilvB and ilvH probes detected exclusively the 8.5-kb mRNA encompassing the entire operon. As shown above, the ilvC probe detected the 8.5-, 5.8-, and 1.2-kb mRNAs. The leuA probe hybridized to the 8.5- and 5.8-kb mRNAs and, with much lower efficiency than the ilvC probe, to the 1.2-kb mRNA. Finally, the leuC- and leuD-specific probes detected only the two large mRNA species.
FIG. 5.
Detection of mRNAs specified by the ilv-leu operon. (A) Northern analysis. RNA was prepared from B. subtilis 168 growing exponentially in minimal medium. Six slots of a 0.6% RNA gel were loaded in parallel with 5 μg of this preparation. After electrophoresis and blotting, the membrane was cut into six strips, which were subsequently hybridized to probes with specifity for ilvB, ilvH, ilvC, leuA, leuC, and leuD. The reassembled chemiluminographs are shown. (B) Secondary structure (ΔG = −16 kcal/mol) in the 5′-terminal region of leuA as predicted by the Zuker algorithm (43). (C) Transcriptional organization of the ilv-leu operon as derived from the Northern analysis. The lengths of the different transcripts are indicated. The thickness of the arrows reflects the abundance of the transcripts.
These results are summarized in Fig. 5C. The 8.5-kb mRNA initiated at the promoter upstream of ilvB is terminated at the two transcriptional terminators downstream of leuD. The 5′ end of the 5.8- and 1.2-kb mRNAs is predicted to be located close to the 5′ end of ilvC or the 3′ end of ilvH. Finally, the 3′ end of the 1.2-kb mRNA is predicted to be located in the 5′-proximal part of the leuA coding region. Indeed, a significant secondary structure can be derived from the DNA sequence at the 5′ end of leuA (Fig. 5B). This putative stem-loop structure (ΔG = −16 kcal/mol) is located around 120 bases downstream of the leuA start codon. Whereas this secondary structure could explain the generation of the 3′ end of the 1.2-kb mRNA by partial transcriptional termination or by stalling of exoribonucleases, the origin of the 5′ end of 1.2- and 5.8-kb mRNA remained to be detected. As with the ilvA-ypmP operon, this 5′ end could be generated by endoribonucleolytic mRNA processing or by the activity of an internal promoter.
Detection of an mRNA-processing event within the ilv-leu operon.
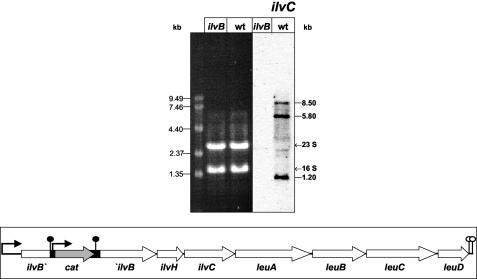
To differentiate between the activity of an internal promoter and an mRNA-processing event, uncoupling of all downstream located genes from the ilvB promoter was achieved by insertion of the chloramphenicol resistance cassette into the 5′ half of the ilvB coding region as described in Materials and Methods (Fig. 6). B. subtilis 168 and the isogenic ilvB mutant strain were grown in minimal medium in the presence of CAA to overcome the auxotrophy of the mutant strain. For RNA preparation, cells were harvested during exponential growth (OD500 of 0.5). In a Northern experiment, these RNA preparations were hybridized with an ilvC-specific probe (Fig. 6). Whereas the corresponding RNA gel showed the good quality of the RNA preparations, the luminograph of the Northern hybridization unambiguously demonstrated the absence of ilvC-specific mRNAs in the mutant strain. This result revealed that the complete ilv-leu operon is inactivated in the ilvB mutant. Consequently, the 5.8- and 1.2-kb mRNAs are generated by endoribonucleolytic mRNA processing of the 8.5-kb mRNA and not by the activity of an internal promoter.
FIG. 6.
Northern analysis of the ilvB::cat mutant strain UM102. RNA was prepared from UM102 (ilvB) and B. subtilis 168 (wt) growing exponentially in minimal medium in the presence of CAA (0.2%). This RNA preparation (5 μg per lane) was electrophoretically separated in a 0.6% agarose gel, which is shown on the left side. The corresponding chemiluminograph after hybridization of the blotted RNA with an ilvC probe is depicted on the right side. The drawing shows the chromosomal organization of the ilv-leu locus in UM102.
