Abstract
In Alzheimer's disease, progressive supranuclear palsy, and a number of other neurodegenerative diseases, the microtubule associated protein tau aggregates to form intracellular neurofibrillary tangles and glial tangles, abnormal structures that are part of disease pathogenesis. Disorders with aggregated tau are called tauopathies. Presently, there are no disease-modifying treatments for this disease class. Tau is encoded by the MAPT gene. We propose that reducing MAPT expression and thus the amount of tau protein made could prevent aggregation, and potentially be an approach to treat tauopathies. We tested 31 morpholinos, complementary to the sense strand of the MAPT gene to identify oligonucleotides that can downregulate MAPT expression and reduce the amount of tau protein produced. Oligonucleotides were tested in human neuroblastoma cell lines SH-SY5Y and IMR32. We identified several morpholinos that reduced MAPT mRNA expression up to 50% and tau protein levels up to ~80%. The two most potent oligonucleotides spanned the 3′ boundary of exons 1 and 5, masking the 5′-splice sites of these exons. Both morpholinos induced skipping of the targeted exons. These in vitro findings were confirmed in mice transgenic for the entire human MAPT gene and that express human tau protein. These studies demonstrate the feasibility of using modified oligonucleotides to alter tau expression.
Introduction
A defining neuropathologic feature of Alzheimer's disease (AD), progressive supranuclear palsy, frontotemporal lobar dementia-tau type (FTLD-T), and some other neurodegenerative disorders is abnormal intracellular aggregates of tau protein, as neurofibrillary tangles and in some cases, glial tangles. Collectively, these disorders are known as tauopathies.1 Highly penetrant missense mutations in tau-encoding MAPT gene cause FTLD-T2,3,4 proving that aggregated tau is pathogenic. In vitro experiments show that these mutations increase aggregation rates.5 Some of these mutations alter the amino acid sequence of tau. Others are intronic, altering the alternative splicing of exon 10, and changing the isoform ratios of tau protein.3,4,6 Both types of mutations result in glial tangles and/or neurofibrillary tangles. Also, common genetic variations in the MAPT genomic region increase risk for developing progressive supranuclear palsy7,8 and Parkinson's disease.9 Thus, altered tau protein or its regulation can cause neurodegeneration.
Tau is a microtubule-associated protein that stabilizes microtubules, facilitating axonal transport. MAPT knock-out mice are viable and mostly normal10,11 showing limited changes in axonal structure, muscle strength, and behavior. Because these studies were performed in mice completely lacking tau, the observed changes could be due to developmental effects of no tau at conception. In mice, tau can be replaced in part by another microtubule-associated protein, MAP1B.12 Whether this is true in humans is not known. Mouse tauopathy models where the human tau cDNA sequence with an FTLD-T mutation is overexpressed develop aggregated tau pathology.13,14,15,16 Studies in these models show that injection of aggregated tau causes tau aggregation at, and beyond the injection site.17,18 Thus, in tauopathies, tau aggregation may spread from cell to cell with seeding of new aggregates dependent on cytoplasmic tau concentrations. AD mouse models where the amyloid precursor protein is overexpressed develop amyloid plaques, learning deficits, hyperactivity, long-term potentiation deficits, and axonal transport defects, but no neurofibrillary tangles. These deficits in amyloid precursor protein transgenic mice are ameliorated in animals lacking endogenous tau.19,20,21,22 Thus, lowering endogenous tau may be a treatment paradigm for tauopathies including AD.
One strategy for altering gene expression and the synthesis of a specific protein is the use of modified oligonucleotides complementary to sequences in a target gene that interfere with RNA transcription, processing, or translation. Antisense oligonucleotide approaches are being used in trials to treat Duchenne muscular dystrophy (DMD). This approach involves designing oligonucleotides to induce skipping of exons that contain a premature stop codon.23 These oligonucleotides target intron–exon boundary splice site sequences and presumably act by blocking access of splicing factors to the targeted splice site. When a mutated exon is skipped, the resulting protein is sufficiently active for improved muscle function.24 Antisense oligonucleotides that induce skipping of exon 51 in dystrophin transcripts are in phase 2/3 clinical trials.25,26 Preliminary results indicate exon skipping is achieved, and that muscle function is improved,26 although one of these trials failed to show efficacy.
Here, we tested modified oligonucleotides to reduce MAPT mRNA expression and tau protein levels. Structural changes include morpholine rings linked through phosphodiamidate bonds to the nitrogen moieties of nucleic acids. Oligonucleotides complementary to the sense strand of MAPT were tested in the human neuroblastoma cell lines IMR32 and SH-SY5Y to identify candidate targets for treatment of tauopathies. Within MAPT, we targeted sequences bracketing the start codon, splice acceptors and donors, splicing branch points, polypyrimidine track-related sequences, and splicing enhancer and inhibitor sequences. Targeting the ATG codon would potentially block translation initiation. The remaining oligonucleotides were designed to interfere with splicing to induce exon skipping. Oligonucleotides that induce exon 1 skipping will result in an mRNA without the start ATG, and thus MAPT mRNA would not be translated. Oligonucleotides that induce exon skipping of exons 1, 5, or 7 would result in a frame shift and a downstream premature stop codon, resulting in nonsense-mediated decay, a reduction in MAPT mRNA, and a reduction in tau protein. The two most potent oligonucleotides masked the 5′-splice sites of exons 1 and 5. We also show that the latter oligonucleotide is able to reduce human tau mRNA and protein levels in muscle in a mouse transgenic for the entire human MAPT gene. This work shows the feasibility of using morpholino oligonucleotides to reduce human tau expression in vivo.
Results
We designed morpholino oligonucleotides complementary to MAPT transcripts with the goal of reducing the amount of tau protein produced by cells (Table 1; Figure 1). MAPT exons 0, 1, 4, 5, 7, 9, and 10 were targeted. Exons 0 and 1 contain the 5′ untranslated sequences of MAPT mRNA. The translation initiation ATG is in exon 1. Exons 1, 4, 5, 7, and 9 are constitutive exons found in all six brain MAPT isoforms.27,28,29 We also targeted exon 10, an alternatively spliced exon present in three of the six brain isoforms. When exon 10 is included in MAPT transcripts, the tau isoform produced has four microtubules-binding repeats (4R tau). When exon 10 is missing, the protein produced has only 3 repeats (3R tau).27,28,29 Mutations that increase exon 10 inclusion cause frontotemporal dementia—tau type.2,3,4 Since excess 4R tau may be toxic, we tested oligonucleotides that would potentially block exon 10 inclusion (E10.1–E10.8).
