Abstract
Self-amplifying replicon RNA (RepRNA) possesses high potential for increasing antigen load within dendritic cells (DCs). The major aim of the present work was to define how RepRNA delivered by biodegradable, chitosan-based nanoparticulate delivery vehicles (nanogel-alginate (NGA)) interacts with DCs, and whether this could lead to translation of the RepRNA in the DCs. Although studies employed virus replicon particles (VRPs), there are no reports on biodegradable, nanoparticulate vehicle delivery of RepRNA. VRP studies employed cytopathogenic agents, contrary to DC requirements—slow processing and antigen retention. We employed noncytopathogenic RepRNA with NGA, demonstrating for the first time the efficiency of RepRNA association with nanoparticles, NGA delivery to DCs, and RepRNA internalization by DCs. RepRNA accumulated in vesicular structures, with patterns typifying cytosolic release. This promoted RepRNA translation, in vitro and in vivo. Delivery and translation were RepRNA concentration-dependent, occurring in a kinetic manner. Including cationic lipids with chitosan during nanoparticle formation enhanced delivery and translation kinetics, but was not required for translation of immunogenic levels in vivo. This work describes for the first time the characteristics associated with chitosan-nanoparticle delivery of self-amplifying RepRNA to DCs, leading to translation of encoded foreign genes, namely influenza virus hemagglutinin and nucleoprotein.
Introduction
Dendritic cells (DCs) play critical roles in immune defense development, due to their highly diverse endocytic networks,1,2,3,4 promoting adaptive immune responses.4,5,6,7,8 Yet DCs show limited intracellular uptake of nucleic acids.4 Synthetic nanoparticulate delivery has shown promise for facilitating intracellular uptake of RNA molecules,4,9,10,11,12,13,14 proving particularly useful for delivering oligoRNA such as siRNA. Nevertheless, this does not guarantee similar success with the more complex self-replicating (self-amplifying) replicon RNA (RepRNA). Its size being over 100 kb, compared with the tens of bases for oligoRNAs, will influence the interaction with synthetic nanoparticulate delivery vehicles; there will also be fewer molecules of RepRNA per nanoparticle. More importantly are the different intracellular sites to be targeted for functional readout. RepRNA must translate by interacting with the ribosomal machinery, unlike siRNA, which interacts with RNA interference pathway components. Both the translation and subsequent replication of RepRNA render it particularly sensitive to RNase, which can easily destroy ribosomal entry or gene translation. The latter is especially problematic for RepRNA translating as a polyprotein.
RepRNA offers the capacity for gene delivery, including genes encoding vaccine antigens such as those of influenza virus.4,14,15,16,17,18 Both the number of mRNA molecules and the half-life of translation are enhanced through delivery of RepRNA.4,14 This is due to RepRNAs behaving like viruses; they are defective virus genomes—essential genes encoding viral structural proteins are deleted—which still translate and replicate without producing progeny virus.4,14,15,16,17,18,19,20 Thus, RepRNA provides the template for increasing the number of RNA molecules translating, which in turn increases the rounds of antigen production.
RepRNA delivery has focused on virus replicon particles (VRPs),15,16,17,19,20 but VRP delivery is dependent on the cell tropism of the particle, which is not readily modified for controllable targeting of DCs. VRPs also suffer from species or individual restriction for infecting cells, while anti-VRP host immunity can prevent delivery. Electroporation of replicons has been reported,21 but again does not guarantee DC targeting.
We therefore sought to determine if synthetic nanoparticulate vehicles could promote RepRNA delivery to DCs, due to their application with mRNA.4,11,13 Our approach adapted a process for synthetic, particulate vehicle delivery to the RepRNA.14 The aim was to define RepRNA delivery to DCs promoting RNA translocation from endocytic vesicles into the cytosol for translation. This is an important consequence considering that DCs possess the major capacity to process and present translated antigens,22 leading to activation of immune defense.4,5,6,7,8 DCs also transport antigen into lymphoid tissues and organs;4,23 properties epithelial cells do not possess. Importantly, the DCs must survive RepRNA delivery. This requires application of noncytopathogenic RepRNA14,22,24 rather than the frequently employed cytopathogenic alphavirus replicons—noncytopathogenic RepRNA permits prolonged retention of the RNA and translated antigens for interaction with the immune system.4,7
Accordingly, our major aim was to define how RepRNA delivered by chitosan-based delivery vehicles (nanogel-alginate (NGA)) interact with DCs. This is the first description of RepRNA delivery to DCs by synthetic nanoparticulate vehicles, which provided both RNase protection and delivery. The outer alginate coat of these vehicles promoted interaction with the DCs, but it was not essential—this was also noted with the chitosan core lacking an alginate coat. Cytosolic translocation was likely dependent on this cationic chitosan core, for which the additional presence of cationic lipids proved advantageous with low RNA concentrations. A major aim of the study was to characterize DC handling of nanoparticle-delivered RepRNA, relating this ultimately to RepRNA translation. Analyses were primarily in vitro, to facilitate detailed study of the DCs and RNA translation, which is difficult to monitor directly in vivo. Nevertheless, it was considered important to confirm that the in vitro data were pertinent, for which reason in vivo studies were performed to confirm that translation of nanogel-delivered RepRNA was not simply an in vitro phenomenon.
Results
NGA-RepRNA complexes
It was important to ascertain that RepRNA efficiently interacted with the chitosan during NGA formation. The ΔErns replicon construct (Figure 1a) was labeled with fluorescein isothiocyanate (FITC) for this purpose. Flow cytometry forward and side scatter settings were adjusted to detect the particular light scattering properties of the NGA (Figure 1b, NGA alone). This was not an attempt to define size, but simply visualize the particles, which were clearly distinguishable from diluent alone (MQ-water) (Figure 1b). Dy490-labeled RepRNA alone (Figure 1b; Replicon alone) provided a clear shift in FL-1, but no change in side (or forward) scatter profiles over diluent alone. When RepRNA was incorporated with chitosan during NGA formation, the RepRNA fluorescence signal clearly associated with the particles (Figure 1b; NGA (2 μg RNA) to NGA (16 μg RNA)).
Figure 1.
Replicon constructs and replicon interaction with chitosan. (a) Gene arrangement of the classical swine fever virus (CSFV) genome parent for the replicon RNA (RepRNA) constructs (CSFV parent), showing the Erns gene deletion for the ΔErns replicon. Insertion of the influenza virus hemagglutinin (HA) and nucleoprotein (NP) genes at the 3′ end of the Npro gene are shown as HA replicon and NP replicon (the luciferase RepRNA has the luciferase gene inserted at the same position). Insertion of the influenza virus HA at the 3′ end of the C gene is shown as C-HA-C replicon. The inserted EMCV IRES ensured reinitiation of translation of the RepRNA after the inserted influenza virus genes. (b) Encapsulation efficiency for RepRNA interacting with chitosan during the formation of the chitosan cores of the nanoparticles. The RepRNA was labeled with FITC. Chitosan-based nanoparticles (NGA) are detected by their side scatter (or forward scatter, data not shown) profiles in the Flow Cytometer; RepRNA is detected in the FL-1 channel. The signal level with diluent alone (MQ-water) or NGA alone set the gating for positive FL-1 signals. Different concentrations of RepRNA were associated with the NGA (from 2 to 16 μg RepRNA in 17 μl 0.1% TPP plus 250 μl chitosan, made up to 1 ml prior to analysis). (c) Physical characteristics of the NGA, alone or carrying oligoRNA or RepRNA cargoes, measured in water (the standard diluent for NGA production) or in Dulbecco's modified Eagle's medium (DMEM) (the medium in which the NGA were employed for interaction with dendritic cells (DCs)). Measurement of the hydrodynamic diameter (Z-average size, dHZ), polydispersity index (PDI) and surface charge (ζ-potential) was by dynamic light scattering at 25 °C with a scattering angle of 173°. (d) Capacity of chitosan to protect fluorochrome-labeled RNA probes from RNAse-dependent release of the fluorochrome, using the RNaseAlert Kit. Interaction of chitosan with the RNA in either water or Tris buffer was compared with RNA alone (in Tris buffer is shown; in water gave the same results) and RNA with TPP in water or Tris buffer. (e) Influence of chitosan on protecting the integrity of a Dy-781 labeled 50-mer RNA probe against RNase digestion, using the procedure described by Python et al.49 TPP, tripolyphosphate.
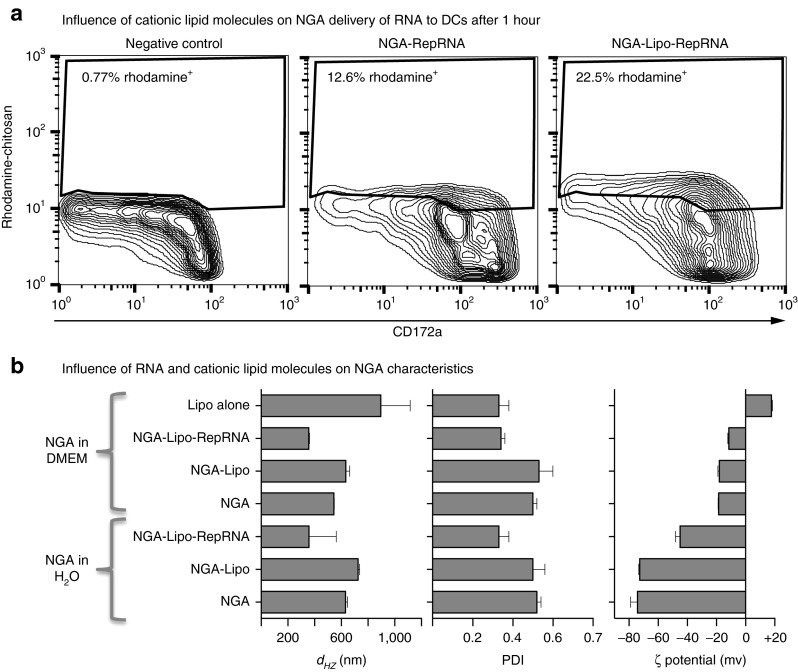
The influence of RepRNA on NGA size and ζ-potential was assessed, in comparison with oligoRNA. These were determined in water and Dulbecco's modified Eagle's medium (DMEM), the latter being more pertinent to interaction with DCs. Suspension in DMEM did modify the image of NGA obtained in water, as already reported25 (Figure 1c). In DMEM, RepRNA slightly increased particle dHZ and polydispersity index (PDI), while oligoRNA had little or variable effect. ζ-potential was unaffected by RepRNA, while oligoRNA led to a more negative ζ-potential.