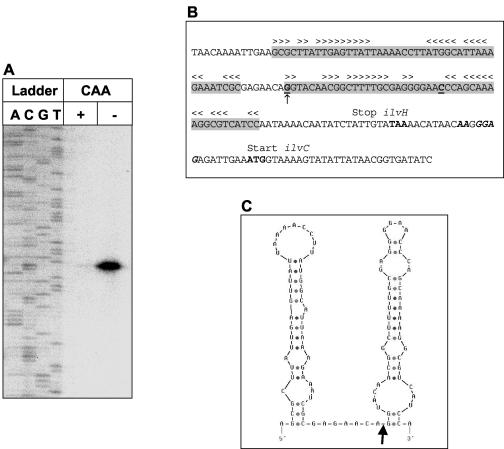
Mapping of the mRNA-processing site upstream of ilvC by primer extension.
To determine the mRNA cleavage site upstream of ilvC, a primer extension experiment was carried out using RNA prepared from B. subtilis 168 grown in minimal medium in the presence or absence of CAA. These RNA preparations were hybridized to a radioactively labeled primer complementary to the ilvC-specific mRNA, 113 bases downstream of the ilvC start codon. The result of the primer extension experiment is shown in Fig. 7A. One single mRNA 5′ end was mapped, corresponding to a G residue located 92 bases upstream of the ilvC start codon within the ilvH coding region, 66 bases upstream of the ilvH stop codon (Fig. 7B). The sequencing reaction of the ilvH-ilvC PCR product revealed an additional C residue 46 bp upstream of the 3′ end of the ilvH coding region which was lacking in the published sequence (25). This was confirmed by sequencing two independently generated PCR products. This insertion (indicated in Fig. 7B) changes the C-terminal amino acid sequence of IlvH from SKRRHPIKQYLLYKT to QQKASSNKTISIV, resulting in a protein shortened by 2 amino acids.
FIG. 7.
Mapping of the mRNA-processing site upstream of ilvC. (A) Primer extension analysis. The reactions were performed using 5 μg of RNA prepared from B. subtilis 168 growing exponentially in minimal medium in the presence (+) or absence (−) of CAA (0.2%). Lanes A, C, G, and T show the dideoxy sequencing ladder obtained with the same primer as used for the primer extension reactions. (B) Sequence of the chromosomal region surrounding the mapped processing site. Regions predicted to specify secondary structures on the mRNA level are highlighted in grey, and stem-loop structures are depicted as arrowheads. The C residue absent in the published genome sequence is underlined and in boldface, and the G residue representing the mapped 5′ end is marked by an arrow. The ilvH stop codon and the ilvC start codon are shown in boldface, and the ilvC ribosome-binding site is shown in italic. (C) Secondary-structure prediction for the mRNA surrounding the processing site according to the Zuker algorithm (43). The mapped mRNA cleavage site is indicated by an arrow.
The signal representing the mapped 5′ end exhibited the expected regulatory pattern (Fig. 7A). To test for a nonspecific reverse transcriptase product, the primer extension was performed in parallel using ilvH-ilvC RNA generated by in vitro transcription. In this control reaction, no significant signal appeared at the position of the mapped 5′ end (data not shown). To obtain more information about the structural determinants of the mapped processing site, the surrounding mRNA region was folded in silico by using the Zuker algorithm (43). This reveals that the mRNA cleavage site is located between an A and a G residue directly upstream of a relatively stable complex secondary structure (ΔG = −19.0 kcal/mol), at the distal 3′ site of a single-stranded mRNA stretch of 7 bases (Fig. 7C). The single-stranded region is flanked at the 3′ side by the above-mentioned secondary structure and at the 5′ side by an additional, less stable secondary structure (ΔG = −3.0 kcal/mol) at the 5′ side. After cleavage, the 5′ ends of the resulting processing products are located within a base-paired double-stranded region. Since it is known that 5′ base-paired ends of mRNA confer protection against degradation in E. coli as well as B. subtilis (10), it could be predicted that the 5.8- and 1.2-kb processing products are more stable than the 8.5-kb precurser. This assumption was further strengthened by the observation that the processing products are present in the cell in larger amounts than is the full-length precursor mRNA, as revealed by the Northern analyses. Therefore, it was tempting to speculate that the physiological function of the processing event was the generation of mRNAs with higher stabilities. This prediction was tested by half-life determination of the 8.5-, 5.8-, and 1.2-kb mRNAs.