Table 1. Morpholino oligonucleotides designed for this study.

Figure 1.
Localization of morpholino oligonucleotides in relation to MAPT exons. Boxes represent MAPT exons and solid lines show the location of the morpholino oligonucleotides used (see Table 1 for more detail).
Most oligonucleotides tested were designed to induce exon skipping, either by hybridizing to intronic branch point sequences (E1.7, E4.2, E4.3, E5.4), or by targeting splice sites directly, bracketing intronic and exonic sequences (e.g., E1.1). E9.1 and E9.2 targeted exon splicing enhancers in exon 9, a constitutively spliced exon. Alternative splicing of exon 10 is controlled by exonic splicing enhancer and silencer elements and by intronic sequences downstream of the 3′ end of the exon. Exonic splicing regulatory sequences were targeted by E10.3, which is complementary to an SC-35-like splicing enhancer element and E10.4, which is complementary to both an A/C rich splicing enhancer element and an exonic polypurine enhancer6,30,31 (Table 1). Splicing regulatory sequences downstream of E10 include an intronic splicing silencer and an intron splicing modulator.31,32 These elements were targeted by E10.5, E10.6, E10.7, and E10.8 (ref. 31,32). Oligonucleotide E1.2 was designed to block translation by hybridizing to the ATG start codon and surrounding sequence in exon 1.
We initially used a 96 well format to test pairs of oligonucleotides targeting more than one sequence for a given exon. Human neuroblastoma cell lines SH-SY5Y and IMR32 were nucleofected with the oligonucleotides and harvested after 6 days. Promising pairs were then tested in combination and singly in both cell lines. MAPT mRNA levels were measured by reverse transcription quantitative real-time polymerase chain reaction (RT-PCR). Tau protein, the product of the MAPT gene, was measured by an enzyme-linked immunosorbent assay (ELISA) and by immunoblotting. Both cell lines produce predominantly 3R tau with 4R mRNA accounting for 13.3–23.5% of the total MAPT mRNA produced (Supplementary Table S1) as previously observed with these cell lines.33 This isoform distribution was confirmed by immunoblotting (Figure 2a).
Figure 2.
MAPT expression was decreased after treatment with exon 1-targeting morpholino oligonucleotides. Panel a is immunoblot analysis of tau protein levels in morpholino-treated cultures. Phosphatase-treated lysates of IMR32 were subjected to electrophoresis and immunoblot analysis using antibody Tau-13 (#sc-21796) at 1:1,000. Untreated (mock) cultures predominantly express 0N3R isoform, with lesser but detectable levels of 0N4R and 1N3R isoforms. Following morpholino treatment, 0N3R isoform levels were substantially reduced while the remaining isoforms were not detected at all. Protein loads/lane: 15.6 μg, mock; 18.5 μg, E1.4; 16.8 μg, E5.3; 16.7 μg, control morpholino (cont MO). Panels b and c show levels of MAPT transcripts in IMR32 and SH-SY5Y cultures, respectively, after mock nucleofection or treatment with the different oligonucleotides. Panels d and e show tau protein levels determined by ELISA in IMR32 and SH-SY5Y cultures, respectively, after mock nucleofection or treatment with the different oligonucleotides. Symbols indicating significance levels of oligonucleotide–treated cell transcript of protein levels relative to mock-treated control levels: *P < 0.01; **P = 0.01; #P < 0.001; †P < 0.05; ##P < 0.005. Error bars indicate standard error of the mean (SEM), n = 3–5.
Effects of oligonucleotides on total MAPT mRNA levels and total tau protein
We measured the effects of 31 oligonucleotides on MAPT mRNA levels. Oligonucleotides targeting exons 0, 4, 9, and 10 did not significantly reduce transcript levels (Supplementary Figures S1–S4). As described below, some of the oligonucleotides targeting exons 1, 5, and 7 did significantly reduce MAPT transcript and tau protein levels.
Exon 1. We examined the effects of seven antisense morpholino oligonucleotides that target exon1 in IMR32 cells (Figure 2b). Oligonucleotides tested include E1.7, which is complementary to the exon 1 branch point and polypyrimidine tract sequences, E1.1, which spans the splice acceptor sequence at the 5′-end of the exon and the ATG initiation codon, and E1.3–E1.6, targeting the splice donor sequence at the 3′-end of the exon. These oligonucleotides were designed to inhibit inclusion of exon 1 in MAPT transcripts. We also tested an oligonucleotide which did not hybridize to a splice site but that was complementary to the ATG start codon and would potentially block translation (E1.2). When tested individually, only oligonucleotides that targeted the splice donor site significantly reduced MAPT transcript levels. The most potent was E1.4, which is complementary to the last 7 nucleotides of exon 1 and the first 18 nucleotides of intron 1. This oligonucleotide reduced MAPT mRNA expression in IMR32 cultures by 50% (SEM = 4.13%, n = 4). We also tried a combination of oligonucleotides that targeted the splice donor (E1.4) and the splice acceptor site along with the ATG codon (E1.1), or the branchpoint sequence E1.7, or all three together. Only the combination of all three oligonucleotides was more effective than E1.4 alone. Similar results were obtained using SH-SY5Y cells (Figure 2c).
We sequenced RT-PCR products from both IMR32 and SH-SY5Y cells (Supplementary Figures S5 and S6). The sequence from mock-treated cells (no oligonucleotide control) showed that exon 1 was spliced to exon 4 and alternatively spliced exons 2 and 3 were excluded (0N tau). RT-PCR products from cells treated either with E1.4 alone or E1.1 + E1.4 showed two products containing exon 0 and exon 4 (Supplementary Figure S6). One was the normal 0N transcript with exon 1 spliced to exon 4. The second smaller products showed exon 0 spliced directly to exon 4, consistent with the skipping of exon 1. This latter product is consistent with a transcript lacking the ATG codon and would not be expected to produce tau protein. When total tau protein was measured using an ELISA, E1.4 reduced protein levels by 77% (tau protein in “no-oligo” controls = 357.6 ± 37 pg tau/μg protein versus 79.88 ± 20.8 pg tau/μg protein in E1.4-treated cultures; n = 5) in IMR32 and 48% (88.24 ± 8.5 pg tau/μg protein versus 45.7 ± 13.2 pg tau/μg protein; n = 3) in SH-SY5Y cells (Figure 2d,e). This result was confirmed by immunoblot experiments (Figure 2a) in IMR32 cultures.
