Abstract
Antisense-based molecules targeting HIV-1 RNA have the potential to be used as part of gene or drug therapy to treat HIV-1 infection. In this study, HIV-1 RNA was screened to identify more conserved and accessible target sites for ribozymes based on the hepatitis delta virus motif. Using a quantitative screen for effects on HIV-1 production, we identified a ribozyme targeting a highly conserved site in the Gag coding sequence with improved inhibitory potential compared to our previously described candidates targeting the overlapping Tat/Rev coding sequence. We also demonstrate that this target site is highly accessible to short hairpin directed RNA interference, suggesting that it may be available for the binding of antisense RNAs with different modes of action. We provide evidence that this target site is structurally conserved in diverse viral strains and that it is sufficiently different from the human transcriptome to limit off-target effects from antisense therapies. We also show that the modified hepatitis delta virus ribozyme is more sensitive to a mismatch in its target site compared to the short hairpin RNA. Overall, our results validate the potential of a new target site in HIV-1 RNA to be used for the development of antisense therapies.
Keywords: HIV-1, reverse transcriptase, ribozyme, short hairpin RNA, viral kinetics
Introduction
Over 30 small molecules are available for the treatment of Human Immunodeficiency Virus 1 (HIV-1) infection, targeting the viral enzymes reverse transcriptase (RT), protease and integrase, as well as the cellular entry coreceptor, CCR5.1 Although treatment of HIV-1 with combination small molecule therapy is effective in preventing acquired immune deficiency syndrome, it cannot eradicate the virus and is associated with a number of short- and long-term side effects.2 Alternative therapeutic strategies for long-term viral suppression with low adverse effects are needed and small RNAs represent a growing class of molecules with the potential to complement or replace current therapies. They are being evaluated for use in ex vivo gene therapy3 and with advances that have been made in their systemic delivery,4 may soon be evaluated for use in combination drug therapy. Many small RNAs, including antisense oligonucleotides (ASONs), ribozymes (Rzs), decoys, aptamers, small nuclear (sn) RNAs, and small interfering (si) or short hairpin (sh) RNAs have been designed with diverse target sites in the HIV-1 replication cycle.5 Antisense-based RNAs (ASONs, Rzs, snRNAs, sh/si RNAs) can be designed to target HIV-1 RNA, and several therapeutic candidates have been described.
Rzs targeting HIV-1 RNA have been made by modifying hammerhead, hairpin6 and bacterial RNase P7 motifs. The HDV Rz represents an alternative small Rz motif, that has evolved to function in human cells and has the potential to be used for the development of therapeutic Rzs.8 To improve the specificity of the HDV Rz for its target RNA, the SOFA (Specific On/oFf Adaptor) module was engineered9,10 (Figure 1a). Several SOFA-HDV-Rzs have been identified with the potential to target human,11,12 viral9,13,14 and bacterial15 RNAs, including three Rzs that we have evaluated targeting the overlapping Tat/Rev coding sequence of HIV-1 RNA.16
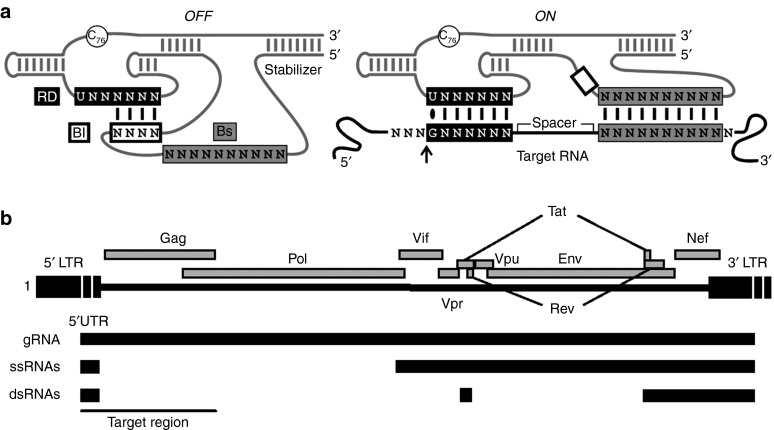
Figure 1.