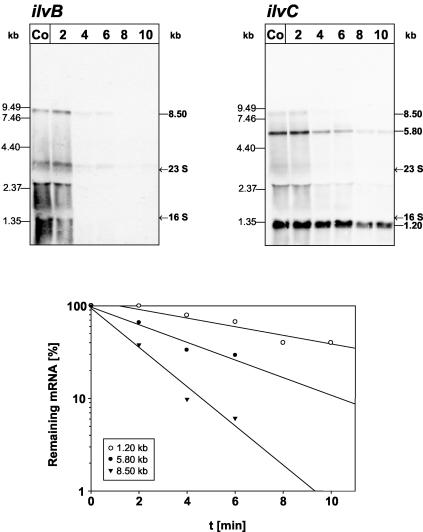
Half-life determination of the three mRNA species of the ilv-leu operon.
Rifampin was added to exponentially growing B. subtilis 168 cells to prevent the initiation of transcription. Samples for RNA preparation were removed before and at different times after the rifampin addition. These RNA samples were hybridized in Northern experiments with ilvB- and ilvC-specific probes (Fig. 8). Quantification of the resulting luminographs allowed the calculation of specific half-lifes: for the 8.5-kb primary transcript, a half-life of 1.2 min was determined, whereas the 5.8- and 1.2-kb processing products exhibited half-lifes of 3.0 and 7.6 min, respectively. Thus, the postulated differential mRNA stabilities resulting from the different 5′ ends of the primary transcript and the processing products were verified.
FIG. 8.
Half-life determination of the different mRNAs of the ilv-leu operon. RNA (5 μg per lane) was prepared from B. subtilis 168 growing exponentially in minimal medium before (Co) and at different times (given in minutes) after the addition of rifampin. After RNA electrophoresis in 0.6% agarose gels and blotting, the membranes were hybridized to ilvB- and ilvC-specific probes. The half-lifes of the 8.5-, 5.8-, and 1.2-kb mRNA were determined by linear regression analysis plots of the percentage of remaining mRNA versus the time. The half-life data were obtained from three experiments using independently prepared RNA.
The mRNA analyses showed that the ilvB and ilvH genes encoding the acetolactate synthase subunits are expressed only as part of the 8.5-kb mRNA and specify the smallest amounts of mRNA of all genes in the operon. The mRNA-processing event upstream of ilvC, which generates the 5.8- and 1.2-kb mRNAs, should also result in the generation of a 5′-proximal processing product of around 2.7 kb containing ilvB and the truncated ilvH. It is possible that the faint band above the nonspecific 23 S rRNA signal, which was detected with the ilvB-specific probe, represents this processing product (Fig. 1, 2, 3, 5, and 8). However, the detection of this mRNA was not reliably reproducible since the band was absent in several RNA preparations, pointing to a very low mRNA stability. The leuA, leuB, leuC, and leuD genes, encoding 2-isopropylmalate synthase, 3-isopropylmalate dehydrogenase, and the 3-isopropylmalate dehydratase subunits, respectively, are expressed as part of the 8.5- and 5.8-kb mRNA. Consequently, these genes specify more mRNA than do ilvB and ilvH. The largest amount of mRNA is specified by ilvC, encoding ketol-acid reductoisomerase, which is expressed as part of three mRNA species. In addition to the 8.5- and 5.8-kb mRNAs, ilvC specifies the 1.2-kb mRNA which represents the most abundant transcript. These observations raised the question whether the different amounts of mRNA are also reflected in the amounts of proteins synthesized. Therefore, the relative amounts of the proteins encoded by the operon were compared.
Determination of the relative protein amounts of the enzymes encoded by the ilv-leu operon.