Chitosan also protected labeled RNA probes against RNase (Figure 1d). This was confirmed with gel analysis of RNA integrity (Figure 1e). It was not possible to employ NGA due to the damage to the RNA cargo when attempting to release it for analysis, but the properties of chitosan were expected to be reflected in the NGA.
NGA-mediated delivery of RNA to DCs
Initial characterizations using fluorochrome-labeled oligoRNA (Dy781-O1-RNA) probes showed that RNA alone did not associate with DCs, but required NGA delivery (Figure 2a); lipofectamine served as a transfection control. NGA delivery to monocyte-derived DCs (MoDCs) was in a kinetic manner (Figure 2a). Variation in the signal showing delivery varied between experiments (Figure 2a compared with Figure 2b); this was noted using DCs prepared on different days from the same animals as well as cells from different blood donors (data not shown). Analyzing whole cell pellets in place of cell lysates improved visualization of delivery, particularly at later time points (Figure 2b), confirming the kinetic nature over a 24-hour period.
Figure 2.
Chitosan-based nanoparticle delivery to dendritic cells (DCs). The results shown are representative of 3 (microscopy) and 5–10 (RNA blots) individual experiments. (a) Left blot: NGA delivery of a Dy781-labeled oligoRNA probe to MoDCs in comparison with lipofectamine 2000 transfection and RNA alone—RNA measured in cell lysates from MoDCs treated with the NGA for different periods of time. Right blot: Similar to the left blot, but comparing cell lysates with supernatant medium from cells before lysis—kinetics of RNA association with MoDCs; lipofectamine and electroporation are used as transfection controls. (b) As in (a), but comparing the signal obtained from lysates with cell pellets prior to lysis. (c) Uptake of chitosan-based nanoparticles and delivered RNA by DCs. NGA interaction with MoDCs at 2 and 16 hours was observed by confocal microscopy using rhodamine labeled chitosan for generating the NGA. The pattern of interaction with the DCs is compared with that obtained using the chitosan cores of NGA (no alginate coating) and NGA-Lipo, in which lipofectamine 2000 was mixed with the chitosan prior to addition of the TPP to form the cores of the nanoparticles. The scale bar is 20 or 10 μm with the extreme right image in each row. (d) Comparison of NGA with chitosan cores and NGA-Lipo for the kinetics of RNA delivery to MoDCs, using the same experimental conditions for (b), except only cell pellet analysis is shown.
Modified NGA interaction with DCs
Lipofectamine 2000 (Lipo) was employed as a positive transfection control, but was also tested for its influence when incorporated together with chitosan during NGA formation (NGA-Lipo). Due to their more cationic nature, chitosan cores (without the alginate coating) were also assessed. Using labeled chitosan, NGA delivery was frequently observed as a vesicular distribution in MoDCs (Figure 2c). NGA-Lipo formulations, and particularly chitosan cores, aggregated upon interaction with MoDCs (Figure 2c), which proved somewhat detrimental to DC integrity for the cores, but not NGA-Lipo. Nevertheless, cells were observed at 16 hours with discrete vesicular inclusions, similar to those obtained with the NGA delivery.
Use of Dy781-O1-RNA cargoes confirmed that NGA, NGA-Lipo, and chitosan cores would deliver to MoDCs (Figures 2d). A more rapid delivery was apparent with the NGA-Lipo (observed after 1–2 hours), but by 16–24 hours, all three delivery systems provided similar results.
Comparative delivery of oligoRNA and RepRNA
NGA delivery of oligoRNA and RepRNA was compared using MoDCs and blood DCs. The latter offered additional information from the intensity of CD172a expressions—CD172ahi cells are dominated by monocytes, CD172alo cells are dominated by DCs,26,27 although DC subsets are found in both populations. Within 1 hour, similar percentages of the cells were positive for NGA-delivered RepRNA and oligoRNA (Figure 3a). Both CD172ahi and CD172alo cells were positive for RepRNA, whereas oligoRNA was primarily with CD172ahi cells. By 16 hours, the RepRNA distribution in both CD172ahi and CD172alo cells was confirmed, while oligoRNA remained with the CD172ahi population. Moreover, the percentage of oligoRNA+ cells was similar at 1 and 16 hours, whereas the percentage of RepRNA+ cells doubled, and the relative fluorescence signal increased (Figure 3a).
Figure 3.
Comparative delivery of replicon RNA (RepRNA) and oligoRNA to CD172a+ blood cells (monocytes plus dendritic cells (DCs)). The results shown are representative of three individual experiments. (a) NGA delivery of Dy490-RepRNA or Alexa488-oligoRNA to CD172a+ cells (4 μg/ml RNA prepared as in Figure 1b and 2a in Dulbecco's modified Eagle's medium (DMEM), added directly to the cells) after 1 or 16 hours incubation at 39 °C in DMEM supplemented with 10% (v/v) porcine serum to maintain cell viability. The y-axis shows the intensity of the labeled RNA signal, while the x-axis shows the intensity of the CD172a labeling (the blood DCs are mostly present in the CD172alo population, but the conditions to maintain cell viability will permit the monocytes to begin differentiation). The gated population shows the RNA+ cells, based on a negative gate created with the negative (NGA alone) sample. (b) As in (a), using different concentrations of fluorescein (FITC)-labeled RepRNA delivered to the cells by NGA as in Figure 1b, prepared in DMEM; the amount of RepRNA shown is the concentration per milliliter when added to the cells.
Modified NGA delivery efficiency for RepRNA
NGA were prepared with 4 μg as well as the 2 μg FITC-labeled RepRNA most often used, to facilitate addition of different amounts of NGA-associated RepRNA to a fixed number (2 × 105) of CD172a+ cells (Figure 3b). Detectable RepRNA+ cells increased in a RepRNA concentration-dependent manner, with maximum levels observed using 4 μg RepRNA. This concentration-dependent delivery appeared to be saturable, but there was a distribution shift with the higher concentrations of RepRNA associating with CD172alo cells more than the lower concentrations.
NGA-Lipo formulations were compared with NGA for Dy490-UTP-labeled RepRNA delivery, again using CD172a+ blood cells. Although RNA delivery was enhanced by NGA-Lipo (as observed with oligoRNA delivery; Figure 2d), this was most obvious when the NGA delivery level was low (0.2 μg/RepRNA; Figure 4a). This was not due to the presence of free Lipo in the preparations—Lipo alone has higher dHZ than NGA-Lipo, and positive rather than negative ζ-potential (Figure 4b). Moreover, association of Lipo in the NGA formulation only slightly increased dHZ with no influence on PDI or ζ-potential in DMEM (Figure 4b). The added presence of RepRNA did render a less negative ζ-potential on the NGA-Lipo, and also reduced the dHZ and PDI (Figure 4b).
Figure 4.
Influence of cationic lipid molecules on NGA delivery of RNA related to the NGA characteristics. (a) As in Figure 3a, but showing the enhanced level of Dy-490 labeled replicon RNA (RepRNA) delivery after 1 hour at 39 °C offered by NGA-Lipo when NGA delivery provides a low percentage of RNA+ cells (2 μg/ml RepRNA prepared as in Figure 1b, diluted 1:10 to have 0.2 μg/ml when added to the cells). (b) As in Figure 1c, but comparing NGA and NGA-Lipo carrying RepRNA, as well as Lipofectamine 2000 alone.
Intracellular RepRNA delivery
RepRNA translation requires cytosolic translocation following delivery to cells. Consequently, the CD172a-labeling was employed as a DC surface indicator to characterize intracellular uptake of NGA-delivered RepRNA. Within 1 hour at 39 °C, the RepRNA signal (red) was observed on the cytosolic side of the CD172a-labeled surface (blue) (Figure 5a). This contrasted with incubation at 4 °C—RepRNA remained closely associated with the CD172a-labeled surface (Figure 5a). The confocal microscopy also showed stronger signals and more positive cells for RepRNA than oligoRNA (Figure 5a), and the RepRNA signal increased from 1 to 18 hours.
Figure 5.
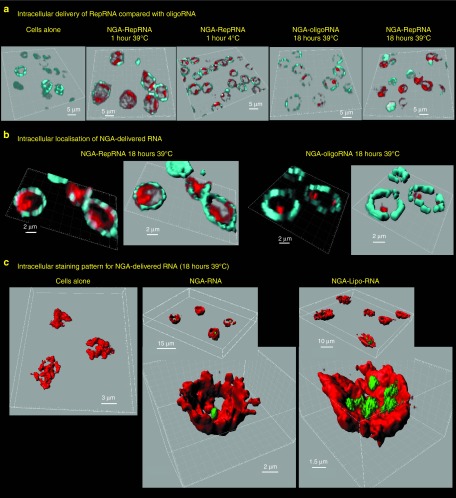
Intracellular delivery of replicon RNA (RepRNA) and oligoRNA by NGA and NGA-Lipo. (a) Confocal microscopy images from 3D scans of narrow sections through the centre of blood CD172a+ cells. NGA was used to deliver rhodamine-labeled RepRNA or Alexa488-labeled oligoRNA, prepared as in Figure 1b to give 2 μg/ml when added to the cells; incubations were 1 hour at 4 °C or 1 and 18 hours at 39 °C as shown. The cell surface was labeled with anti-CD172a antibody. Cell surface labeling is colored turquoise, while the RNA signal is colored red, to facilitate comparative observations. All micrographs are 3D Blend images with threshold subtraction and gamma correction set as in the “Cells alone” control; the 3D mages have been turned to provide a comparative view through the cells. Scale bars are all 5 μm. (b) As in (a), for 18 hours at 39 °C, but showing a zoom together with resolution format to provide optimum voxel sizes for the fluorochromes employed. maximum intensity projection and Blend versions of the 3D images are provided for both RepRNA and oligoRNA delivery observed after 18 hours incubation. In the Blend 3D images, the surface module of the Imaris software was employed with the CD172a labeling to create an enhanced depiction of the cell surface in this narrow section through the centre of the cells. Scale bars are 2 μm. (c) Similar to (b) 18 hours at 39 °C, but using 0.2 μg RepRNA/ml when added to the cells, with which NGA delivery was weak; this allowed observation of an enhanced delivery by NGA-Lipo. In this experiment, the cell surface is colored red, while the RepRNA is green. Again, all settings are from those set with the Cells alone control, and the surface labeling was enhanced using the surface module of the Imaris software. The scale bars are 3 μm for “cells alone”, 2 μm for “NGA-RNA” with 15 μm for the insert, and 1.5 μm for “NGA-Lipo-RNA” with 10 μm for the insert.