Exon 5. We tested oligonucleotides that targeted exon 5 splice sites. These morpholinos were complementary to the branchpoint sequence (E5.4), the splice acceptor site (E5.1), and the splice donor site (E5.2 and E5.3) (Table 1). Only E5.3, when tested individually, significantly reduced total MAPT transcript levels in both cell types tested to 28.5–46% (Figure 3a,b). This morpholino was complementary to the last 6 nucleotides of exon 5 and the first 19 nucleotides of intron 5. Transcript levels were also significantly reduced when both the acceptor and donor sequences were targeted (E5.1 + E5.2). E5.3 also reduced total tau protein levels in IMR32 cells (62%; 357.6 ± 37 pg tau/μg protein versus 133 ± 29 pg tau/μg protein; n = 3) and in SH-SY5Y cells (58%, 88.24 ± 8.5 pg tau/μg protein versus 23.7 ± 6.5 pg tau/μg protein; n = 3) (Figure 3c,d; see also Figure 2a). The largest reduction in MAPT mRNA observed with E5.3 was at 10 μmol/l, the highest concentration tested, though some reduction was observed with 5 and 1 μmol/l (Supplementary Figure S7).
Figure 3.
MAPT expression was decreased after treatment with exon 5-targeting morpholino oligonucleotides. Panels a and b show levels of MAPT transcripts in IMR32 and SH-SY5Y cultures, respectively, after mock nucleofection or treatment with the different oligonucleotides. Panels c and d show tau protein levels determined by enzyme-linked immunosorbent assay in IMR32 and SH-SY5Y cultures, respectively, after mock nucleofection or treatment with the different oligonucleotides. Symbols and abbreviations are as in Figure 2. n = 3 in each morpholino-treated group.
We sequenced RT-PCR products from both IMR32 and SH-SY5Y cells treated with a combination of E5.1 and E5.2. Two sequences were observed that spanned exon 4 to exon 7. One, the normal product, included exons 4, 5, and 7. The second smaller sized product showed exon 4 spliced to exon 7 (Supplementary Figure S8a,b) indicating that E5.1 and E5.2 blocked inclusion of exon 5 into the final MAPT transcript. The same results were observed in cells treated with E5.3 oligo (Supplementary Figure S8c).
Exon 7. We tested three morpholino oligonucleotides targeting the splice acceptor (E7.1, E7.2) and the splice donor site (E7.3) (Supplementary Figure S9). These oligonucleotides did not significantly reduce MAPT transcript levels in IMR32 cells (Supplementary Figure S9a), but E7.3 did in SH-SY5Y cells by 30% (Supplementary Figure S9b; SEM = 5%, n = 3). E7.1 targets the exon 7 splice acceptor site and is complementary to the last 2 nucleotides of intron 6 and the first 23 nucleotides in exon 7 (Table 1). E7.3 did reduce protein expression in IMR32 cells, but the difference was not significant (Supplementary Figure S9c; P = 0.07). However, in SH-SY5Y cultures, consistent with decrease in MAPT transcripts, E7.3 oligonucleotide also decreased tau protein levels by 67% (Supplementary Figure S9d; 88.24 ± 8.5 pg tau/μg versus 29 ± 10.2 pg tau/μg; n = 3; P = 0.005).
Effects of morpholino oligonucleotides on inclusion of exon 10
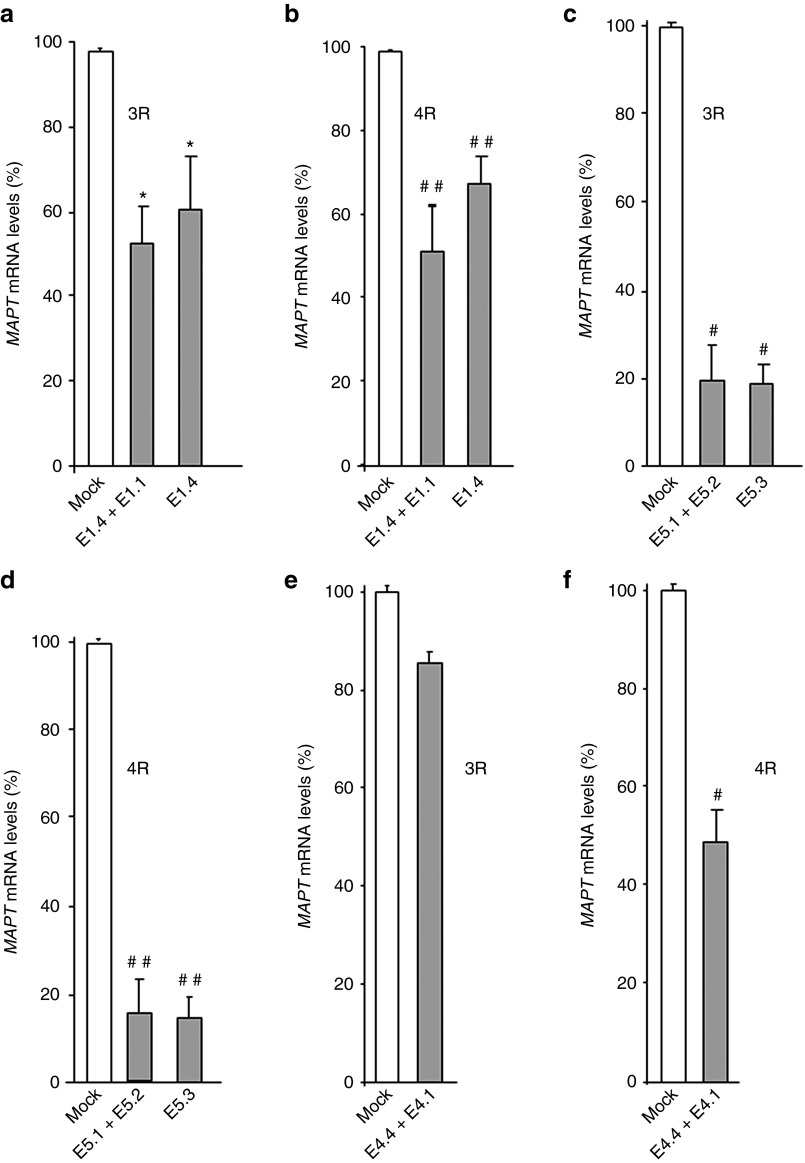
Exon 10 is alternatively spliced and is found in three of the six tau isoforms found in brain. None of the oligonucleotides directly targeting exon 10 reduced total MAPT mRNA levels nor did they alter the amount of exon 10+ (4R) message (Supplementary Figure S4). We also examined exon 10 inclusion in SH-SY5Y cells using morpholinos that targeted exons 1, 4, and 5 (Figure 4). For exon 1, oligonucleotides E1.4 and E1.1 + E1.4 that reduced total MAPT transcript levels (Figure 2a,b) reduced both 3R and 4R transcripts equally (Figure 4a,b). The same was true for exon 5 oligonucleotides E5.3 and E5.1, which reduced total MAPT transcript levels (Figure 3a,b) and transcripts encoding 3R and 4R isoforms (Figure 4c,d).