To determine the relative cellular amounts of the proteins involved in branched-chain amino acid biosynthesis, protein extracts were prepared from B. subtilis 168 grown in minimal medium. These extracts were separated by 2-D polyacrylamide gel electrophoresis and stained with SYPRO Ruby protein gel stain (Fig. 9). Following quantification of the 2-D gel images, the relative amounts of the IlvB, IlvC, LeuA, LeuB, LeuC, and LeuD proteins could be calculated; IlvH was not detectable in the 2-D gels in the pH range from 4 to 7 due to its alkaline isoelectric point (pI). However, since IlvB and IlvH form a heteromer and are translationally coupled (the ilvB stop codon and the ilvH start codon share two bases), it can be postulated that the two proteins are present in similar amounts. The averaged quantification data revealed the following relative proteins amounts: 1.2 ± 0.16 (IlvB), 11.76 ± 1.0 (IlvC), 1.72 ± 0.20 (LeuA), 1.72 ± 0.22 (LeuB), 1.12 ± 0.25 (LeuC), and 1.0 ± 0.23 (LeuD). Thus, the large amount of ilvC-specific mRNA, which is caused primarily by the remarkably long half-life of the 1.2-kb processing product, is indeed reflected by a correspondingly large amount of the IlvC protein. Whereas the monomeric species of the other proteins encoded by the ilv-leu operon were detected in comparable amounts, the abundance of IlvC was around 10-fold higher. However, there was no difference between the amounts of the proteins encoded by ilvB showing the smallest mRNA amount of the operon and leuA, leuB, leuC, and leuD, exhibiting mean mRNA amounts. In addition to the proteins encoded by the ilv-leu operon, the IlvA and IlvD proteins were analyzed. Whereas the amount of IlvD (1.28 ± 0.23) was comparable to IlvB, LeuA, LeuB, LeuC, and LeuD, the IlvA protein was present in significantly smaller amounts (0.36 ± 0.03).
FIG. 9.
SYPRO ruby-stained cytosolic proteins of B. subtilis 168 separated by 2-D protein gel electrophoresis. The protein extract (100 μg) prepared from cells growing exponentially in minimal medium was separated in a pH gradient of 4 to 7. The labeled spots represent proteins involved in the biosynthesis of branched-chain amino acids.
DISCUSSION
In the present study, the transcriptional organization and regulation of the genes encoding the branched-chain amino acid biosynthetic enzymes were analyzed. As already reported by Molle et al. (33), the ilv-leu operon and the ilvA, ilvB, and ybgE genes were shown to be controlled by the global transcriptional regulator, CodY. However, it is still not known why the branched-chain amino acid biosynthetic pathway is coregulated with early-stationary-phase and sporulation genes in response to the nutritional state of the cell. Possibly, CodY regulation of branched-chain amino acid biosynthesis occurs because spore formation requires de novo biosynthesis of fatty acids (36). Valine, leucine, and isoleucine are the precursors for biosynthesis of iso- and anteiso-branched fatty acids, which represent the major fatty acid species of the membrane lipids in Bacillus species (11). The absence of de novo fatty acid biosynthesis at the onset of sporulation results in a block of σE-directed gene expression in the mother cell, caused by the failure of pro-σE processing to its active form (36). It was suggested that de novo fatty acid biosynthesis contributes to the specific phospholipid composition of the mother cell, which is predicted to be essential for activation of the pro-σE processing protease, SpollGA (36). Therefore, CodY-mediated induction of branched-chain amino acid biosynthesis genes would ensure the availability of precursors for de novo fatty acid biosynthesis at the onset of sporulation.
The most interesting results of this study were obtained in the context of the posttranscriptional regulation of the ilv-leu operon. This operon is transcribed from a single promoter which is located upstream of ilvB. The corresponding transcriptional start site was mapped 482 bases upstream of the ilvB start codon (14). The large leader region mediates regulation of the ilv-leu operon by antitermination via the T-box mechanism (14, 30). A full-length 8.5-kb primary transcript is synthesized by transcriptional initiation at the ilvB promoter and termination at one of the two terminator structures (ΔG = −19.8 and −12.7 kcal/mol), which can be derived from the sequence immediately downstream of leuD. The processing event upstream of ilvC generates different processing products: a 5′-proximal product of around 2.7 kb containing ilvB and the truncated ilvH is postulated to exhibit a very low stability. Removal of the 5′ portion of the 8.5-kb mRNA generates the new 5′ end of the 5.8-kb as well as the 1.2-kb mRNA, which results in significantly more stable processing products.