Imaris software Surface Module was employed to enhance the CD172a-defined cell surfaces, to confirm the intracellular localization of delivered RNA (Figure 5b). The bright (red) staining was reminiscent of vesicular structures, but the form and edges of those with RepRNA were not so readily discernible, often “coalescing”. Moreover, an additional more diffuse (and weaker) staining pattern of RepRNA was observed, closely associated with the more densely staining structures, reminiscent of cytosolic translocation events.28,29 Accordingly, images were acquired by scanning through a larger depth of the cell (Figure 5c). These experiments also compared NGA and NGA-Lipo delivery (0.2 μg/RepRNA to facilitate observation of NGA-Lipo enhanced delivery). RepRNA+ inclusions (green) together with associated diffuse staining were again observed (Figure 5c). These events were most numerous and visible with the NGA-Lipo delivery of 0.2 μg/RepRNA (Figure 5c).
DC detection of and response to NGA formulations
Considering that DCs were capable of both interacting with and internalizing NGA carrying RepRNA, analyses were performed to determine if the cells were capable of detecting the presence of the NGA. The DCs showed no apparent modulation in the presence of RepRNA free of any particulate formulation (data not shown); nor did the cells respond to that RNA, which reflects their poor capacity for internalizing free RepRNA. Accordingly, migration assays were performed as described in Materials and Methods to determine if DCs actually detect the presence of the chitosan nanoparticulate vehicles. In order to prevent random interaction between the cells and the particles, NGA and DC were separated by transwell filters; only the cells that traversed the filters into the NGA-containing compartment were measured. The results (Figure 6a) are shown as the number of cells migrating and the migration index (number of cells migrating to the NGA formulations relative to the medium control—random migration). Both the complete CD172+ cell population (monocytes plus DCs) and the CD14− subset (CD14− DCs) migrated toward the NGA formulations. This was often more prominent than migration toward the LPS ligand for TLR4 serving as a positive control. The additional presence of the RepRNA (NGA RepRNA) or RepRNA and cationic lipid (NGA-Lipo RepRNA) in the nanoparticle structure did not interfere with the migration characteristics.
Figure 6.
Dendritic cell sensing of NGA formulations. (a) CD172a+ sorted cells (0.5 × 106 cells/well; prelabeled with anti-CD172a and anti-CD14 antibodies) were placed in a 3 µm filter transwell inserts, and added into wells of 24-well companion plates preseeded with the NGA formulations (10% v/v in phenol red-free DMEM) or controls as shown on the x-axis. Following incubation at 39 °C for 3 hours, the cells which had migrated through the filters into the chamber containing the NGA formulations were assessed by flow cytometry. Quantification of the migrated cells was performed by counting a reference of 5000 CountBright beads to obtain the number of migrating cells/cm3. This was related to the number of migrating cells obtained with the medium control to give the migration index. (b) CD172a+ sorted cells were incubated either alone (“negative”), with NGA (“NGA alone”), with NGA carrying unlabeled or labeled RepRNA (“NGA-RepRNA unlabeled”; “NGA-RepRNA FITC-labeled”), or with NGA-Lipo carrying labeled RepRNA (“NGA-Lipo RepRNA”). Following incubation in DMEM/1% (v/v) porcine serum for 2 or 16 hours at 39 °C, the forward and side scatter profiles (FSC, SSC) of the cells was determined by flow cytometry. The results show an example for the 16 hours incubation time point from 10 separate experiments; similar results were obtained with the 2 hours incubation time point.
In contrast to this apparent ability of the DCs (and monocytes) to detect the presence of the NGA formulations, there was no detectable activation of the cells. This was monitored in terms of flow cytometric forward and side scatter properties (FSC versus SSC; Figure 6b), as well as NFκB nuclear translocation and IL-6 induction (data not shown).
Translation of delivered RepRNA in vitro
With the images in Figure 5 suggesting cytosolic translocation,28,29 essential for RepRNA translation, nanoparticle delivery of RepRNA encoding luciferase (RepLuc) was assessed. For these analyses, a delivery of 0.2 μg RepRNA was again employed, to facilitate detection of enhanced delivery by NGA-Lipo particles (see Figures 4a and 5c).
Not unexpectedly, VRP delivery to MoDCs provided the strongest luciferase signal (Figure 7a)—VRPs, like the virus, target DCs for replication.22 Analysis of early kinetics showed that NGA-Lipo and chitosan core formulations provided a 24-hour readout in MoDCs, yielding similar luciferase translation kinetics from as early as 2 hours postdelivery (Figure 7a, left graph). These luciferase levels continued to increase by 24 hours (Figure 7a, left graph), although not surprisingly they did not reach those obtained with VRPs. NGA delivery with this low 0.2 μg RepRNA did not provide luciferase readout within 24 hours, in agreement with the poor signals obtained with labeled RNA (0.2 μg RepRNA) delivery (Figures 3b and 4a).
Figure 7.
NGA delivers replicon RNA (RepRNA) for translation in vitro. (a) Kinetics of luciferase-encoding RepRNA (RepLuc) translation in MoDCs, compared with RepRNA delivered as virus replicon particles (VRPs) or transfected (lipofectamine 2000). NGA were compared with modified NGA (NGA-Lipo with lipofectamine 2000) and chitosan cores (no alginate coating). A final concentration of 0.2 μg RepRNA/ml added to the cells was employed with the NGAs; VRPs were used at three infectious VRPs per cell. Translation of the luciferase gene was determined during the first 24 hours (left graph) as well as after 24, 48, and 96 hours (right histogram) as described.22 A representative of five individual experiments is shown. (b) Concentration dependency of delivered RepRNA translation measured in terms of the replicon NS3 expression, to control for continued translation beyond the foreign gene insert. Similar methods to (a) were employed. NS3 expression was monitored using anti-NS3 monoclonal antibody, after fixing and permeabilising cells 72 hours postdelivery. NS3 detection was by flow cytometry (y-axis) with labeling of cell surface CD172a (x-axis) as in Figure 3b. Positive controls were electroporation (RepRNA electro), lipofection (RepRNA lipo), and VRP delivery (VRP). A representative of three individual experiments is shown.
Looking beyond 24 hours to determine durability of translation, indicative of replication (Sutter et al., personal communication), VRPs again provided the strongest signal, maintained over 96 hours as evidence of replication (Figure 7a, right histogram). Lipofection gave lower and variable (among experiments) readouts (data not shown), which were not maintained over 96 hours (Figure 7a, right histogram). NGA delivery was more consistent than lipofection across experiments (data not shown); a readout was noted at 48 hours and maintained at 96 hours (Figure 7a, right histogram). NGA-Lipo delivery yielded clear luciferase signals from 24 hours, as noted for the early kinetics; these were also maintained over 96 hours (Figure 7a, right histogram). Interestingly, chitosan core delivery again provided moderate luciferase readouts, from 24 hours (Figure 7a; no 96 hours sample was available due to core-induced cell damage).
RepRNA delivery was also assessed for translation of the NS3 viral protein necessary for RepRNA replication. Both MoDCs and blood-derived DC/monocytes were employed—Figure 7b shows results with blood cells. NGA delivery yielded an NS3 readout; in agreement with the results on RNA delivery (Figure 3b), this was dependent on the concentration of RNA delivered. The 0.2 μg/ml again did not provide a detectable readout; efficiency of translation was discernible in a concentration-dependent manner from 0.4 μg/ml, reaching maximum at 2–8 μg/ml delivered RepRNA. The NS3 translation from NGA-delivered RepRNA was on a par to that for VRP when high levels of RepRNA were delivered, but more efficient than lipofection.
Translation of delivered RepRNA in vivo
While the in vitro data clearly demonstrated that nanoparticulate delivery of RepRNA to DCs led to RepRNA translation, it was considered important to confirm that such translation was pertinent within an in vivo environment. For this purpose, we considered that induction of antibody would prove an informative readout, due to the fact that the B-lymphocytes require intact antigen rather than processed peptides (as for the T lymphocytes). Moreover, it is most difficult to accurately detect RepRNA translation directly in DCs in vivo, possibly due to the low numbers of DCs involved. Induction of antibody provided the more demanding test for the RepRNA, reflecting that the RNA did indeed successfully translate its encoded foreign genes as intact antigen, which in turn was released to interact with the B-lymphocytes. Thus, the in vivo analyses focused on antibody induction (Figure 8a).
Figure 8.
NGA delivers replicon RNA (RepRNA) for translation in vivo. (a) Mice and rabbits were injected subcutaneously at 0, 14, and 28 days as described in Materials and Methods (left panel). RepRNA (0.2 μg HA-RepRNA in 50 μl plus 0.2 μg NP-RepRNA in 50 μl) was delivered in NGA, and compared with the RepRNA alone (Rep) or virus replicon particles (VRP) delivery (105.7 VRP units for each RepRNA in 50 μl). Mice received 50 μl of each RepRNA mixed together, while rabbits received twice this dosage; all vaccines were adjuvanted in PEGylated MALP-2 adjuvant. Serum samples were assessed for anti-hemagglutinin (HA) and anti-nucleoprotein (NP) antibody by enzyme-linked immunosorbent assay (ELISA) at 42 days after the first injection. Administration of NGA-RepRNA was repeated, but using the C-HA-C-RepRNA and NP-RepRNA, as well as comparing with delivery using chitosan cores and NGA-Lipo, prepared as for (Figure 7a. Mice and rabbits were injected at days 0, 35, and 62 (arrows). Anti-HA and anti-NP antibody titers were estimated by ELISA at the times shown. (b) The immunization of mice as shown in (a) was repeated, using NGA alone, NGA-RepRNA, or NGA-Lipo-RepRNA. Splenocytes were prepared and used in vitro as either unstimulated controls or for restimulation with recombinant HA or NP as described in Materials and Methods. Multiple labeling was employed to distinguish CD4+ from CD8+ T lymphocytes gating on memory subsets, as described in Materials and Methods. T-lymphocyte activation was determined with respect to CD25 upregulation or reduction of the CFSE label in prelabeled cells (indicative of cell division). The CD25 upregulation was measured in terms of mean fluorescence intensity change, relative to that obtained with unstimulated splenocytes, to obtain a stimulation index (for the unstimulated splenocytes, this was set as 1). Cell proliferation was measured in terms of reduced CFSE signal as % cells gated as CSFElow.