Figure 4.
Effects of morpholino oligonucleotide treatment on MAPT transcripts lacking exon 10 (3R) or transcripts containing exon 10. Transcripts lacking exon 10 (3R) were assayed using a probe that hybridizes to the MAPT mRNA exon 9 to exon 11 junction sequence. Transcripts containing exon 10 (4R) were measured using a probe that hybridizes to the MAPT mRNA exon 9-exon 10 junction sequence. All experiments are in SH-SY5Y cultures. Panels are: a and b, exon 1 oligonucleotides for 3R and 4R transcripts, respectively; c and d, exon 5 oligonucleotides for 3R and 4R transcripts, respectively; and e and f, exon 4 oligonucleotides for 3R and 4R transcripts, respectively. Symbols are as in Figure 2. n = 3 in each morpholino-treated group.
Morpholinos masking polypyrimidine track (E4.2 and E4.3), as well as additional oligo targeting the splice donor site, E4.5, also did not cause an appreciable reduction in total transcript levels (data not shown). Oligonucleotides targeting the splice acceptor and splice donor sequences to exon 4, E4.1 and E4.4, respectively (Table 1) did not significantly reduce total MAPT transcript levels (Supplementary Figure S3) though there was a trend toward reduced expression. However, two RT-PCR products were evident in cells treated simultaneously with E4.1 and E4.4, and when we sequenced the smaller product, exon 1 was spliced to exon 5 (Supplementary Figure S10), the expected result if exon 4 were skipped and alternatively spliced exons 2 and 3 were also skipped. Interestingly, this pair of oligonucleotides reduced 4R but not 3R transcripts (Figure 4e,f). Presumably, we did not observe a reduction in total MAPT transcript levels with E4.1 + E4.4 because the 4R isoform is only a minor component of the total MAPT produced (Supplementary Table S1).
In vivo activity of E5.3 on MAPT expression and tau protein levels
To determine whether anti-MAPT oligonucleotides can affect gene expression in vivo, we used mice transgenic for the human MAPT gene. The transgene used is a 201 kb fragment of human genomic DNA that contains the entire MAPT gene and some flanking sequence.34,35 The mice produce all six isoforms of the human tau protein and are null for the mouse Mapt gene.35 We injected the gastrocnemius muscles with either E5.3 or the control oligonucleotide or no injection. E5.3 was selected because it was one of the most potent oligonucleotides tested. Muscles were harvested 6 days later and assayed for MAPT mRNA and tau protein. The dose injected was the same as concentration used in in vitro experiments. Histological analysis showed that these injections caused minimal tissue damage (Supplementary Figure S11). We found that muscles (left) injected with E5.3 showed significantly less MAPT mRNA compared to noninjected muscles. Noninjected muscles had comparable levels of mRNA and protein when compared to the control oligonucleotide (Figure 5a,b). E5.3 reduced tau protein from 18.1 ± 3.6 to 5.3 ± 0.85 pg tau/µg protein (P = 0.02, Figure 5c,d). These results are comparable to those observed in neuroblastoma cell lines (Figure 3). Thus, morpholino oligonucleotides can reduce human MAPT expression in vivo.
Figure 5.
Effects of morpholino E5.3 on MAPT expression in transgenic mice. (a) MAPT transcript levels for mice injected with control morpholino (left) or no injection (right); (b) MAPT transcript levels after administration E5.3 oligonucleotide or no injection (##P < 0.005). (c) tau protein levels for mice injected with control morpholino (left) or no injection (right); (d) tau protein levels after administration E5.3 oligonucleotide or no injection (†P < 0.05). Each experimental condition is the average of four mice. Abbreviations and symbols are as in Figure 2.
Discussion
The work presented here demonstrates the feasibility of reducing human tau protein levels using modified oligonucleotides complementary to MAPT expressed mRNA sequences. The primary strategy used here is to design morpholino oligonucleotides that induce exon skipping. The most effective oligonucleotides targeted intron–exon boundaries, presumably by blocking access of splicing factors to the targeted exon. Other strategies included targeting intronic branchpoint sequences, polypyrimidine track sequences, exonic splicing enhancers, and intronic enhancer sequences were less successful. Exon skipping of exon 1 generates an mRNA lacking an ATG transcription start site and thus preventing normal translation. Exon skipping of either exons 1, 5, or 7 will result in changing the open reading frame of the mRNA leading to a premature stop codon and possible nonsense-mediated decay. The exon-skipping strategy used here to reduce protein levels differs from the exon skipping work with the DMD gene. In the latter case, exon skipping is used to by-pass exons with premature stop codon DMD mutations, but retain an open reading frame to produce a functional protein.23,24
One of the most potent oligonucleotide was E1.4, a morpholino complementary to the splice donor site of exon1. This oligonucleotide induces exon 1 skipping and results in mRNA lacking exon 1, with exon 0 spliced directly to exon 4. The transcript produced in the presence of this oligonucleotide lacks the translation start ATG sequence, and the reading frame is changed leading to a premature stop codon at amino acid 20 encoded by exon 4 and another stop codon at amino acid 151 encoded by exon 9. E1.4 reduces MAPT mRNA levels by 50–55 percent and tau protein synthesized (Figure 2). The reduced mRNA levels, suggest that the introduction of a premature stop codon may lead to nonsense mediated decay. Any message that is not degraded would not be translated. The end result is tau protein is reduced to 20–50% of normal expression in the two cell lines. Although it is possible that an alternative ATG site could be used, we did not detect a shorter than normal form of tau by immunoblotting (Figure 2), though protein produced at a very low level would possibly not be detected.