Possibly, partial transcriptional termination occurs at the stem-loop structure in the 5′-terminal region of the leuA coding sequence (Fig. 5B). This would result in a primary transcript of around 3.9 kb, which should be detectable using ilvB, ilvH, and ilvC probes. However, a corresponding band was not detected, suggesting a very short half-life of this postulated primary transcript. Rapid processing at the site upstream of ilvC may prevent the accumulation of detectable amounts of mRNA. Alternatively, the secondary structure in the 5′-terminal region of leuA may not function primarily as a transcriptional terminator but as a 3′ mRNA-stabilizing element. Accordingly, the 1.2-kb processing product is generated by endoribonucleolytic cleavage of the 8.5-kb precurser at the processing site upstream of ilvC and exonucleolytic degradation of the 8.5- and 5.8-kb mRNAs, which is impeded at the stem-loop structure downstream of ilvC. The latter assumption is further strengthened by the fact that despite sharing an mRNA 5′ end, the 5.8-kb processing product has a significantly shorter half-life (3.0 min) than does the 1.2-kb product (7.6 min). The strong secondary structures formed by the terminators downstream of leuD can be predicted to protect the distal 3′ end of the 8.5- and 5.8-kb mRNAs against exoribonucleolytic degradation. Since exoribonucleases require unpaired single-stranded 3′ ends to attach to the mRNA, recognition sites for endoribonucleases able to generate such 3′ ends have to be postulated in the region between the secondary structure downstream of ilvC and the farthest 3′ end of the operon. Finally, we cannot exclude the possibility that this secondary structure functions as both a 3′ mRNA stabilizer and a transcriptional terminator.
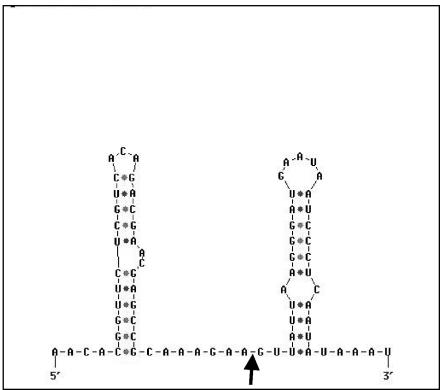
The processing site upstream of ilvC is of especial interest. The secondary-structure prediction for this mRNA region resembles the one of the processing site mapped in the cggR-gapA-pgk-tpi-pgm-eno operon (gapA operon) at the farthest 3′ end of the cggR coding region (27). As shown in Fig. 7C and 10, mRNA cleavage occurs in both cases between A and G residues within single-stranded regions flanked by stem loop structures, near the downstream stem-loop. Single-stranded, mostly AU-rich RNA regions which are stabilized by adjacent stem-loop structures represent recognition sites for the most important E. coli endoribonuclease RNase E (8). An enzymatic activity similar to RNase E was described in B. subtilis (9), although the corresponding gene is absent from the genomic sequence (25). This postulated RNase E-like B. subtilis enzyme, whose gene still remains to be identified, might be responsible for the endoribonucleolytic processing of the gapA and ilv-leu operons.
FIG. 10.
The mRNA secondary structures surrounding the processing site mapped in the gapA operon (27). The mapped mRNA cleavage site is indicated by an arrow.
In both cases, the processing events generate products with different mRNA stabilities. Due to its very low stability, the 5′-proximal ilvBH processing product of the ilv-leu operon could not be be detected reliably whereas the 3′-proximal 1.2- and 5.8-kb processing products exhibited higher stabilities (half-lives of 7.6 and 3.0 min, respectively). The 5′-proximal processing product of the cggR-gapA-pgk-tpi-pgm-eno operon, which encompasses the leader region and a 3′-truncated cggR coding sequence, exhibited a half-life of 1.5 min, whereas the 3′-proximal 1.2-kb gapA-processing and 6.2-kb gapA-pgk-tpi-pgm-eno-processing products showed half-lives of 3.5 and 3.0 min, respectively. The more stable 3′-proximal processing products of the ilv-leu and gapA operons have a common structural feature: the processing events generate 5′ ends which are characterized by complex secondary structures. In the case of the gapA operon, this 5′ end represents an overhang of two bases upstream of the stem-loop structure (Fig. 10), whereas in the case of the ilv-leu operon, the 5′ end is included in the base pairing of the stem-loop structure (Fig. 7C). The steric shielding of 5′ ends results in mRNA stabilization in E. coli as well as in B. subtilis (8, 10). In E. coli, this can be explained by the fact that the crucial step in the initiation of mRNA degradation, the binding of RNase E, occurs at the 5′ end (28). Efficient attachement of RNase E requires a single-stranded, non-base-paired 5′ end (28). If 5′ ends are protected either by the presence of a stable secondary structure or by the frequent binding of ribosomes to an adjacent Shine-Dalgarno sequence, impaired attachment of RNase E results in increased mRNA stability (8, 10). Although the identity of the RNase E-like enzyme(s) in B. subtilis is still unknown, the fact that similar structural determinants at the 5′ end confer mRNA stability in E. coli and B. subtilis suggests that the mRNA degradation systems might be comparable (10). Most probably, the increased stabilities of the 3′-proximal processing products of the gapA and ilv-leu operons result from steric exclusion of an enzymatic apparatus which initiates mRNA degradation by binding to 5′ ends.