This confirmation of the in vitro translation was assessed in vivo by injecting both mice and rabbits with RepRNA encoding influenza virus hemagglutinin (HA) (H5) or nucleoprotein (NP) (Figure 1; “HA replicon”, “NP replicon”). Comparators were truly “naked” RepRNA and VRPs carrying the same replicon. The “naked” RepRNA never provided any readout, as exemplified in Figure 8a (left panel, “Rep”). VRPs induced anti-HA and anti-NP responses in both species (Figure 8a, left panel, “VRP”). NGA delivery in mice provided no anti-HA response, but a strong anti-NP response, while in rabbits NGA delivered RepRNA induced both anti-HA and anti-NP antibodies (Figure 8a, left panel, “NGA”). Overall, anti-NP responses were stronger than anti-HA, relating to in vitro levels of HA and NP translation by these HA-RepRNA and NP-RepRNA (Suter et al, unpublished data).
The HA gene of the HA-RepRNA (Figure 1, “HA replicon”) was inserted at the same position as the NP gene. Modified insertion of the HA gene to be downstream of the C gene (Figure 1, “C-HA-C replicon”) was found to give better translation of the HA in vitro (Suter et al, unpublished data). Thus, the in vivo translation experiments were repeated using this C-HA-C RepRNA. In addition, due to the results seen in Figure 7a, NGA delivery of 0.2-μg RepRNA was compared with chitosan cores and NGA-Lipo. Now, NGA delivery led to both anti-HA and anti-NP responses in mice and rabbits (Figure 8a, second panel “NGA RepRNA”). The RepRNA translation was able to induce responses within 7 days, which clearly benefited from a second injection for translation in vivo. With the exception of only the occasional animal, the titers were reasonably well clustered for each time point.
Despite the in vitro aggregation problem with chitosan core-RepRNA delivery, this also promoted translation in vivo (Figure 8a, third panel “Chitosan Cores RepRNA”), as had been witnessed in vitro while the cultures remained viable. While the titers from individuals were reasonably well clustered for each time point, there was more variation for the anti-HA readouts.
NGA-Lipo delivery did not always display the advantages observed in vitro. With mice, the translation in vivo did not appear to be superior to that for NGA (Figure 8a, right panel “NGA-Lipo RepRNA”). In contrast, NGA-Lipo delivery did promote more efficient translation in rabbits, particularly notable early after the first injection in terms of increased titers. This was observed with delivery of both the C-HA-C RepRNA and NP-RepRNA, so was not particular to the replicon construct, but more to the delivery system.
In order to confirm these observations on antibody induction, splenocytes were taken from mice at the end of the experiment to assess T-lymphocyte activity using in vitro restimulation with HA and NP antigen (Figure 8b). Both CD25 expression and reduced carboxyfluorescein diacetate succinimidyl ester (CFSE) signal from preloaded cells (due to cell division distributing the dye to daughter cells) were employed. Multiparameter flow cytometry permitted gating on CD3+ CD44high CD62Llow memory T-lymphocytes and CD3+, CD44low, CD62Lhigh naive T-lymphocytes within both the CD4+ Th-lymphocyte and CD8+ Tc-lymphocyte populations. Proliferating CFSElow cells were scored as a percentage of the total cells within that gate. For cells upregulating CD25 expression, this was measured as mean fluorescence intensity (MFI), from which a stimulation index was calculated relative to the MFI obtained with unstimulated cells.
Cells from certain nonvaccinated individuals have been found to show a degree of response (possibly aspecific or through naive cells) in vitro. Accordingly, the control animals receiving NGA alone (did not induce detectable antibody titers; data not shown) were employed as reference points for the assays—unstimulated, HA and NP restimulations (Figure 8b; “NGA alone”). The cells from most animals in this group did not respond in vitro, and the few that showed increased CD25 expression generally gave similar responses when unstimulated. Lack of HA and NP recognition was even more apparent with CFSE measurement—any apparent increase in CFSElow cells was similar to the levels obtained when unstimulated.
Splenocytes from animals receiving the NGA RepRNA (Figure 8b; “NGA-RepRNA”) gave clear responses to the HA and NP restimulation. This was most obvious with the CD25 stimulation index. Interestingly, the CD8+ population proved to be the most responsive, particularly to NP restimulation. Similar images of responsiveness to the HA and NP restimulations (relative to the unstimulated cultures) were obtained with cells from the NGA-Lipo RepRNA-treated animals (Figure 8b; “NGA-Lipo RepRNA”); the anti-NP response measured in terms of CFSElow cells was inferior to that observed with cells from the NGA RepRNA-treated animals (Figure 8b; “NGA RepRNA”). Nevertheless, the combined observations on CD25 expression and CFSElow cells confirmed that the T-lymphocytes in the mice had also responded to HA and NP antigen, which could only have arisen following RepRNA translation in vivo.
Discussion
Application of biodegradable, nanoparticulate delivery vehicles has been widely employed with subunit and recombinant antigens as well as protein-based drugs.4,30,31,32 However, such delivery is limited by the quantity of antigen or drug transported by the delivery vehicle, and the duration of that materiel within the targeted cell.4 Due to its replicative nature, self-amplifying RepRNA offers high potential for increasing antigen load within cells and maintaining prolonged antigen presence.4,14,15,16,17,20 Consequently, DCs would prove an important target for RepRNA delivery.
While alphavirus replicons have been studied in depth during the past 20 years,15,16,17,18 most reports have employed cell lines. Their cytopathogenic nature is also problematic, precluding long-term retention of the replicon and translated antigen by DCs, impacting on the duration of antigen production and immune stimulation. This presents a difficulty considering the characteristics of DCs—slow processing and retention of antigen for prolonged interaction with the adaptive immune system. Many of the critical issues concerning the manner of RepRNA delivery and interaction with DCs have not been answered by the most frequently employed mode of replicon delivery—replicons packaged as VRPs.15,16,17,19,20,22,24 This packaging requires complementing cells to supply the gene deleted from the replicon, but VRPs may preferentially target cells other than DCs. VRP application must also counter problems of species restriction and anti-VRP immunity. Synthetic particulate delivery avoids such drawbacks associated with VRPs, offering advantages for RepRNA delivery.4,13,14 Application with noncytopathogenic RepRNA would also provide conditions more appropriate to DC requirements, as described in the application of synthetic particulate delivery vehicles for RepRNA delivery employing a noncytopathogenic RepRNA derived from classical swine fever virus.4,14
There is no direct evidence to date that synthetic, nanoparticulate delivery vehicle will target large molecules such as RepRNA to DCs. These molecules have particular requirements for their delivery, leading into endocytic processes and routes promoting cytosolic translocation;4 only then can the RNA access the appropriate machinery of the cell for translation and replication. Such processes are unrelated to those required for delivery of antigen, DNA, and RNA interference. Accordingly, we sought to define how synthetic chitosan-based nanoparticle (NGA) delivery vehicles carrying RepRNA interact with DCs, to ascertain the prospects for cytosolic delivery of self-amplifying RepRNA leading to translation.
An initial aim was to determine if RepRNA and chitosan-nanoparticles were compatible. RepRNA associated efficiently with NGA, the chitosan offering protection against RNAse. The association had little influence on the NGA physical properties. Hydrodynamic diameter, PDI, and ζ-potential of the NGA were measured in a physiological buffer (DMEM), more relevant to the physiological environments in which DCs would interact. The observations that the NGA characteristics were different in a physiological buffer compared to water relates to similar observations reported by Schütz et al.25 This is the first description defining the interaction of RepRNA with synthetic nanoparticles. Moreover, the interaction did not require the addition of protein or lipids, which can pose problems due to their immunogenic capacities.
It was important to determine if this efficient association of RepRNA with NGA particles would promote interaction with DCs. This was essential; free RepRNA was shown in several experiments to be incapable of interacting directly with DCs, relating to similar deficiencies observed with mRNA.33,34,35 RepRNA delivery unequivocally required the application of the NGA particles. Both the particles and the RNA cargoes were visualized within the DCs as vesicular-like structures. Although this was considered to be a consequence of the high capacity for the alginate coat to interact with DCs (unpublished data), the alginate was not obligatory. Chitosan cores (no alginate coating) also interacted efficiently with the DCs. However, the cores showed high levels of aggregation detrimental to both uptake by the DCs and cell integrity, which may relate to amine protonation on chitosan requiring reduced pH.36 With the cores stabilized at physiological pH by the alginate coating of NGA particles, this capacity of chitosan for amine protonation would become pertinent following endocytosis. As the endocytic vesicles containing the NGA particles interacted with endosomes, vacuolar H+-ATPases would initiate the proton pump for acidification of the vesicles. Under such conditions, the amines of the chitosan may become more available for protonation, which in turn would create a “proton sponge” effect.4,13,37,38 Such effects destabilize the acidifying endocytic vesicles, in turn facilitating cytosolic translocation of the vesicular contents, which is an essential step for the RepRNA to reach the site for translation.
In this context of NGA protonation destabilizing the endocytic vesicles, we applied additional cationic components within the NGA structure to determine any potential benefit for the RepRNA delivery to DCs. Lipofectamine 2000 (Lipo) has been described by the manufacture as a cationic lipid-based method for nucleic acid delivery. Accordingly, the influence of Lipo on RepRNA delivery was assessed using NGA particles, in which the Lipo was incorporated with chitosan during NGA formulation (NGA-Lipo). This increased the aggregation of the particles, but was less detrimental to the cells than the cores. Moreover, RNA was again delivered into DC vesicular structures. Such effects were only observed when Lipo was added with the chitosan during incorporation of RNA into the particles. If the Lipo were added with the RNA rather than chitosan, or together with alginate during coating, it had no influence; nor did admixing preformed NGA carrying RepRNA with Lipo.