Schematic representation of the SOFA-HDV-Rz and the HIV-1 RNA region used to identify SOFA-HDV-Rz target sites. (a) The SOFA-HDV-Rz is illustrated in both its OFF and ON conformations. In the OFF conformation, the SOFA blocker (Bl) base pairs with the last four nucleotides (nts) of the recognition domain (RD). When the SOFA biosensor (Bs) base pairs with a specific target sequence, the RD is released from the Bl sequence and binds at three to five nts upstream from the Bs binding site in the ON conformation. The first nt in the target site (n + 1) must be a G, forming a wobble base pair with the RD U. The cleavage site is indicated with an arrow and the nt C76, which can be mutated to disable the catalytic activity of the SOFA-HDV-Rz, is shown as a circle in the Rz backbone. (b) The full-length genomic (g), singly-spliced (ss), and doubly-spliced (ds) RNA species of HIV-1 are illustrated. Reading frames for all HIV-1 proteins are shown above the different RNAs and the 5′ region, used to identify SOFA-HDV-Rz target sites, is underlined.
Optimal hammerhead Rz target sites in HIV-1 RNA have been identified using libraries of Rzs with randomized binding arms17,18 and a library of Rzs targeting highly conserved sequences.19 Using different methods and datasets to estimate sequence conservation, sets of optimal siRNAs20 or shRNAs21,22 have been identified and two of these studies have reported their conservation estimates in 19–21 nucleotide (nt) frames.20,22 Estimates have also been reported at the nt level to identify or characterize Rz,23 snRNA,24 and shRNA25 target sites. In this study, we generated conservation estimates at the nt level to identify a set of optimal SOFA-HDV-Rz target sites from the beginning of the 5′ untranslated region (UTR) to the end of the Gag coding sequence (Figure 1b). Out of 18 new SOFA-HDV-Rzs, one Rz targeting a novel site in the Gag coding sequence was particularly effective at reducing viral production. An shRNA targeting the same site was shown to be an extremely potent inhibitor of viral production and evidence that the identified target site is accessible to inhibition in diverse HIV-1 strains is provided.
Results
Identification of SOFA-HDV-Rz target sites in HIV-1 RNA
HIV-1 sequence conservation was estimated to identify target sites that are relevant for the majority of HIV-1 strains. Estimates at the nt level were made (Supplementary File S1) using all complete sequences available in the Los Alamos National Laboratory (LANL) database (1850 at the time of analysis, subtype distribution shown in Supplementary Figure S1). These estimates were used to identify highly conserved SOFA-HDV-Rz target sites (Figure 2a) that were identical in HIV-1 strain NL4-3 (Figure 2b). The Ribosubstrates informatics tool26 was used to exclude SOFA-HDV-Rzs targeting 12 highly conserved and 19 moderately conserved regions in HIV-1 RNA, due to their potential to target human RNAs.
Figure 2.
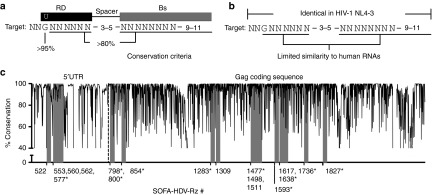
SOFA-HDV-Rz target site identification. (a) Criteria used to identify SOFA-HDV-Rz target sites in HIV-1 RNA based on our conservation estimates at the nucleotide (nt) level are illustrated. The number of nts between the RD and the Bs (spacer, 3–5), and the length of the Bs (9–11), were adjusted to avoid poorly conserved positions or to reduce potential off-target effects on human RNAs. A spacer of 4 nt and Bs length of 10 nt were used as the default positioning. (b) Target sites were excluded if they were not identical in HIV-1 NL4-3 or if the corresponding Rz had potential target sites in human RNAs using a cut-off score of 20 in the Ribosubstrates tool.26 (c) Sequence conservation estimates in the 5′ region of HIV-1 RNA are shown for each nt position in HIV-1 NL4-3 with the selected Rz binding sites shaded in grey. SOFA-HDV-Rzs were named according to the first nt in their binding site. The dashed line represents the separation between the 5′UTR and Gag ORF. SOFA-HDV-Rz target sites that were moderately conserved, but did not meet our conservation criteria, are indicated with an asterisk (*).