A further common feature of the processing sites of these operons is the relatively large distance between the 5′ secondary structures and the ribosome-binding sites of the first genes of the generated processing products: the distances between the mapped 5′ ends and the start codons of gapA and ilvC are 65 and 91 bases, respectively, and the distances between the 5′ stem-loops and the Shine-Dalgarno sequences (AAAGGAGGAAAC and CAAGGGAGAGAT, consensus positions underlined) are 20 and 31 bases. These relatively large leader regions might reflect the necessity of preventing interference of the stem-loop structures with ribosomal subunits: as a 30S ribosomal subunit bound to a Shine-Dalgarno sequence covers a region of around 30 nucleotides (42), too close a neighboring 5′ stabilizing stem-loop could decrease the translation initiation rate by reducing the efficiency of ribosome binding.
The high stability of the monocistronic 1.2-kb ilvC processing product caused by the combination of the stem-loop structures at the 5′ and 3′ ends is reflected at the protein level: the amount of IlvC is around 10-fold greater than that of the other proteins encoded by the ilv-leu operon. Thus, different amounts of protein are achieved by adjusting the differential stabilities of mRNAs specified by the same operon. There are two further examples of polycistronic B. subtilis operons which regulate the amounts of their encoded proteins by differential mRNA stabilities: the gapA operon and the dnaK operon. For the above-mentioned gapA operon, the low stability of the mRNA species which encodes the repressor protein CggR results in a 100-fold-smaller amount of the monomeric form of CggR compared to the product of the second gene of the operon, gapA (31). In this case, the physiological function of this molar ratio is obvious: the gene product of gapA, the glyceraldehyde-3-phosphate dehydrogenase, is a metabolic enzyme required in high copy numbers in the cell. In contrast, the repressor protein CggR binds to only a single target sequence located upstream of cggR (12) and is consequently required only in very small quantities.
The mRNA species which contain the first gene of the heptacistronic hrcA-grpE-dnaK-dnaJ-orf35-orf28-orf50 operon also exhibit a very low stability, resulting in a molar ratio of the monomeric forms of the proteins HrcA and DnaK of 1:100 at 37°C and 1:40 at 48°C (21). The physiological function of this molar ratio resembles that postulated for the gapA operon: the Hsp70 chaperone of B. subtilis, DnaK, is required in large amounts under normal growth conditions and is present in even greater amounts after a temperature upshift. The HrcA protein is the repressor of the class I heat shock operons hrcA-grpE-dnaK-dnaJ-orf35-orf28-orf50 and groESL (41). Because there are only two target sites of this repressor, the required cellular quantities are clearly smaller than those of DnaK. As in the case of the gapA operon, an mRNA-processing event generates segments of differential mRNA stability. Cleavage in the intercistronic region between hrcA and grpE results in a monocistronic upstream hrcA-processing product of low stability and a more stable downstream processing product carrying dnaK. However, the mapped cleavage site is located within a secondary-structure element and may therefore be recognized by an endoribonuclease different from the one which most probably cleaves the mRNA at the 3′ end of cggR and the 5′ end of ilvC (21).
For the ilv-leu operon, the rationale for the different protein amounts has not yet been identified. On the one hand, the ketol-acid reductoisomerase encoded by ilvC may represent a multimeric protein, and the larger amounts of monomeric subunits would result in a comparable molar ratio of the functional enzymes. Indeed, the IlvC proteins of Salmonella enterica serovar Typhimurium and Spinacia oleracea were shown to form homotetramers (4, 19). However, since the B. subtilis IlvC lacks several large stretches of amino acids found in these enzymes, a reliable prediction of a multimeric structure is not possible. B. subtilis ketol-acid reductoisomerase may exhibit a low specific activity compared to other enzymes encoded by the ilv-leu operon, and the cell compensates for this lower efficiency by having a larger amount of protein. Indeed, the purified E. coli enzyme shows a ca. 20-fold-lower specific activity than that of the purified acetolactate synthase encoded by ilvBH (7, 18). Although it cannot be assumed that the B. subtilis and E. coli enzymes have the same properties, the different protein amounts of the branched-chain amino acid biosynthetic enzymes could be explained by different activities of the enzymes encoded by the ilv-leu operon.