Further analyses employing both MoDC and CD172a-sorted blood cells—CD172ahi cells (dominated by monocytes) and CD172lo cells (majority of DCs)26,27—uncovered differences in RepRNA and oligoRNA delivery. The latter preferentially targeted CD172ahi cells, whereas RepRNA delivery associated with both CD172hi and CD172lo cells. It is not clear why there should be this difference, because cell targeting was mediated by the NGA (free RNA did not interact discernibly with DCs or monocytes). Differences in recycling within the cells of delivered oligoRNA and RepRNA might be an explanation. If the CD172ahi cells initially interacted more efficiently with NGA before the CD172lo cells, a different recycling for RepRNA could explain their increased appearance in CD172lo cells. It is possible that the monocytes were retaining both forms of RNA, whereas the DCs showed higher capacity for retaining the RepRNA. With the DCs being the more efficient antigen-presenting cell, and tending more to retain material, such characteristics of RepRNA delivery could prove beneficial for translation. Indeed, the RepRNA+ signal, but not oligoRNA+ signal, increased with time. This is also consequential for the important differences in the mode and intracellular sites of function for RepRNA and oligoRNA such as siRNA. RepRNA has an obligate requirement to interact with the ribosomal translation machinery; oligoRNA would interact with RNA-detecting entities such as TLR3/7 and helicases or the RNA interference pathway components (for siRNA). Clearly, it cannot be assumed that effective delivery of oligoRNA such as siRNA will be reproduced with RepRNA delivery—these different forms of RNA have their own particular requirements and specialized cell compartments for appropriate interaction leading to functionality.
Preliminary studies on RNA association with the NGA particles had provided a recommended dosage of 0.2 μg–0.4 μg/ml RNA (2–4 μg RNA plus 250 μl 0.1% w/v chitosan, in 1 ml, diluted 1:10) to ensure stability (P Käuper and Medipol SA, personal communication). However, this yielded variable results for RNA delivery when assessed with different preparations of DCs. Deviation in the functional characteristics of different DC isolations, from the same or different donors, may have contributed to these variations. Nevertheless, low concentration RNA cargoes did facilitate detection of enhanced delivery by NGA-Lipo particles. The lower dHZ and PDI of NGA-Lipo carrying RepRNA may have had an influence—this was not due to free Lipo, which showed higher dHZ and positive ζ-potential. DCs do prefer smaller particles,39,40,41 and the surface charge of particles can also influence interaction with DCs.39 These studies identified another important contributory factor, which was the amount of RepRNA delivered by NGA. Analyses on increasing the amounts of RepRNA cargo demonstrated that stable NGA particles with their cargo could be generated. Under these conditions, 4 μg/ml RNA was optimal for observing delivery to the cells (and indeed translation therein), whereby the NGA-Lipo no longer offered observable advantages.
Following delivery, it was essential that DCs internalized RepRNA. Labeling CD172a to define the DC surface confirmed that NGA-delivered RepRNA (and oligoRNA) was internalized, in a time- and temperature-dependent manner. RepRNA accumulated in brightly stained vesicular structures, typical of endocytic elements such as macropinosomes.1,2,3,4,42 As time progressed, the appearance of an additional weaker RepRNA signal adjacent to these bright structures—particularly notable using NGA-Lipo with 0.2 μg/ml RNA cargo—was suggestive of cytosolic translocation, as reported for oligoRNA delivery to Hela cells.28,29 Although these authors did not study DCs or replicons, they described a similar accumulation in vesicular compartments over time. Adjacent weaker signals were confirmed to indicate cytosolic translocation.
For RepRNA, observable cytosolic translocation alone is inadequate. The translocation must lead to RepRNA translation—this contrasts with siRNA, which must interact with different intracellular compartments involved in RNA interference. Not surprisingly, the VRP-positive controls were the most effective at promoting translation. VRPs possess the same envelope as the virus, and therefore mimic virus infection; the efficiency comes from VRPs using the same cell receptors and efficient mechanism for cytosolic genome delivery as the virus.43 The need for VRPs can be seen by the incapacity of free RepRNA to interact with or translate in DCs. This fits with the RNase sensitivity of the replicon, and the difficulties that RNA has for crossing cell membranes.13,33,35 Synthetic delivery vehicles were not expected to provide this same efficiency—they lack the VRP capacity for interacting with cell receptors promoting the viral cytosolic translocation mechanism. Nevertheless, NGA and NGA-Lipo delivery, as well as chitosan cores, did promote translation in DCs. Translation by NGA delivered RepRNA was not observed until after 24 hours, whereas NGA-Lipo and chitosan core delivered RepRNA showed an initial kinetics of translation beginning by 2 hours and increasing over 24 hours. This was tested using the lower 0.2 μg RNA/ml with which NGA-Lipo-enhanced delivery was observable. It was interesting that the difference in translation kinetics between NGA delivery and NGA-Lipo delivery related to differences observed in microscopic detection of the weak signals indicative of cytosolic translocation. Importantly, the translation was maintained over the 96-hour period of observation, which reflects replication of the replicon (Suter et al, unpublished data).
This successful NGA delivery of RepRNA to DCs leading to translation was confirmed in vivo. Again, naked RepRNA was incapable of translating, and therefore, no immune response against the encoded antigens could be induced. Considering the delivery vehicles, it was interesting that chitosan cores were as efficient as the NGA for delivery RepRNA for translation in vivo. In addition, NGA-Lipo delivery did not display the same capacity for enhancement over NGA delivery observed in vitro. This may reflect a question of detectability with the in vitro observations—the lower 0.2 μg RNA/ml payload was employed in vivo, for relating to the NGA-Lipo-enhanced delivery observable in vitro. Nevertheless, the in vitro translation studies had demonstrated that the increased efficiency of delivery by NGA-Lipo was reflected in a more rapid translation, and the NGA delivered RepRNA did eventually translate to the same levels as the NGA-Lipo-delivered material (and also the chitosan core delivered RepRNA). It therefore seems likely that all three forms of the delivery vehicle were efficient at delivering the RepRNA for translation. Although there was a difference in the initial kinetics of translation, the production of the encoded antigens reached similar immunogenic levels, as witnessed by the in vivo readout.
To date, synthetic particle delivery of RNA has focused on siRNA and mRNA. There are no reports describing RepRNA interaction with DCs. Our present work characterized for the first time chitosan-based nanoparticulate vehicle delivery of self-amplifying RepRNA to DCs, promoting RepRNA translation in vitro and in vivo. The RNase-sensitive RepRNA is protected by the delivery vehicle, which in turn promotes intracellular delivery to the DCs, thus overcoming the inability of “naked” RepRNA to survive in biological environments and cross the cell membrane barrier. We are currently defining how targeting different DC receptors influences the efficiency of this delivery by synthetic, biodegradable nanoparticulate vehicles, relating to the endocytic processes and cytosolic translocation of the RepRNA for successful translation.
Materials and methods
Chemicals. Crustacean shell-origin Chitosan (low-viscosity chitosan, Primex, Siglufjordur, Iceland) was kindly provided purified through steps of acidic dissolution, 0.1 μm filtration, precipitation and dialysis by Medipol (Medipol SA, Lausanne, Switzerland). Pentasodium triphosphate, purum p.a. > 98% (TPP) was purchased from Sigma-Aldrich (Steinheim, Germany). Sodium alginate (Na-alg, Keltone LVCR, ISP, San Diego, CA) was kindly provided purified by Medipol SA. RNAse-free water was purchased from Ambion (Life Technologies Europe B.V., Zug, Switzerland). The chitosans as well as 0.1% (w/v) solutions of TPP and 0.1% (w/v) solutions of Na-alg were prepared under sterile conditions, then sterile filtered through hydrophilic membranes (0.2 μm, Minisart type, Sartorius AG, Gottingen, Germany). A batch of the chitosan was fluorescently labeled (1:50) with carboxyrhodamine (5(6) carboxytetramethyrhodamine, Fluka, Buchs, Switzerland) using standard carbodiimide conjugation.
Self-replicating RepRNA constructs. The self-replicating RepRNA is derived from full-length cDNA clone pA187-1 (ref. 44) from the noncytopathogenic classical swine fever virus. The cDNA was engineered to lack genes encoding viral structural proteins, but carrying NotI endonuclease restriction sites facilitating introduction of foreign genes of interest.22,24 We have generated RepRNA-encoding luciferase,22 as well as the HA and NP proteins of influenza virus using the same techniques. The constructs used on the present work are shown in Figure 1a, with the full classical swine fever virus genomic sequence for comparison. These are derived from cDNA clones using the published sequences of the full-length cDNA clone pA187-1 (ref. 14,44). Viral cDNA lacking the Erns gene is used for the replicon (Figure 1); bicistronic viral cDNA is used. The first cistron contains the Npro gene fused in frame to the foreign gene (e.g., influenza virus NP or HA gene) followed by the encephalomyocarditis virus internal ribosomal entry site (EMCV IRES) that initiates translation of the remaining viral polyprotein (C to NS5B).14,22 From this replicon, both Npro and the foreign gene are expressed in their authentic form. The activities of Npro, autoprotease and inhibition of type 1 IFN induction or production, are maintained.
For generating the RepRNA packaged into viral particles (VRPs), the defective replicon genomes are electroporated into 5 × 106 SK-6 cells expressing Erns (SK6 (Erns)) (refs. 22, 24). Following incubation at 37 °C for 48 hours, the VRPs produced are harvested by freezing and thawing of the cultures. The titer of VRPs is measured by infecting SK-6 cells with different dilutions of the VRP preparation, removing the inoculum after 1 hour, culturing for 48 hours at 37 °C, and the number of cells expressing the RepRNA-encoded antigens enumerated.