Consistent with previous studies,20,22 several highly conserved target sites were identified in the 5′LTR U5 region within the 5′UTR (Figure 2c). The Gag coding sequence had much lower overall conservation; however, four highly conserved and, with some exceptions to our conservation criteria (Figure 2a), nine moderately conserved target sites were identified in this region (Figure 2c). Of the Rzs that we have previously evaluated targeting the Tat/Rev exon1 coding sequence of HIV-1 RNA,16 the target sites for Tat1 and Tev1 were highly and moderately conserved, respectively. Conservation exceptions and sequences of all target sites used in this study are illustrated, along with the DNA sequences of the corresponding SOFA-HDV-Rz variable regions, in Supplementary Table S1.
SOFA-HDV Rz screen for inhibition of HIV-1 production
The effect of each SOFA-HDV-Rz expressing plasmid on HIV-1 production was evaluated by cotransfection with HIV-1 molecular clone pNL4-3 in HEK293T cells, using conditions similar to those reported for other Rzs7,18,27 and shRNAs.21,22 HIV-1 RT activity was measured to estimate the production of virus released into the medium of transfected cells and effects of Rzs were normalized to cotransfection of pNL4-3 with an empty Rz expression plasmid. An irrelevant Rz targeting Hepatitis B Virus RNA (SOFA-HDV-RzHBV, adapted from SOFA-δRz-3039) was used as a negative control and previously described SOFA-HDV-Rzs Tat1 and Tev116 were used as positive controls. Compared to SOFA-HDV-RzHBV, Rzs targeting both the 5′UTR and Gag coding sequences significantly inhibited viral production, with the top candidate (SOFA-HDV-Rz-1498) targeting the Gag coding sequence (Figure 3).
Figure 3.
Inhibition of HIV-1 production by SOFA-HDV-Rzs. HEK293T cells were seeded in 24-well plates and cotransfected with HIV-1 pNL4-3 plasmid DNA (75 ng) and one of the indicated psiRNA SOFA-HDV-Rz expression plasmids (750 ng). Viral production was estimated 48 hours following transfection by measuring the activity of HIV-1 RT in culture supernatants. Each replicate was expressed as a percentage of the value obtained for cotransfection with the empty Rz expression plasmid tested in parallel (Relative RT activity). Rzs were evaluated in at least three independent experiments with one to three replicate transfections, data are expressed as the mean ± SEM (n = 5–10). Graph Pad Prism was used to calculate P values for the effects of each HIV-1 specific SOFA-HDV-Rz compared to the irrelevant control (-RzHBV). Results from un-paired t-tests are shown above each SOFA-HDV-Rz that demonstrated a significant inhibition of viral production compared to the control (*P < 0.05, **P < 0.01, ***P < 0.001).
Antisense and mismatched variants of SOFA-HDV-Rz1498 are not effective inhibitors of HIV-1 production
To evaluate the antisense effect of SOFA-HDV-Rz1498, we generated an inactive variant (SOFA-HDV-Rz1498A76) in which a C to A mutation at position 76 in its backbone disables its cleaving capability28 (Figure 4a). SOFA-HDV-Rz1498A76 did not significantly inhibit HIV-1 production at similar expression levels to SOFA-HDV-Rz1498 (Figure 4b), suggesting that Rz catalytic cleavage is primarily responsible for the effects of SOFA-HDV-Rz1498. No effect on the infectivity of virus from SOFA-HDV-Rz1498 expressing cells was observed (Supplementary Figure S2), suggesting that the Rz reduces the amount of virus produced but does not affect the quality of the virions.
Figure 4.
Effects of SOFA-HDV-Rz1498 variants on HIV-1 production. (a) Schematic representation of the SOFA-HDV-Rz1498 target site (T) and variants (Rz). SOFA-HDV-Rz1498A76 has a C to A mutation in the Rz backbone, -Rz1498Bs1 and -Rz1498Bs2 have one or two nucleotide (nt) variants in the biosensor (Bs), indicated in lower case. (b) Effects of each SOFA-HDV-Rz1498 variant on viral production in HEK293T cells were evaluated exactly as in Figure 3. Rzs were evaluated in at least three independent experiments with one to three replicate transfections (reported as mean ± SEM, n = 6–10). The relative expression of Rz and 5S RNA loading control for the different conditions are shown below for one of two independent experiments performed in HEK293T cells seeded in a 12-well plate and cotransfected with twice the amount of DNA used for the evaluation of viral production in 24-well plates. (c) Single turnover in vitro cleavage activities for SOFA-HDV-Rz1498, -Rz1498Bs1, and -Rz1498Bs2 were determined at different incubation times with a small substrate RNA (Rz>>substrate). Cleavage % was measured by dividing cleaved products by cleaved + uncleaved products, quantified from bands on a gel. A nonlinear regression one phase exponential association equation with least squares (ordinary) fit was determined using Graph Pad Prism for the different Rzs. All data points represent two independent experiments and are reported as mean ± SEM (n = 2). The average rate constants (kobs) and maximum cleavage values (Fmax) for the SOFA-HDV-Rzs are reported in a table.