In summary, the genes for branched-chain amino acid biosynthesis are regulated at different levels. First, the promoters preceding the four trancriptional units involved in this pathway (ilvA-ypmP, ilvBHC-leuABCD, ilvD, and ybgE) are under CodY control (33). Second, expression of the ilv-leu operon is regulated by tRNA-mediated transcriptional antitermination in response to leucine availability (14, 30). Finally, as demonstrated in the present work, different amounts of the enzymes encoded by the ilv-leu operon are adjusted by mRNA processing and differential segmental mRNA stability of the resulting processing products.
Acknowledgments
We are indebted to Abraham L. Sonenshein for providing strains PS29 and PS37 and to Christian Detsch, Holger Ludwig, Anastasija Matin, Mareike Stieg, and Jörg Stülke for providing strains carrying the translational fusions of ilvA, ilvB, ilvD, and ybgE to lacZ. We thank Abel Ferrandéz for sharing unbublished results, Claudia Böhnisch for help with the Northern analyses, and Rüdiger Bode for continuous support.
This work was supported by grants from the EU consortium QLG2-CT-1999-01455.
REFERENCES
- 1.Aganostopoulos, C., and I. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson, M. R., L. V. Wray, Jr., and S. H. Fisher. 1990. Regulation of histidine and proline degradation enzymes by amino acid availability in Bacillus subtilis. J. Bacteriol. 172:4758-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belasco, J. G., J. T. Beatty, C. W. Adams, A. von Gabain, and S. N. Cohen. 1985. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell 40:171-181. [DOI] [PubMed] [Google Scholar]
- 4.Biou, V., R. Dumas, C. Cohen-Addad, R. Douce, D. Job, and E. Pebay-Peyroula. 1997. The crystal structure of plant acetohydroxy acid isomeroreductase complexed with NADPH, two magnesium ions and a herbicidal transition state analog determined at 1.65 A resolution. EMBO J. 16:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mäder, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 6.Chopin, A., V. Biaudet, and S. D. Ehrlich. 1998. Analysis of the Bacillus subtilis genome sequence reveals nine new T-box leaders. Mol. Microbiol. 29:662-664. [DOI] [PubMed] [Google Scholar]
- 7.Chunduru, S. K., G. T. Mrachko, and K. C. Calvo. 1989. Mechanism of ketol acid reductoisomerase — steady-state analysis and metal ion requirement. Biochemistry 28:486-493. [DOI] [PubMed] [Google Scholar]
- 8.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acid Res. Mol. Biol. 62:55-108. [DOI] [PubMed] [Google Scholar]
- 9.Condon, C., H. Putzer, D. Luo, and M. Grunberg-Manago. 1997. Processing of the Bacillus subtilis thrS leader mRNA is RNase E-dependent in Escherichia coli. J. Mol. Biol. 268:235-242. [DOI] [PubMed] [Google Scholar]
- 10.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67:157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Mendoza, D., G. E. Schujman, and P. S. Aguilar. 2002. Biosynthesis and function of membrane lipids, p. 405-414. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus and its closest relatives. ASM Press, Washington, D.C.