Oligonucleotides targeting exons 5 and 7 also effectively reduced both mRNA and protein levels (Figure 3 and Supplementary Figure S9). Again, the most effective morpholino spanned the 3′ end of the exon and the beginning of the next intron. The most effective morpholino was E5.3, which reduced tau protein to 42% of normal levels in SH-SY5Y cells (Figure 3d). The resulting mRNA contained exon 4 spliced to exon 7 using the normal spice sites for these two exons (Supplementary Figure S8), and no cryptic splice site mRNA species were detected.
Mutations that change isoform ratios cause FTLD-T, either by increasing or decreasing inclusion of exon 10. The cell lines used here make mRNA both lacking exon 10 (3R tau) and with exon 10 (4R tau) (Supplementary Table S1). The most effective oligonucleotides tested here (E1.4 and E5.3) reduced expression of both 3R and 4R mRNA (Figure 4) showing that the 3R/4R isoform ratio is not significantly altered by these two morpholinos. Interestingly, while oligonucleotides targeting exon 4 splice sites did not significantly reduce total MAPT mRNA levels (Supplementary Figure S3), the amount of 4R mRNA was reduced (Figure 4f). Exclusion of exon 4 does not alter the reading frame of MAPT, but could change the kinetics of overall MAPT RNA processing such that the downstream alternatively spliced exon 10 is less likely to be included. Other more direct mechanisms are also possible.
In the neuroblastoma cell lines used here, morpholinos targeting exon 10 sequences did not reduce inclusion of this exon. This result in surprising because the sequences known to regulate exon 10 splicing are located close to or within exon 10 (refs. 30,31,32,36). Also, Kalbfuss et al.37 observed that antisense oligonucleotides complementary to exon 10 splice sites reduced exon 10 inclusion in mRNA from a minigene containing MAPT exon 10, and in endogenous tau mRNA from PC12 cells. In both these systems, exon 10+ was the predominant mRNA expressed. The minigene contained mutations that increased exon 10 inclusion, and rodent cells (PC12) normally produce predominantly 4R (exon 10+) isoforms. In the work reported here using human neuroblastoma cell lines, 3R tau (exon 10-) is the predominant mRNA species. Thus the regulation of exon 10 inclusion in cells making mostly 4R tau mRNA may differ from the regulation in cells that mostly exclude exon 10, and the sequences and splicing factors that act when exon 10 inclusion is favored may not function the same when exon 10 is primarily excluded.
As a proof of concept, we show that the most potent oligonucleotide used here can reduce human MAPT expression in vivo in muscle. Reduction of tau levels in the brain is a potential therapeutic for multiple tauopathies including AD. Certainly the fact that mice lacking MAPT are relatively normal argues that reduction of MAPT expression in an adult could be well tolerated.10,11 In fact, knockout of the mouse Mapt gene reduces susceptibility to induced seizures.38 When antisense oligonucleotides against Mapt sequences are introduced into mouse brain, Mapt mRNA is reduced as is mouse tau protein, and susceptibility to induced seizures is decreased. This work also shows the feasibility of reducing brain tau as a therapeutic for at least seizures if not AD.
The primary barrier to using morpholinos for treatment is achieving adequate levels of modified oligonucleotides in the brain. Several strategies for getting oligonucleotides to cross the blood brain barrier have been described,39,40,41 though these approaches have not yet been tried in man. Other approaches include infusion of oligonucleotides into the cerebrospinal fluid42 or by use of implanted pumps.43 While oligonucleotide-based therapies are attractive in that any expressed sequence could be targeted, delivery of adequate levels to oligonucleotides to the target tissue needs to be addressed.
Materials and methods
Cells. Human neuroblastoma cell lines, SH-SY5Y and IMR32, were used in this study. IMR32 cells (ATCC# CCL-127) were maintained in Eagle's minimum essential medium (ATCC# 30-2003) containing 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Gibco). SH-SY5Y, a kind gift from Dr Virginia M.-Y. Lee (Center for Neurodegenerative Research, University of Pennsylvania), were maintained in 1:1 mix of Ham's F12 and Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% nonessential amino acids (Gibco). Both cell lines were maintained in a humidified incubator with 5% CO2.
Designing antisense morpholino oligonucleotides. The morpholino oligonucleotides used were designed to hybridize to exon splice sites, branchpoint sequences, and regulatory sequences in or close to MAPT exons. Splice site morpholino oligonucleotides were picked based on splice-site predictions from SROOGLE44 and on previously determined intron–exon boundaries.27,34 Wherever possible, we used a “sliding window design” to generate a panel of antisense oligonucleotides for blocking the same splice site. Exon 10 oligonucleotides were designed to hybridize to regulatory sequences in the exon6,30,31 and in intronic sequences that immediately follow this exon.32 BLAST analyses were performed for each morpholino oligonucleotide sequence to avoid off-target hybridization. Controls included “mock” nucleofection with no oligonucleotide added and a negative control morpholino oligonucleotide (Gene-Tools) that targets mutated β-globin transcripts. This oligonucleotide only elicits an effect in hematopoietic cells from β-thalassemia subjects. This control oligonucleotide did not affect tau levels in either IMR32 or SH-SY5Y cells or in transgenic mice (Figures 2 and 5 and Supplementary Figure S11, respectively).