SOFA-HDV-Rz1498 variants with either a single or double mutation in their biosensor (Bs) sequence were also generated to evaluate the potential for SOFA-HDV-Rz1498 to tolerate mismatches with its target (SOFA-HDV-Rz1498Bs1 and SOFA-HDV-Rz1498Bs2, Figure 4a). Neither variant inhibited HIV-1 production (Figure 4b), suggesting that the effect of SOFA-HDV-Rz1498 is highly sensitive to mismatches with its target. The mismatched Rzs had similar in vitro cleavage rate constants (kobs) with significantly reduced maximum cleavage (Fmax) values (Figure 4c), suggesting that part of their failure to inhibit HIV-1 production in cells is related to a reduced capacity to cleave their target.
An shRNA targeting the 1498 site is a potent inhibitor of HIV-1 production and provides an additive effect in combination with SOFA-HDV-Rz1498
To evaluate the potential for other antisense molecules targeting the SOFA-HDV-Rz1498 target site to inhibit HIV-1 production, we designed shRNA1498. According to our conservation estimates, each nt in the shRNA1498 target site was conserved at >80% (Figure 5a, Gag). Compared to a nonsense shRNA (shRNAns) as well as SOFA-HDV-Rz1498 and its controls, shRNA1498 provided a near complete inhibition of viral production (Figure 5b). This inhibition correlated with a decrease in intracellular expression of the HIV-1 Gag polyprotein and one of its processing products, capsid (CA). Unexpectedly, the decrease in CA expression was much more pronounced for both the Rz and shRNA compared to their effects on Gag expression. A similar effect was observed for shRNAs targeting sequences in the 5′UTR and Tat/Rev coding sequences of HIV-1 RNA (Supplementary Figure S3), suggesting that it is not specific to an shRNA targeting the Gag 1498 sequence.
Figure 5.
Effects of shRNA1498 on HIV-1 production (a) Sequences targeted by shRNA1498 and control shRNAs targeting HIV-1 RNA (shRNA522, shRNA553, and shRNA5983) are shown in relation to our conservation estimates at the nt level from Supplementary File S1. (b) Effects of shRNA1498 and a nonsense shRNA (shRNAns) on viral production in HEK293T cells were evaluated exactly as in Figure 3. Rzs and shRNAs were evaluated in at least three independent experiments with one to three replicate transfections (reported as mean ± SEM, n = 6–10). The relative intracellular expression of HIV-1 Gag polyprotein (Gag, p55), matrix-capsid intermediate (MA-CA, p39), and capsid (CA, p24) proteins as well as GAPDH loading control are shown below for one of two independent experiments performed in HEK293T cells seeded in a 12-well plate and cotransfected with twice the amount of DNA used for the evaluation of viral production. Relative band intensities for Gag and CA were calculated using Image J software and are expressed as a fraction of the intensity of Gag in the SOFA-HDV-RzHBV control lane. (c) The potency of shRNAs was evaluated by cotransfecting HEK293T cells seeded in a 24-well plate with 100 ng of pNL4-3 DNA and 1–750 ng of shRNA expressing plasmids. For lower amounts of shRNA plasmid DNA (1–500 ng), cotransfections were topped up to 850 ng total DNA by the addition of an irrelevant plasmid (pBluescript SK+, Stratagene, La Jolla, CA). Relative RT activity measurements were log transformed and a nonlinear regression log(inhibitor) versus response equation with least squares (ordinary) fit was determined using Graph Pad Prism for the different shRNAs. EC50 values from this equation are mentioned in the text. All data-points represent at least two independent experiments with two to three replicates and are reported as mean ± SEM (n = 4–8). (d) Combinations of SOFA-HDV-Rz and shRNA expressing plasmids were evaluated in HEK293T cells seeded in 24-well plates and cotransfected with 100 ng pNL4-3, 10 ng of shRNA expressing plasmid and 1 µg of Rz expressing plasmid. Data were normalized to cotransfection of 100 ng pNL4-3 with 1 µg of the empty Rz/shRNA expression plasmid and are reported as the mean ± SEM from two independent experiments performed in triplicate (n = 6).