Nanogel preparation. Chitosan-TPP nanogel cores (“chitosan cores”) were prepared by the ionic gelation of low-viscous chitosan with TPP, according to Calvo et al. Briefly, one volume of 0.1% (w/v) TPP was added drop-wise under constant stirring into nine volumes of 0.1% (w/v) chitosan (e.g., 100 μl + 900 μl), resulting in spontaneous chitosan-gel formation. The pH was maintained under pH 4 by adding 0.1 N HCl, and the particles thus obtained were stirred for at least 1 hour.
Empty or RNA-loaded chitosan cores (see below) were diluted 1:1 with RNAse-free water, then added drop-wise to an equal volume of 0.15% (w/v) aqueous Na-alg solution under strong agitation. The pH was closely monitored and adjusted to pH 7.0–7.3 with 0.1 mol/l NaOH to generate the final nanogel product. Formulations were stirred for at least 1 hour; then stored at 4 °C overnight prior to use.
RNA encapsulation into chitosan particles. For oligoRNA-loaded particles, either oligoRNA-Alexa488 (Qiagen AllStars NegsiRNA AF488, Hombrechtikon, Switzerland) or Dy781-O1-RNA (Microsynth, Balgach, Switzerland) were added to the TPP solution prior to nanogel formation. The oligoRNA-Alexa488 was dissolved at 25 pmol/ml in the 0.1% (w/v) TPP solution and the Dy781-O1-RNA at 120 pmol/ml. The process of RNA/TPP solution addition to the chitosan was held under the same conditions as described above, with the ratios as (for example) 16 μl plus 34 μl TPP added to 450 μl chitosan, followed by 500 μl H2O.
For RepRNA-loaded particles, either RepRNA labeled with fluorescein or rhodamine Mirus labeling kits (LabForce AG, Nunningen, Switzerland) or RepRNA labeled by incorporating Dy490-UTP (Dyomics GMBH, Jena, Germany) during synthesis were employed. Studies on the translation of the RepRNA after nanogel delivery employed unlabeled RepRNA to ensure that the labeling had no influence on RepRNA translation efficiency. The various forms of the RepRNA were added to the TPP solution prior to nanogel formation, similar to that described for the oligoRNA above (e.g., 2 μg RepRNA in 16 μl plus 34 μl TPP added to 450 μl chitosan, followed by 500 μl H2O). The quantity of RepRNA added to the chitosan allowed for a concentration of 2 μg RepRNA per ml of the final NGA preparation prior to the 1:10 dilution for adding to the cells (unless otherwise stated in the Results section).
Modified nanogel preparation and RNA encapsulation. When particles carrying Lipofectamine 2000 (Life Technologies) were to be prepared (NGA-Lipo), the above method for nanogel preparation was employed, except that the lipofectamine was mixed with the chitosan prior to addition of the TPP (e.g., 50 μl TPP added to 434 μl chitosan plus 16 μl lipofectamine). When RNA was incorporated, this was again applied with the TPP (e.g., 16 μl RepRNA + 34 μl TPP added to 450 μl chitosan/Lipo (434 μl chitosan + 16 μl Lipofectamine).
Nanogel characterization. Hydrodynamic diameter (Z-average size, Z(av) or dHZ), PDI and surface charge (ζ-potential) of the particles were analyzed by dynamic light scattering using a Malvern ZetaSizer (ZEN3600 Nano-ZS; Malvern Instruments, Worcestershire, UK). Measurements were performed at 25 °C at a scattering angle of 173° (see Results).45
Cell culture. Unless specified, all cell culture reagents were purchased from Gibco-BRL (Invitrogen, Life Technologies, Basel, Switzerland). South American Origin fetal bovine serum was acquired from Biowest (TecoMedical, Sissach, Switzerland) and porcine serum from Milan Analytica (Rheinfelden, Switzerland) or (Sigma-Aldrich, Buchs, Switzerland). RPMI 1640, Optimem, Trypsin, 0.05% (w/v) ethylenediaminetetraacetic acid (EDTA) 1× were obtained from Invitrogen (Life Technologies Europe B.V)
Swine kidney SK-6 cells, for producing the VRPs, were kindly provided by Professor Maurice Pensaert (University of Gent, Belgium). They were also used as the reference point for controlling RepRNA replication and translation, due to their high capacity for accommodating both virus and replicon. These cells were cultured in minimum essential medium supplemented with glutamax and 10% (v(/v) horse serum.
Use of animals. All animal studies have been approved by the institute review board and performed under license from the competent cantonal (Canton Bern, Switzerland) and Swiss Federal authorities (license BE 72/12).
Generation of DCs. Porcine DCs were employed for their close relationship with human DCs and availability of large quantities from the same individual, facilitating the level of analyses pursued in the described experiments under Results. MoDCs were obtained from porcine peripheral blood mononuclear cells (PBMC)46 as previously described.47 Briefly, PBMCs were isolated by density centrifugation on Ficoll-Paque PLUS (GE Healthcare, Glattbrugg, Switzerland), and monocytes were enriched from the PBMCs by magnetic cell sorting using the magnetic-activated cell-sorting system (MiltenyiBiotec, Bergisch Gladbach, Germany) and an anti-CD172a mAb (74-22-15, kindly provided by Dr J.K. Lunney, US Department of Agriculture, Beltsville, MD). MoDCs were generated by culturing for 5 days at 39 °C in DMEM supplemented with 10% (v/v) porcine serum, glutamax and recombinant porcine granulocyte macrophage colony stimulating factor (150 ng/ml) plus rpIL-4 (100 U/ml); 39 °C is employed rather than 37 °C due to the natural body temperature of the pig. MoDCs were fed with fresh medium and cytokines on day 2 and collected for use on day 5.
Fresh blood-derived DCs were also employed. CD172alow cDCs+pDCs and CD172ahigh monocytes were isolated using magnetic-activated cell-sorting (Miltenyi Biotec) separation of porcine PBMCs.47 1 × 106cells/ml were cultured in DMEM supplemented with 1% v/v porcine serum and 50 U/ml porcine GM-CSF, again at 39 °C due to the natural body temperature of the pig.48 The animal experiment was approved (authorization N° 18/04 delivered by the canton Bern) and carried out in accordance with the laws on care and use of laboratory animals in Switzerland.
A total of 10 blood donor animals were employed. The use of pigs allowed cells to be obtained at regular intervals from each individual. This permitted the identification of variability in responsiveness of the cells in terms of interaction with the particles and internalization of the RepRNA when DCs were obtained from the same individual. Such variation did not alter the overall interpretation of the data, and was actually not different from the variation observed with cells derived from the different blood donors. These observations provided a greater confidence in the results and their value to the understanding of DC interactions with particles and the synthetic delivery of RepRNA.
RNase resistance assay. For the assay of RNase protection by chitosan, the procedure was as described by Python et al.49 A 50-mer RNA oligonucleotide probe complementary to nucleotides 12242–12193 of the vA187-1 genome sequence (GenBank accession number X87939.1) and carrying a Dyomics 781 modification at the 5′ end (Dy781-O1-RNA) was synthesized by Dr Fabian Axthelm (Microsynth AG, Balgach, Switzerland). The Dy781-O1-RNA probe was mixed at 40 nmol/l final concentration with minimum essential medium containing 3 × 10−5 to 3 × 10−8 U RNase A/ml as digestion control, with 50 mmol/l TrisHCl pH 7.4 as negative control, and with the chitosan or TPP samples to be tested for RNase activity; all were incubated for 1 hour at 37 °C. The treated probes were mixed with 2 volumes of 97% Formamide (Sigma-Aldrich) and separated on a 10% (w/v) polyacrylamide and 35% (w/v) urea gel in 133 mmol/l TrisHCl, 45.5 mmol/l boric acid, and 3.2 mmol/l EDTA. Image acquisition was performed with the Odyssey Infrared Imaging System (LI-COR, Bad Homburg, Germany).
An alternative assay used the RNaseAlert Kit (Ambion, Life Technologies, LuBio Science, Lucerne, Switzerland), following the manufacturer's instructions. The labeled RNA probe for detecting RNase was again associated with either chitosan or TPP, in water or Tris buffer, as mentioned in the above RNase assay.
RNA upload efficacy in particles. In order to verify the effective RNA loading in particles, samples obtained from mixing Dy781-O1-RNA with water (or TPP) or Dy781-O1-RNA incorporated into particles were distributed on Ultrafree Biomax 100 kDa centrifugal devices (Biomax Millipore, Fischer, Switzerland) and centrifuged at 1,000 ×g for 40 minutes. TPP or water were also added to the filters after centrifugation to resuspend the original samples and recover any Dy781-O1-RNA remaining on the filters. The samples obtained before and after centrifugation were finally analyzed on urea page 10% gels (100V/1 hour) and observed for infrared signaling analyzed with an Odyssey Scan (Li-COR).
Infrared imaging assay for nanogel delivery of Dy781-O1-RNA. At 16 hours before nanofection, cells were seeded at a density of 100,000 cells/well in 96-well plates. Dy781-O1-RNA alone or incorporated into particles were added to the cells and incubated at 37 °C (cell lines) or 39 °C (MoDCs) for the times shown in Results, then washed three times with cold phosphate-buffered saline (PBS)/EDTA and finally lysed with RA1 Lysis Buffer 1× (Macherey Nagel, Oensingen, Switzerland) on ice. Controls employed Dy781-O1-RNA electroporated or lipofected (Lipofectamine 2000) into the cells. The cells were observed for infrared signaling with an Odyssey Scan (LI-COR) before and after lysis.
Nanofection of cells determined by flow cytometry. Cells were seeded overnight at a density of 200,000 cells/well in 24-well or 12-well plates (Nunc, Wiesbaden, Germany) in phenol red-free DMEM. Alexa488-oligoRNA, Dy490-labeled RepRNA or fluorescein-labeled RepRNA encapsulated in particles, or RNA alone, were added to the cells, and incubated at 37 °C (cell lines) or 39 °C (MoDCs; relating to the body temperature of the pig donors) for various periods of times as shown in the results; controls employed electroporation and lipofection (Lipofectamine 2000) of the same RNA. After incubation, the cells were washed three times with cold PBS/EDTA, and their surface CD172a molecules labeled with mAb 74-22-15 followed by the appropriate conjugate as shown in Results. Following a final wash, the cells were fixed with 4% (w/v) paraformaldehyde (PFA, Sigma-Aldrich) prior to measurement on a FACS Calibur analytical FCM (Becton Dickinson, Basel, Switzerland) with CellQuest Pro software (Becton Dickinson). Analyses employed FlowJo versions 9 and 10 software (Treestar, San Carlos, CA).