Nucleofection. Morpholino oligos were introduced in neuroblastoma cells with the Amaxa Nucleofector-II device (Lonza, Walkersville, MD). For initial testing of morpholino efficacy, nucleofection was carried out in 96-well plates. Nucleofector solution SF was selected after preliminary experiments, based on high viability and efficient transfection. For nucleofection of neuroblastoma cell lines in six-well plates, Amaxa recommended kits V and L were used for introducing antisense morpholinos in SH-SY5Y and IMR32 cells respectively. On the day of nucleofection, 60–80% confluent cultures were trypsinized, and cells plated in each well of six-well plates. At 24 hours postnucleofection, sample of nucleofected cells were analyzed by flow cytometry to ascertain % of cells positive for carboxyfluorescein-tag on the morpholino oligonucleotides. In all cases, 97–99% of cells were fluorescent (data not shown).
Harvesting cells from nucleofected cultures and further processing. Cells were harvested at day-6 postnucleofection for analysis of RNA and protein. At day 6, there was no significant difference in viability of mock and morpholino-treated cultures using an 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide assay45 (data not shown). Lysis and further processing was performed using the Illustra TriplePrep kit (GE Healthcare, Fairfield, CT) per manufacturer's instructions. These were used in Taqman analyses and ELISA. Lysates were homogenized by passing five times though a 22g-needle attached to 1cc syringe. Homogenates were stored at −80 °C till extraction of RNA and protein, performed as per manufacturer's instructions. Briefly, antisense-treated cells were lysed in kit-supplied buffer, supplemented with 1% 2-mercaptoethanol. After removing gDNA, acetone was added to precipitate RNA, subsequently bound to silica columns and later collected in Elution buffer. Flow-through at this step contained cellular proteins. Kit-supplied precipitation solution was used to pellet the proteins, subsequently solubilized in 2D-DIGE buffer (7 mol/l urea, 2 mol/l thiourea, 4% (v/v) 3-[(3-cholamidopropyl)dimethylammonio]-1- propanesulfonate (CHAPS), 30 mmol/l Tris-HCl, pH 8.5). For immunoblots, tau levels were analyzed in cytoplasmic lysates prepared in the following buffer: 125 mmol/l Tris-HCl (pH 6.8), 200 mmol/l NaCl, 2.5 mmol/l MgSO4, 0.1% Triton-X, and 0.5% NP-40. Immediately before addition to cell pellets, lysis buffer was supplemented with protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Tau isoforms expressed in neuroblastoma lysates were identified by running recombinant standard (Sigma, St Louis, MO #T7951).
Reverse transcription–quantitative RT-PCR analysis of MAPT transcripts. RT-PCR experiment design and reporting of results is based on Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines.46 The concentration and purity of isolated RNA was determined using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA). Representative RNA samples from different preparations were also assessed for integrity of isolated RNA using an Agilent 2100 bioanalyzer. RNA template in each experimental paradigm was reverse transcribed to cDNA using First-Strand cDNA Synthesis Kit (GE Healthcare) or the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Changes in MAPT expression were detected quantitative real-time PCR was performed using a 7900HT Fast Real Time PCR System (Applied Biosystems) with TaqMan Fast Universal PCR Master Mix and Applied Biosystems TaqMan Gene Expression Assay Mix: ID# Hs00902194_m1. This probe binds the junction of MAPT exons 12–13; constitutive exons present in all isoforms and therefore this assay helps determine total levels of all MAPT transcripts. For 3R isoform detection, assay Hs00902192_m1 was used with a probe that binds to the exon 9 – exon 11 junction fragment. For 4R isoform detection, assay Hs00902312_m1 was used with a probe that binds exon 9-exon 10 junction sequence).
RT-PCR data analysis. Fold changes were calculated by the 2−ΔΔCt compared to mock-transfected (“no oligo”) cultures. For this control, ΔΔCt equals zero and 20 equals one, so that the fold change in gene expression relative to the mock-transfected control equals one by definition.47 Data were normalized to endogenous reference genes (18S rRNA, β-actin, GAPDH) and plotted as % change relative to the mock or “no oligonucleotide” control in figures.
PCR and sequencing analysis to confirm exon skipping. To verify morpholino oligos did exclude targeted exons, cDNA from treated cells was amplified with PCR primers complementary to MAPT sequences outside the skipped exon (Supplementary Table S2). Subsequent analyses revealed shorter transcript fragments in oligo-treated cultures, when compared to mock-cultures. Parameters for PCR amplification were: 95 °C/15 minutes; 30 cycles of 94 °C/30 seconds, 60 °C/30 seconds, 72 °C/1 minute; 72 °C/10 minutes. PCR products were gel-isolated and sequenced on ABI 3130xl genetic analyzer. Sequence outputs were blatted on the UCSC Genome Browser to ensure mapping to MAPT.
Protein analysis. Total cellular protein extracted was quantified with Qubit assay (Invitrogen, Carlsbad, CA). Cell lysates were dephosphorylated using λ-phosphatase (New England Biolabs, Ipswich, MA) as previously described.48 Immunoblotting was performed using previously published protocols.35 Primary antibody for detecting differences in tau levels post-antisense treatment was purchased from Santa Cruz Biotechnology (Dallas, TX) (sc-21796, at 1:1,000). For detection, secondary antibody #NA931V (GE Healthcare) was used at 1:20k. Both antibodies were diluted in 5% nonfat milk in 1× phosphate-buffered saline (Gibco, Carlsbad, CA) supplemented with 0.1% Tween-20. B-actin (Cell Signaling, Beverly, MA) was used as loading control. Morpholino-induced changes in tau protein were quantified with ELISA (Invitrogen) per manufacturer's instructions.
Intramuscular injections in transgenic mice expressing human tau. The intramuscular (i.m.) injections were used a proxy model to explore whether morpholino oligonucleotides that effectively reduced tau protein levels in vitro would work in vivo. The transgenic mouse used was described previously (http://jaxmice.jax.org/strain/017459.html)35 and harbors a point mutation in intron 10 (E10+14) that causes frontotemporal dementia. Morpholino E5.3 was injected intramuscularly in the left gastrocnemius muscles. Control group included animals injected with negative control morpholino, saline, and with vehicle—Kollliphor P407 (formerly called Pluronic F-127), a kind gift from BASF, White Plains, NY. P407 is a nonionic polymer previously used successfully in DMD mouse models to facilitate intracellular delivery of antisense oligonucleotides following i.m. administration.24 Animals were injected with 20 µg/µl morpholino and P407 at a final concentration of 40 µg/µl, diluted in saline, in a total volume of 30 µl per muscle. Vivo-morpholinos were injected at the same volume at 3 µg/µl.