To compare the potency of shRNA1498 to other candidate shRNAs, we designed shRNA522 and shRNA553, modeled after previously characterized siRNAs20 and shRNAs22 targeting the 5′UTR (Figure 5a, 5′UTR) and shRNA5983, modeled after a construct in clinical development targeting the Tat/Rev exon1 coding sequence29 (Figure 5a, Tat/Rev). All four shRNAs inhibited HIV-1 production (Figure 5c). The potency of shRNA1498 was comparable to that of shRNA553 and shRNA5983, with 50% effective concentrations (EC50s) for shRNA plasmids below 5 ng of input DNA, whereas shRNA522 was much less potent with an EC50 value of 702 ng.
To evaluate the potential for shRNA1498 and shRNA5983 to be used in combination with SOFA-HDV-Rz1498, we cotransfected HEK293T cells with HIV-1 pNL4-3 and different combinations of Rzs and shRNAs (Figure 5d). To quantify the effect of the combinations, we chose an input level of shRNA DNA that did not completely inhibit viral production in Figure 5c. In combination with both shRNA1498 and shRNA5983, SOFA-HDV-Rz1498 provided an additional inhibition of HIV-1 production compared to the control Rz, SOFA-HDV-Rz-HBV. The level of inhibition was similar to its effect when cotransfected alone (50%) (Figure 5b), suggesting that the Rz can provide an additive effect in combination with both an shRNA targeting the same site (shRNA1498) and an shRNA targeting a different site (shRNA5983).
SOFA-HDV-Rz1498 and shRNA1498 inhibit viral production from diverse HIV-1 strains
As the Gag 1498 target site was accessible to inhibition by both a Rz and an shRNA in HIV-1 strain NL4-3, we next evaluated whether this inhibition extended to diverse viral strains representing subtype B (Mal30 and AD831), C (Indie-C132 and MJ433), D (94UG11434) and circulating recombinant form (CRF) 02_AG (97GH-AG135). SOFA-HDV-Rz1498 inhibited HIV-1 production from viral strains (Mal, AD8, MJ4, and 97GH-AG1) with nt variants in proximity to their target sites compared to NL4-3 (Figure 6), suggesting that the structure of the target site is equally accessible to the Rz in these strains. Consistent with results using SOFA-HDV-Rz binding site variants (Figure 4, -Bs1 and -Bs2), SOFA-HDV-Rz1498 did not inhibit HIV-1 production from the strains Indie-C1 and 94UG114, which harbor a single nt variant within their Bs binding sites (Figure 6). In contrast, shRNA1498 inhibited HIV-1 production from all strains suggesting that it can tolerate a single nt mismatch in its binding site at position 17 and can inhibit HIV-1 production in diverse strains.
Figure 6.

Inhibition of HIV-1 production from diverse viral strains by SOFA-HDV-Rz1498 and shRNA1498. (a) The sequence in and around the shRNA1498 and SOFA-HDV-Rz1498 target site is shown for HIV-1 NL4-3 (M19921), MAL (K03456), AD8 (AF004394), Indie-C1 (AB023804.1), MJ4 (AF321523), 94UG114 (U88824.1), and 97GH-AG1 (AB049811.1). The overlapping target site for shRNA1498 and SOFA-HDV-Rz1498 is highlighted in blue, with both the RD (7 nucleotide (nt)) and Bs (10 nt) binding sites underlined. Nt variations compared to HIV-1 NL4-3 are highlighted in red and the start and end positions for each sequence are shown according to their annotation in Genbank. (b) Effects of SOFA-HDV-Rz1498, shRNA1498 and their controls on HIV-1 production from different molecular clones in HEK293T cells were evaluated exactly as in Figure 3, with at least two independent experiments, each including two to three replicate transfections (reported as mean ± SEM, n = 4–10).