Nanofection of cells determined by confocal microscopy. Cells were seeded overnight at a density of 200,000 cells/well in fibronectin-coated Lab-Tek II (Nunc) in phenol red-free DMEM, then RNA-loaded particles (Alexa488-labeled oligoRNA, Dy490-labeled RepRNA, rhodamine-labeled RepRNA) were added to the cells and incubated at 37 °C (cell lines) or 39 °C (MoDCs) during 2 or 24 hours. After incubation, the cells were washed three times with cold PBS/EDTA, fixed with 4%(w/v) PFA and mounted in Mowiol mounting solution. The slides were observed by using a Leica DMRXA TCS-SL spectral confocal microscope and Leica LCS software (Leica Microsystems AG, Heerbrugg, Switzerland).
MoDCs or freshly isolated blood DCs were seeded overnight (0.2 million cells/well) in 8-well fibronectin-coated Lab-Tek II chambers (Nunc) containing 10% v/v porcine serum and 50 U/ml of porcine GM-CSF (porcine experiments). For two-color analysis, cells were incubated with primary antibody against CD172a (74-22-15) for 20 minutes on ice. Then they were fixed with 4% (w/v) PFA for 10 minutes and incubated with secondary conjugated antibodies (Alexa546 or Alexa633; Molecular Probes, Leiden, Netherlands) in PBS for 20 minutes. After washing twice in PBS, slides were mounted in Mowiol and analyzed by confocal microscopy.
Confocal microscopy employed a Leica TCS-SL confocal microscope and software (Leica Microsystems AG, Glattbrugg, Switzerland). All images were acquired using a 63× oil-immersion objective, with settings to give high-resolution images acquired at optimum voxel size and automatic threshold applied. The images were analyzed using Imaris 7.5 and 7.6 software (Bitplane AG, Zurich, Switzerland); background subtraction, threshold applications, gamma correction, and maxima settings were employed to maintain a negative image in the negative control cells, such that no false-positive emissions were present. In certain images, the anti-CD172a labeling was employed to delineate the cells surface, for which purpose the surface module of the Imaris 7.6 software was applied.
Migration assay. The ability of CD172a+ cells to detect the presence of nanogels was assessed by nanogel-dependent migration of the cells. A 24-well microchemotaxis chamber technique was employed, consisting of transparent 3 µm filter inserts and the corresponding 24-well companion plates (6.5 mm Transwell with 3.0 µm Pore Polyester Membrane Insert, Product #3472; Corning, Basel, Switzerland). Nanogels were diluted to 10% (v/v) in phenol red-free DMEM, and loaded into the lower compartment of the assay system. The 3.0 µm pore membrane inserts were then added into each well, and the plates incubated at 39 °C for 30 minutes. Following this incubation step, the upper compartment of the filter insert was loaded with 100 µl of CD172a-sorted cells (prelabeled with anti-CD172a and anti-CD14 antibodies) resuspended in phenol red-free DMEM to have 0.5 × 106 cells/well. The system was incubated at 39 °C for 3 hours, after which the filter inserts were removed from the wells. The remaining contents of the wells, which contained the migrated cells, were transferred into FACS tubes, washed twice with CellWash (BD Biosciences, Allschwil, Switzerland), and resuspended in 100 µl of CellWash. Prior to flow cytometric analysis as described above, 50 µl of CountBright solution (Invitrogen, Lubio, Switzerland) was added to each sample. Quantification of the migrated cells was performed by counting a reference of 5000 CountBright beads.
Luciferase assay. The translation of the luciferase gene when encoded by the RepRNA was measured using the assay described by Suter et al.22 for expression levels of luciferase. DCs to which the RepRNA had been delivered were washed once with PBS, lysed with Firefly Luciferase Assay Lysis Buffer (Biotium, Hayward, CA) at room temperature for 15 minutes and stored at −70 °C. Luciferase activity was quantified with a Centro LB 960 luminometer (Berthold Technologies, Regensdorf, Switzerland) using a Firefly Luciferase Assay Kit (Biotium) containing D-luciferin as a substrate.
Assessment of nanogel delivery of RepRNA in vivo. RepRNA was incorporated into NGA, NGA-Lipo, or chitosan cores as described above and 100 μl were used per dose for vaccination of Balb/c mice and New Zealand white rabbits. Each dose comprised 0.2 μg in 50 μl HA-RepRNA or C-HA-C RepRNA plus 0.2 μg in 50 μl NP-RepRNA; mice received one dose for reach RepRNA mixed together, while rabbits received twice this dose. The nanogel vaccines were adjuvanted with PEGylated MALP-2 adjuvant as described.50 Animals were vaccinated subcutaneously at the intervals shown in the Results section, and bled from the tail vein (mice) or ear (rabbits), also at the intervals shown in the Results section. Anti-HA and anti-NP titers were assessed by indirect enzyme-linked immunosorbent assay using recombinant HA (H5 Vietnam 2004; related to the HA gene incorporated into the RepRNA, which was derived from a Swiss isolate of highly pathogenic avian influenza virus (Suter et al., unpublished data) and NP as antigen.
These same antigens were employed at 1 µg/ml concentration in splenocyte restimulation assays. Splenocytes were prepared from the vaccinated mice 2–3 weeks after the final booster vaccination, using disruption through a plastic mesh sieve followed by lysis of the erythrocytes. The washed splenocytes were resuspended to have 107 cells/ml. Following prelabeling with CFSE (Biolegend, Fell, Germany) to monitor cell division, the cells were washed to remove unincorporated CFSE and then seeded in RPMI 1640 supplemented with 10% (v/v) fetal calf serum at 1 ml/well of 24-well plates, together with either no antigen (unstimulated) or 1 µg/ml recombinant HA or NP. After incubation at 37 °C for 7 days (refeeding at 35 days by replacing half of the medium), the cells were harvested and subjected to multicolor labeling for flow cytometric analysis. The labeling permitted gating of CD4+ and CD8+ T-lymphocytes as memory (CD3+, CD44high, CD62Llow) and naive (CD3+, CD44low, CD62Lhigh) populations. Proliferating cells were identified by their reduced CFSE signal (CFSElow), and scored as a percentage of the total cells within the above gates. A similar estimation was made for cells upregulating CD25 expression, measured as MFI; a stimulation index was calculated relative to the MFI obtained with unstimulated cells (the MFI for unstimulated cells was therefore 1).
Acknowledgments
This work was supported by the EU FP6 project PANFLUVAC (grant number 044115), EU FP7 project UniVax (grant number 601738), the Marie-Curie Action FP7-PEOPLE-2009-IAAP, “Replixcel” (grant number 7270014), as well as the ERA-NET projects HCVAX (EuroNanoMed project funded by the Swiss National Science Foundation) (grant number 31NM30-136034/1) and HCRus (funded by the University of Geneva, Switzerland) (grant number ERANetRUS 081). We are most grateful to Brigitte Herrmann for help with the various cell lines and PBMCs employed for this work, and to Andreas Michel and Hans-Peter Lüthi for animal care and bleeding of the donor animals to provide for preparation of PBMCs. For the initial methodologies from which we derived the methods for preparing the particulate delivery vehicles carrying the replicons, and provision of chitosan and alginate, we are most grateful to Nathaniel Rossi and Peter Käuper (Medipol SA). Application of nanoparticulate delivery vehicles for the delivery of vaccines employing the replicons derived from classical swine fever virus, as used in this paper, has been patent filed in Europe, United States, Canada, and Japan, with priority date of 2008. The filing was by the authors K.C. Mc. and N.R., together with Jon Duri Tratschin, and assigned to their employer, the Institute of Virology and Immunology (formerly Institute of Virology and Immunoprophylaxis). It was licensed to Medipol SA for vaccine delivery purposes, but maintenance of the license has now reverted to the Institute of Virology and Immunology following the closure of Medipol SA. This does not alter the authors' adherence to the policies of sharing data and materials. All authors contributed important elements to the work presented in this paper. K.C.McC., I.B., and P.M. designed the various experiments. R.S. and N.R. designed and produced the different replicons. I.B. performed the RNA detection analyses using Dy790-labeled RNA. P.M. prepared the rhodamine and Dy490-labeled RepRNA and performed most of the Flow Cytometry analyses with this material. P.E. performed the studies using different concentrations of RepRNA. P.M., I.B., and K.C.McC. performed the confocal microscopy, and K.C.McC. performed the Imaris analyses. P.M. made the dHZ, PDI and ζ-potential measurements. P.M. and N.R. performed the RNase assays. I.B. performed the in vitro RepRNA translation assays for luciferase, while P.E. and P.M. measured the NS3 readouts. L.T-H., P.M., P.E., T.D., and N.R. performed the animal experiments, while L.T-H. performed the enzyme-linked immunosorbent assay and T.D. performed the T-lymphocyte assays. K.C.McC, P.M. and P.E. assembled the data, and K.C.McC. both prepared the figures and wrote the manuscript.