Histochemical analyses. Hematoxylin and eosin staining was performed according to standard histological techniques. Samples included noninjected controls, gastrocnemius muscles from: saline-injected, vivo-morpholino injected, morpholino + P407 coinjected tissue. Oligonucleotides injected included both tau-specific and negative control morpholinos. Stained slides were visualized using a Leica DM4000b microscope and photographed with a Leica DFC 450 camera.
Statistical analysis. In case of RT-PCR (TaqMan) and ELISA experiments, changes to tau levels were compared to “mock”, or no-oligo control. Statistical evaluation was performed using unpaired t-tests.
SUPPLEMENTARY MATERIAL Figure S1. Treatment with exon 0-specific antisense oligo did not affect MAPT transcript levels Figure S2. Targeting splice sites to exon 9 with morpholinos did not influence tau levels. Figure S3. Targeting splice sites to exon 4 with morpholinos did not influence tau levels. Figure S4. MAPT transcript levels in cells treated with oligonucleotides targeting exon 10. Figure S5. Untreated neuroblastoma predominantly expressed 0N tau isoform. Figure S6. Exon 1 is omitted after treatment with morpholino oligos targeting splice sites to this exon. Figure S7. Effect of varying concentrations of E5.3 antisense oligo on total MAPT transcripts in IMR32 cells. Figure S8. Exon 5 is skipped after treatment with morpholino oligos targeting splice sites to this exon. Figure S9. MAPT expression was decreased after treatment with exon 7-targeting morpholino oligonucleotides. Figure S10. Exon 4 is excluded after its splice sites are masked. Figure S11. Histological analyses of injected muscles comparing morpholino oligonucleotides of different chemistries Table S1. 4R isoform mRNA content in untreated cells. Table S2. Primers used to sequence MAPT in cDNA isolated from morpholino-treated cultures or corresponding controls.
Acknowledgments
This work was supported by National Institutes of Health grant R37 AG 11762. We thank Beth A Dombroski for her technical assistance. Michele Hawk and Doori Jeong helped with the transgenic mice experiments. We are grateful to Edward Lee for his expertise on histopathological analyses. The authors declare no conflict of interest.
Supplementary Material
Treatment with exon 0-specific antisense oligo did not affect MAPT transcript levels
Targeting splice sites to exon 9 with morpholinos did not influence tau levels.
Targeting splice sites to exon 4 with morpholinos did not influence tau levels.
MAPT transcript levels in cells treated with oligonucleotides targeting exon 10.
Untreated neuroblastoma predominantly expressed 0N tau isoform.
Exon 1 is omitted after treatment with morpholino oligos targeting splice sites to this exon.
Effect of varying concentrations of E5.3 antisense oligo on total MAPT transcripts in IMR32 cells.
Exon 5 is skipped after treatment with morpholino oligos targeting splice sites to this exon.
MAPT expression was decreased after treatment with exon 7-targeting morpholino oligonucleotides.
Exon 4 is excluded after its splice sites are masked.
Histological analyses of injected muscles comparing morpholino oligonucleotides of different chemistries
4R isoform mRNA content in untreated cells.
Primers used to sequence MAPT in cDNA isolated from morpholino-treated cultures or corresponding controls.
References
- Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- Poorkaj P, Bird TD, Wijsman E, Nemens E, Garruto RM, Anderson L, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43:815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc Natl Acad Sci USA. 1998;95:7737–7741. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, et al. Association of missense and 5'-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Hong M, Zhukareva V, Vogelsberg-Ragaglia V, Wszolek Z, Reed L, Miller BI, et al. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science. 1998;282:1914–1917. doi: 10.1126/science.282.5395.1914. [DOI] [PubMed] [Google Scholar]
- D'Souza I, Poorkaj P, Hong M, Nochlin D, Lee VM, Bird TD, et al. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc Natl Acad Sci USA. 1999;96:5598–5603. doi: 10.1073/pnas.96.10.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, Klei L, et al. PSP Genetics Study Group Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8:711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- Simón-Sánchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, et al. Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature. 1994;369:488–491. doi: 10.1038/369488a0. [DOI] [PubMed] [Google Scholar]
- Ikegami S, Harada A, Hirokawa N. Muscle weakness, hyperactivity, and impairment in fear conditioning in tau-deficient mice. Neurosci Lett. 2000;279:129–132. doi: 10.1016/s0304-3940(99)00964-7. [DOI] [PubMed] [Google Scholar]
- Takei Y, Teng J, Harada A, Hirokawa N. Defects in axonal elongation and neuronal migration in mice with disrupted tau and map1b genes. J Cell Biol. 2000;150:989–1000. doi: 10.1083/jcb.150.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Ishihara T, Zhang B, Hong M, Andreadis A, Trojanowski J, et al. Transgenic mouse model of tauopathies with glial pathology and nervous system degeneration. Neuron. 2002;35:433–446. doi: 10.1016/s0896-6273(02)00789-4. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Hong M, Zhang B, Nakagawa Y, Lee MK, Trojanowski JQ, et al. Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform. Neuron. 1999;24:751–762. doi: 10.1016/s0896-6273(00)81127-7. [DOI] [PubMed] [Google Scholar]
- Lewis J, McGowan E, Rockwood J, Melrose H, Nacharaju P, Van Slegtenhorst M, et al. Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat Genet. 2000;25:402–405. doi: 10.1038/78078. [DOI] [PubMed] [Google Scholar]
- Götz J, Chen F, Barmettler R, Nitsch RM. Tau filament formation in transgenic mice expressing P301L tau. J Biol Chem. 2001;276:529–534. doi: 10.1074/jbc.M006531200. [DOI] [PubMed] [Google Scholar]