SOFA-HDV-Rz1498 and shRNA1498 have minimal off-target effects on human RNAs
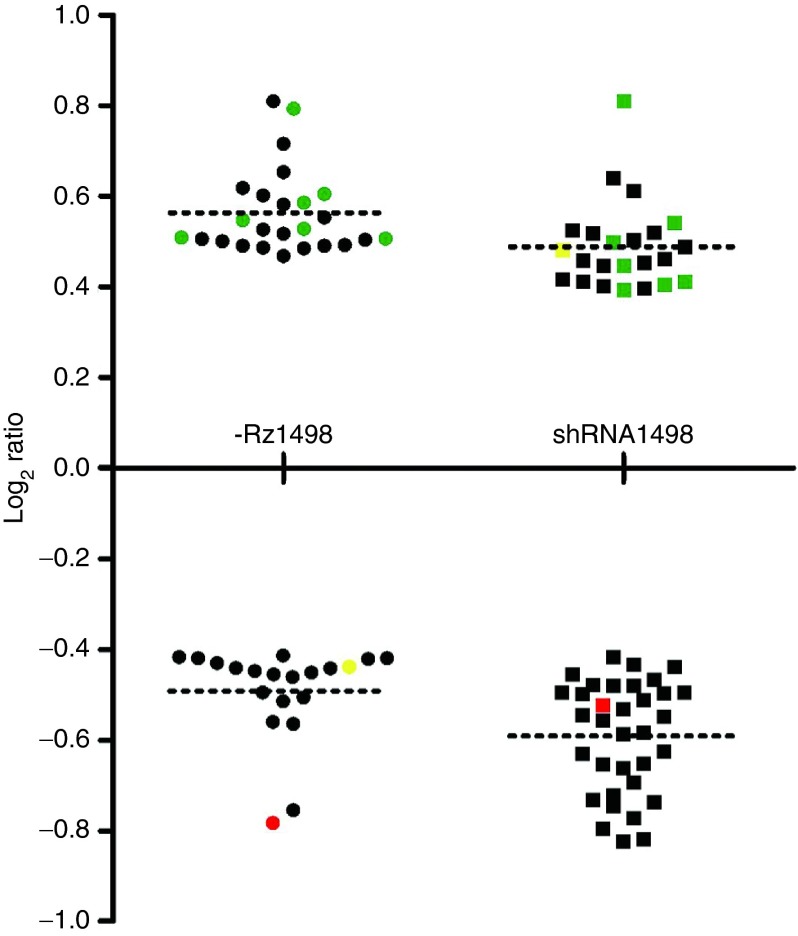
The potential for both SOFA-HDV-Rz1498 and shRNA1498 to affect the expression of human mRNAs was next evaluated in HEK293T cells cotransfected with HIV-1 pNL4-3. Prior to gene expression profiling, the inhibition of viral production was confirmed for each condition (data not shown) and agreed with results presented in Figure 5b. Microarray experiments were performed as triplicate dye swaps and the results were expressed as the log2 ratio of SOFA-HDV-Rz1498 or shRNA1498 compared to the empty vector cotransfected cells (Supplementary Figure S4). All average log2 ratios were low (below 1.0) suggesting that both SOFA-HDV-Rz1498 and shRNA1498 can inhibit HIV-1 production with minimal effects on human mRNA expression. The log2 ratios for mRNAs with the greatest extent of up- or downregulation are illustrated in Figure 7 and listed in Supplementary File S2. Several of these mRNAs were found in both SOFA-HDV-Rz1498 and shRNA1498 conditions, suggesting that part of the observed changes may be target site specific.
Figure 7.
Gene expression changes in cells transfected with SOFA-HDV-Rz1498 and shRNA1498. HEK293T cells were seeded in 12-well plates and cotransfected with HIV-1 pNL4-3 (150 ng) and either SOFA-HDV-Rz1498, shRNA1498 or the empty Rz/shRNA expression plasmid (1.5 µg). Forty-eight hours after transfection, total RNA was harvested and analyzed by micro-array. The log2 ratios of mRNAs with the greatest differential variation between SOFA-HDV-Rz1498 or shRNA1498 transfected cells and the empty expression vector transfected cells are shown. RNAs that were up- or downregulated in both conditions are shown in green and red respectively. One gene that was downregulated by SOFA-HDV-Rz1498 and upregulated by shRNA1498 is shown in yellow. The gene identities and log2 ratio values are provided in Supplementary File S2.