References
- Platta HW, Stenmark H. Endocytosis and signaling. Curr Opin Cell Biol. 2011;23:393–403. doi: 10.1016/j.ceb.2011.03.008. [DOI] [PubMed] [Google Scholar]
- Kumari S, Mg S, Mayor S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 2010;20:256–275. doi: 10.1038/cr.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough KC, Bassi I, Démoulins T, Thomann-Harwood LJ, Ruggli N. Functional RNA delivery targeted to dendritic cells by synthetic nanoparticles. Ther Deliv. 2012;3:1077–1099. doi: 10.4155/tde.12.90. [DOI] [PubMed] [Google Scholar]
- Randolph GJ, Jakubzick C, Qu C. Antigen presentation by monocytes and monocyte-derived cells. Curr Opin Immunol. 2008;20:52–60. doi: 10.1016/j.coi.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Amigorena S. The cell biology of antigen presentation in dendritic cells. Curr Opin Immunol. 2001;13:45–51. doi: 10.1016/s0952-7915(00)00180-1. [DOI] [PubMed] [Google Scholar]
- Jensen PE. Recent advances in antigen processing and presentation. Nat Immunol. 2007;8:1041–1048. doi: 10.1038/ni1516. [DOI] [PubMed] [Google Scholar]
- Vyas JM, Van der Veen AG, Ploegh HL. The known unknowns of antigen processing and presentation. Nat Rev Immunol. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov OO, Mironova NL, Maslov MA, Petukhov IA, Morozova NG, Vlassov VV, et al. Novel cationic liposomes provide highly efficient delivery of DNA and RNA into dendritic cell progenitors and their immature offsets. J Control Release. 2012;160:200–210. doi: 10.1016/j.jconrel.2011.11.034. [DOI] [PubMed] [Google Scholar]
- Midoux P, Pichon C, Yaouanc JJ, Jaffrès PA. Chemical vectors for gene delivery: a current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br J Pharmacol. 2009;157:166–178. doi: 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perche F, Benvegnu T, Berchel M, Lebegue L, Pichon C, Jaffrès PA, et al. Enhancement of dendritic cells transfection in vivo and of vaccination against B16F10 melanoma with mannosylated histidylated lipopolyplexes loaded with tumor antigen messenger RNA. Nanomedicine. 2011;7:445–453. doi: 10.1016/j.nano.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Perche F, Gosset D, Mével M, Miramon ML, Yaouanc JJ, Pichon C, et al. Selective gene delivery in dendritic cells with mannosylated and histidylated lipopolyplexes. J Drug Target. 2011;19:315–325. doi: 10.3109/1061186X.2010.504262. [DOI] [PubMed] [Google Scholar]
- Pichon C, Midoux P. Mannosylated and histidylated LPR technology for vaccination with tumor antigen mRNA. Methods Mol Biol. 2013;969:247–274. doi: 10.1007/978-1-62703-260-5_16. [DOI] [PubMed] [Google Scholar]
- Tratschin JD, Ruggli N, McCullough KC.2008Pestivirus replicons providing an RNA-based viral vector systemEP2130912, PCT/EP2009/003892, WO 2009146867.
- Atkins GJ, Fleeton MN, Sheahan BJ. Therapeutic and prophylactic applications of alphavirus vectors. Expert Rev Mol Med. 2008;10:e33. doi: 10.1017/S1462399408000859. [DOI] [PubMed] [Google Scholar]
- Khromykh AA. Replicon-based vectors of positive strand RNA viruses. Curr Opin Mol Ther. 2000;2:555–569. [PubMed] [Google Scholar]
- Lundstrom K. Alphavirus-based vaccines. Curr Opin Mol Ther. 2002;4:28–34. [PubMed] [Google Scholar]
- Rayner JO, Dryga SA, Kamrud KI. Alphavirus vectors and vaccination. Rev Med Virol. 2002;12:279–296. doi: 10.1002/rmv.360. [DOI] [PubMed] [Google Scholar]
- Pushko P, Parker M, Ludwig GV, Davis NL, Johnston RE, Smith JF. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology. 1997;239:389–401. doi: 10.1006/viro.1997.8878. [DOI] [PubMed] [Google Scholar]
- Pijlman GP, Suhrbier A, Khromykh AA. Kunjin virus replicons: an RNA-based, non-cytopathic viral vector system for protein production, vaccine and gene therapy applications. Expert Opin Biol Ther. 2006;6:135–145. doi: 10.1517/14712598.6.2.135. [DOI] [PubMed] [Google Scholar]
- Johansson DX, Ljungberg K, Kakoulidou M, Liljeström P. Intradermal electroporation of naked replicon RNA elicits strong immune responses. PLoS One. 2012;7:e29732. doi: 10.1371/journal.pone.0029732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter R, Summerfield A, Thomann-Harwood LJ, McCullough KC, Tratschin JD, Ruggli N. Immunogenic and replicative properties of classical swine fever virus replicon particles modified to induce IFN-α/β and carry foreign genes. Vaccine. 2011;29:1491–1503. doi: 10.1016/j.vaccine.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Palucka K, Banchereau J, Mellman I. Designing vaccines based on biology of human dendritic cell subsets. Immunity. 2010;33:464–478. doi: 10.1016/j.immuni.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey CF, Bauhofer O, Ruggli N, Summerfield A, Hofmann MA, Tratschin JD. Classical swine fever virus replicon particles lacking the Erns gene: a potential marker vaccine for intradermal application. Vet Res. 2006;37:655–670. doi: 10.1051/vetres:2006028. [DOI] [PubMed] [Google Scholar]
- Schütz CA, Schmitt F, Juillerat-Jeanneret L, Wandrey C. Fate of hydrophilic nanoparticles in biological environments. Chimia. 2009;63:220–222. [Google Scholar]
- Summerfield A, Guzylack-Piriou L, Schaub A, Carrasco CP, Tâche V, Charley B, et al. Porcine peripheral blood dendritic cells and natural interferon-producing cells. Immunology. 2003;110:440–449. doi: 10.1111/j.1365-2567.2003.01755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield A, McCullough KC. The porcine dendritic cell family. Dev Comp Immunol. 2009;33:299–309. doi: 10.1016/j.dci.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marsico G, Schubert U, et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- Rehman Zu, Hoekstra D, Zuhorn IS. Mechanism of polyplex- and lipoplex-mediated delivery of nucleic acids: real-time visualization of transient membrane destabilization without endosomal lysis. ACS Nano. 2013;7:3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- Collnot EM, Ali H, Lehr CM. Nano- and microparticulate drug carriers for targeting of the inflamed intestinal mucosa. J Control Release. 2012;161:235–246. doi: 10.1016/j.jconrel.2012.01.028. [DOI] [PubMed] [Google Scholar]
- Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- Lai YH, Wang C. Delivery strategies of melanoma vaccines: an overview. Expert Opin Drug Deliv. 2008;5:979–1001. doi: 10.1517/17425247.5.9.979. [DOI] [PubMed] [Google Scholar]
- Ceppi M, Ruggli N, Tache V, Gerber H, McCullough KC, Summerfield A. Double-stranded secondary structures on mRNA induce type I interferon (IFN alpha/beta) production and maturation of mRNA-transfected monocyte-derived dendritic cells. J Gene Med. 2005;7:452–465. doi: 10.1002/jgm.685. [DOI] [PubMed] [Google Scholar]
- Hoerr I, Obst R, Rammensee HG, Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Mockey M, Bourseau E, Chandrashekhar V, Chaudhuri A, Lafosse S, Le Cam E, et al. mRNA-based cancer vaccine: prevention of B16 melanoma progression and metastasis by systemic injection of MART1 mRNA histidylated lipopolyplexes. Cancer Gene Ther. 2007;14:802–814. doi: 10.1038/sj.cgt.7701072. [DOI] [PubMed] [Google Scholar]
- Fang N, Chan V, Mao HQ, Leong KW. Interactions of phospholipid bilayer with chitosan: effect of molecular weight and pH. Biomacromolecules. 2001;2:1161–1168. doi: 10.1021/bm015548s. [DOI] [PubMed] [Google Scholar]
- Hafez IM, Maurer N, Cullis PR. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 2001;8:1188–1196. doi: 10.1038/sj.gt.3301506. [DOI] [PubMed] [Google Scholar]
- Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- He C, Hu Y, Yin L, Tang C, Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- Thiele L, Rothen-Rutishauser B, Jilek S, Wunderli-Allenspach H, Merkle HP, Walter E. Evaluation of particle uptake in human blood monocyte-derived cells in vitro. Does phagocytosis activity of dendritic cells measure up with macrophages. J Control Release. 2001;76:59–71. doi: 10.1016/s0168-3659(01)00412-6. [DOI] [PubMed] [Google Scholar]
- Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- Sharma R, Ghasparian A, Robinson JA, McCullough KC. Synthetic virus-like particles target dendritic cell lipid rafts for rapid endocytosis primarily but not exclusively by macropinocytosis. PLoS One. 2012;7:e43248. doi: 10.1371/journal.pone.0043248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Calvo A, Saiz JC, McCullough KC, Sobrino F, Martín-Acebes MA. Acid-dependent viral entry. Virus Res. 2012;167:125–137. doi: 10.1016/j.virusres.2012.05.024. [DOI] [PubMed] [Google Scholar]
- Ruggli N, Tratschin JD, Mittelholzer C, Hofmann MA. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J Virol. 1996;70:3478–3487. doi: 10.1128/jvi.70.6.3478-3487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütz CA, Juillerat-Jeanneret L, Käuper P, Wandrey C. Cell response to the exposure to chitosan-TPP//alginate nanogels. Biomacromolecules. 2011;12:4153–4161. doi: 10.1021/bm201231x. [DOI] [PubMed] [Google Scholar]
- McCullough KC, Basta S, Knötig S, Gerber H, Schaffner R, Kim YB, et al. Intermediate stages in monocyte-macrophage differentiation modulate phenotype and susceptibility to virus infection. Immunology. 1999;98:203–212. doi: 10.1046/j.1365-2567.1999.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzylack-Piriou L, Balmelli C, McCullough KC, Summerfield A. Type-A CpG oligonucleotides activate exclusively porcine natural interferon-producing cells to secrete interferon-alpha, tumour necrosis factor-alpha and interleukin-12. Immunology. 2004;112:28–37. doi: 10.1111/j.1365-2567.2004.01856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerfield A, Horn MP, Lozano G, Carrasco CP, Atze K, McCullough K. C-kit positive porcine bone marrow progenitor cells identified and enriched using recombinant stem cell factor. J Immunol Methods. 2003;280:113–123. doi: 10.1016/s0022-1759(03)00273-4. [DOI] [PubMed] [Google Scholar]
- Python S, Gerber M, Suter R, Ruggli N, Summerfield A. Efficient sensing of infected cells in absence of virus particles by plasmacytoid dendritic cells is blocked by the viral ribonuclease E(rns.) PLoS Pathog. 2013;9:e1003412. doi: 10.1371/journal.ppat.1003412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsutzky S, Ebensen T, Link C, Becker PD, Fiorelli V, Cafaro A, et al. Efficient systemic and mucosal responses against the HIV-1 Tat protein by prime/boost vaccination using the lipopeptide MALP-2 as adjuvant. Vaccine. 2006;24:2049–2056. doi: 10.1016/j.vaccine.2005.11.025. [DOI] [PubMed] [Google Scholar]