- Clavaguera F, Bolmont T, Crowther RA, Abramowski D, Frank S, Probst A, et al. Transmission and spreading of tauopathy in transgenic mouse brain. Nat Cell Biol. 2009;11:909–913. doi: 10.1038/ncb1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba M, Guo JL, McBride JD, Zhang B, Trojanowski JQ, Lee VM. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer's-like tauopathy. J Neurosci. 2013;33:1024–1037. doi: 10.1523/JNEUROSCI.2642-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, et al. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Delerue F, Bi M, Gladbach A, van Eersel J, et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- Shipton OA, Leitz JR, Dworzak J, Acton CE, Tunbridge EM, Denk F, et al. Tau protein is required for amyloid {beta}-induced impairment of hippocampal long-term potentiation. J Neurosci. 2011;31:1688–1692. doi: 10.1523/JNEUROSCI.2610-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossel KA, Zhang K, Brodbeck J, Daub AC, Sharma P, Finkbeiner S, et al. Tau reduction prevents Abeta-induced defects in axonal transport. Science. 2010;330:198. doi: 10.1126/science.1194653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aartsma-Rus A, van Vliet L, Hirschi M, Janson AA, Heemskerk H, de Winter CL, et al. Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms. Mol Ther. 2009;17:548–553. doi: 10.1038/mt.2008.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA, et al. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med. 2003;9:1009–1014. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N, et al. Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- Andreadis A, Brown WM, Kosik KS. Structure and novel exons of the human tau gene. Biochemistry. 1992;31:10626–10633. doi: 10.1021/bi00158a027. [DOI] [PubMed] [Google Scholar]
- Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc Natl Acad Sci U S A. 1988;85:4051–4055. doi: 10.1073/pnas.85.11.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 1989;8:393–399. doi: 10.1002/j.1460-2075.1989.tb03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza I, Schellenberg GD. Determinants of 4-repeat tau expression. Coordination between enhancing and inhibitory splicing sequences for exon 10 inclusion. J Biol Chem. 2000;275:17700–17709. doi: 10.1074/jbc.M909470199. [DOI] [PubMed] [Google Scholar]
- D'Souza I, Schellenberg GD. Arginine/serine-rich protein interaction domain-dependent modulation of a tau exon 10 splicing enhancer: altered interactions and mechanisms for functionally antagonistic FTDP-17 mutations Delta280K AND N279K. J Biol Chem. 2006;281:2460–2469. doi: 10.1074/jbc.M505809200. [DOI] [PubMed] [Google Scholar]
- D'Souza I, Schellenberg GD. tau Exon 10 expression involves a bipartite intron 10 regulatory sequence and weak 5' and 3' splice sites. J Biol Chem. 2002;277:26587–26599. doi: 10.1074/jbc.M203794200. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Anderton BH, Davis DR, Gallo JM. Tau isoform expression and phosphorylation state during differentiation of cultured neuronal cells. FEBS Lett. 1995;375:243–248. doi: 10.1016/0014-5793(95)01221-y. [DOI] [PubMed] [Google Scholar]
- Poorkaj P, Kas A, D'Souza I, Zhou Y, Pham Q, Stone M, et al. A genomic sequence analysis of the mouse and human microtubule-associated protein tau. Mamm Genome. 2001;12:700–712. doi: 10.1007/s00335-001-2044-8. [DOI] [PubMed] [Google Scholar]
- McMillan P, Korvatska E, Poorkaj P, Evstafjeva Z, Robinson L, Greenup L, et al. Tau isoform regulation is region- and cell-specific in mouse brain. J Comp Neurol. 2008;511:788–803. doi: 10.1002/cne.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Takamatsu J, D'Souza I, Crowther RA, Kawamata T, Hasegawa M, et al. A novel mutation at position +12 in the intron following exon 10 of the tau gene in familial frontotemporal dementia (FTD-Kumamoto) Ann Neurol. 2000;47:422–429. [PubMed] [Google Scholar]
- Kalbfuss B, Mabon SA, Misteli T. Correction of alternative splicing of tau in frontotemporal dementia and parkinsonism linked to chromosome 17. J Biol Chem. 2001;276:42986–42993. doi: 10.1074/jbc.M105113200. [DOI] [PubMed] [Google Scholar]
- DeVos SL, Goncharoff DK, Chen G, Kebodeaux CS, Yamada K, Stewart FR, et al. Antisense reduction of tau in adult mice protects against seizures. J Neurosci. 2013;33:12887–12897. doi: 10.1523/JNEUROSCI.2107-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Ding H, Inoue S, Ljubimov AV, Patil R, Portilla-Arias J, Hu J, et al. Inhibition of brain tumor growth by intravenous poly (β-L-malic acid) nanobioconjugate with pH-dependent drug release [corrected] Proc Natl Acad Sci USA. 2010;107:18143–18148. doi: 10.1073/pnas.1003919107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, et al. Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Hall E, Ast G. SROOGLE: webserver for integrative, user-friendly visualization of splicing signals. Nucleic Acids Res. 2009;37 Web Server issue:W189–W192. doi: 10.1093/nar/gkp320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Biol. 2011;731:237–245. doi: 10.1007/978-1-61779-080-5_20. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Hanger DP, Gibb GM, de Silva R, Boutajangout A, Brion JP, Revesz T, et al. The complex relationship between soluble and insoluble tau in tauopathies revealed by efficient dephosphorylation and specific antibodies. FEBS Lett. 2002;531:538–542. doi: 10.1016/s0014-5793(02)03611-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Treatment with exon 0-specific antisense oligo did not affect MAPT transcript levels
Targeting splice sites to exon 9 with morpholinos did not influence tau levels.
Targeting splice sites to exon 4 with morpholinos did not influence tau levels.
MAPT transcript levels in cells treated with oligonucleotides targeting exon 10.
Untreated neuroblastoma predominantly expressed 0N tau isoform.
Exon 1 is omitted after treatment with morpholino oligos targeting splice sites to this exon.
Effect of varying concentrations of E5.3 antisense oligo on total MAPT transcripts in IMR32 cells.
Exon 5 is skipped after treatment with morpholino oligos targeting splice sites to this exon.
MAPT expression was decreased after treatment with exon 7-targeting morpholino oligonucleotides.
Exon 4 is excluded after its splice sites are masked.
Histological analyses of injected muscles comparing morpholino oligonucleotides of different chemistries
4R isoform mRNA content in untreated cells.
Primers used to sequence MAPT in cDNA isolated from morpholino-treated cultures or corresponding controls.