- 12.Doan, T., and S. Aymerich. 2003. Regulation of the central glycolytic genes in Bacillus subtilis: binding of the repressor CggR to its single DNA target sequence is modulated by fructose-1,6-bisphosphate. Mol. Microbiol. 47:1709-1721. [DOI] [PubMed] [Google Scholar]
- 13.Eymann, C., G. Homuth, C. Scharf, and M. Hecker. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grandoni, J. A., S. A. Zahler, and J. M. Calvo. 1992. Transcriptional regulation of the ilv-leu operon of Bacillus subtilis. J. Bacteriol. 174:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundy, F. J., and T. M. Henkin. 1994. Conservation of a transcription antitermination mechanism in aminoacyl-tRNA synthetase and amino acid biosynthesis genes in gram-positive bacteria. J. Mol. Biol. 235:798-804. [DOI] [PubMed] [Google Scholar]
- 16.Heck, C., R. Rothfuchs, A. Jäger, R. Rauhut, and G. Klug. 1996. Effect of the pufQ-pufB intercistronic region on puf mRNA stability in Rhodobacter capsulatus. Mol. Microbiol. 20:1165-1178. [DOI] [PubMed] [Google Scholar]
- 17.Heck, C., A. Balzer, O. Fuhrmann, and G. Klug. 2000. Initial events in the degradation of the polycistronic puf mRNA in Rhodobacter capsulatus and consequences for further processing steps. Mol. Microbiol. 35:90-100. [DOI] [PubMed] [Google Scholar]
- 18.Hill, C. H., S. S. Pang, and R. G. Duggleby. 1997. Purification of Escherichia coli acetohydroxyacid synthase isoenzyme II and reconstitution of active enzyme from its individual pure subunits. Biochem. J. 327:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofler, J. G., C. J. Decedue, G. H. Luginbuhl, J. A. Reynolds, and R. O. Burns. 1975. The subunit structure of alpha-acetohydroxyacid isomeroreductase from Salmonella typhimurium. J. Biol. Chem. 250:877-882. [PubMed] [Google Scholar]
- 20.Homuth, G., S. Masuda, A. Mogk, Y. Kobayashi, and W. Schumann. 1997. The dnaK operon of Bacillus subtilis is heptacistronic. J. Bacteriol. 179:1153-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homuth, G., A. Mogk, and W. Schumann. 1999. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol. Microbiol. 32:1183-1197. [DOI] [PubMed] [Google Scholar]
- 22.Jäger, S., O. Fuhrmann, C. Heck, M. Hebermehl, E. Schiltz, R. Rauhut, and G. Klug. 2001. An mRNA degrading complex in Rhodobacter capsulatus. Nucleic Acids Res. 29:4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klug, G., C. W. Adams, J. Belasco, B. Doerge, and S. N. Cohen. 1987. Biological consequences of segmental alterations in mRNA stability: effects of deletion of the intercistronic hairpin loop region of the Rhodobacter capsulatus puf operon. EMBO J. 6:3515-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klug, G., and S. N. Cohen. 1990. Combined actions of multiple hairpin loop structures and sites of rate-limiting endonucleolytic cleavage determine differential degradation rates of individual segments within polycistronic puf operon mRNA. J. Bacteriol. 172:5140-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 26.Lopez, J. M., A. Dromerick, and E. Freese. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146:605-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig, H., G. Homuth, M. Schmalisch, F. M. Dyka, M. Hecker and J. Stülke. 2001. Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol. Microbiol. 41:409-422. [DOI] [PubMed] [Google Scholar]
- 28.Mackie, G. A. 1998. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395:720-723. [DOI] [PubMed] [Google Scholar]
- 29.Mäder, U., G. Homuth, C. Scharf, K. Büttner, R. Bode, and M. Hecker. 2002. Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability. J. Bacteriol. 184:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marta, P. T., R. D. Ladner, and J. A. Grandoni. 1996. A CUC triplet confers leucine-dependent regulation of the Bacillus subtilis ilv-leu operon. J. Bacteriol. 178:2150-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meinken, C., H.-M. Blencke, H. Ludwig, and J. Stülke. 2003. Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology 149:751-761. [DOI] [PubMed] [Google Scholar]
- 32.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelchat, M., and J. Lapointe. 1999. Aminoacyl-tRNA synthetase genes of Bacillus subtilis: organization and regulation. Biochem. Cell Biol. 77:343-347. [PubMed] [Google Scholar]
- 35.Ratnayake-Lecamwasam, M., P. Serror, K. W. Wong, and A. L. Sonenshein. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schujman, G. E., R. Grau, H. C. Gramajo, L. Ornella, and D. de Mendoza. 1998. De novo fatty acid synthesis is required for establishment of cell type-specific gene transcription during sporulation in Bacillus subtilis. Mol. Microbiol. 29:1215-1224. [DOI] [PubMed] [Google Scholar]
- 37.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of the β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 39.Versteeg, S., A. Mogk, and W. Schumann. 1999. The Bacillus subtilis htpG gene is not involved in thermal stress management. Mol. Gen. Genet. 261:582-588. [DOI] [PubMed] [Google Scholar]
- 40.Wetzstein, M., U. Völker, J. Dedio, S. Löbau, U. Zuber, M. Schiesswohl, C. Herget, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J. Bacteriol. 174:3300-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan, G., and S.-L. Wong. 1995. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J. Bacteriol. 177:6462-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yusupova, G. Z., M. M. Yusupov, J. H. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]
- 43.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]