SOFA-HDV-Rz1498 and shRNA1498 inhibit HIV-1 replication in a T lymphocyte cell line
To evaluate the potential for SOFA-HDV-Rz1498 and shRNA1498 to inhibit HIV-1 replication, we transfected Jurkat T cells with the same constructs used for their delivery to HEK293T cells and selected stably transfected cells in the presence of Zeocin. All cell lines had a similar distribution of GFP expression from the integrated plasmids and proliferated at similar levels (Supplementary Figure S5). Following infection with HIV-1 NL4-3, both SOFA-HDV-Rz1498 and shRNA1498 expressing cells had a lower level of viral replication compared to cells expressing the control molecules, SOFA-HDV-RzHBV and shRNAns (Figure 8). A moderate suppression of viral replication was also observed in cells expressing the catalytically inactive Rz (SOFA-HDV-Rz1498A76). These results suggest that in addition to its accessibility in HEK293T cells, the Gag 1498 target site is also accessible to inhibition by both a SOFA-HDV-Rz and an shRNA in a T lymphocytic cell line.
Figure 8.
Inhibition of HIV-1 replication by SOFA-HDV-Rz1498 and shRNA1498. (a) Jurkat T cells stably transfected with the indicated SOFA-HDV-Rzs and shRNAs were infected with HIV-1 NL4-3. The average RT activity (cpm/µl) in culture supernatants across four independent infections performed in triplicate (n = 12) is shown for days 10 and 14 following infection. (b) Time course of a representative infection (n = 3) followed out to 32 days after infection.
Discussion
Rzs targeting HIV-1 RNA were among the first gene therapy agents tested in a clinical setting,36 and the only agent tested in a phase II vector controlled trial.37 While no toxicity has been observed in this study, the moderate effect reported emphasizes the need to find more inhibitory molecules for use in gene therapy. Approaches that have been described to identify new Rz candidates include the use of (i) RNA Polymerase III (Pol III) promoters to achieve higher levels of expression,38,39 (ii) cellular screens to identify optimal target sites,18,19 (iii) chimeric Rzs to enhance anti-viral effects,18,27,40,41 and (iv) alternative motifs such as modified RNase P and HDV Rzs.7,16 In this study, we screened SOFA-HDV-Rzs, expressed from the RNA Pol III H1 promoter, targeting the 5′UTR and Gag coding sequence of HIV-1 RNA for effects on viral production. Several candidate Rzs with the potential to target a broad range of HIV-1 strains were identified (Figure 2) and one of these (SOFA-HDV-Rz1498) was a particularly effective inhibitor of HIV-1 production (Figure 3). In agreement with other Rz screens,17,18,19 where only a small number of Rzs out of a library of candidates were effective, the majority of SOFA-HDV-Rzs evaluated did not strongly inhibit HIV-1 production. One explanation for this could be that Rzs may be particularly sensitive to the accessibility of their target sites and that most target sites in HIV-1 RNA do not permit Rzs to bind and form the appropriate structure required for catalysis. Other explanations include a failure of most Rzs to fold correctly or to localize with their target RNA.
According to the secondary structure published for the full length genomic RNA of HIV-1 NL4-3,42 the target site we identified in the Gag coding sequence lies predominantly in a 27 nt single-stranded loop enumerated 1033 to 1059 based on its position in NL4-3 HIV-1 RNA (1487 to 1513 in NL4-3 DNA). In addition to being accessible to inhibition by a SOFA-HDV-Rz, the 1498 target site was also accessible to an shRNA, with similar potency in an HIV-1 production assay compared to previously described shRNA candidates (Figure 5c). Although both SOFA-HDV-Rz1498 and shRNA1498 target the same site, our results suggested that they could be used together to inhibit viral production with an additive effect comparable to the combination of SOFA-HDV-Rz1498 with shRNA5983 targeting a different site (Figure 5d). These results are compatible with SOFA-HDV-Rz1498 and shRNA1498 acting in different cellular compartments to provide a nonredundant effect on HIV-1 production. Consistent with this, Rzs expressed from the RNA Pol III promoter U6 were shown to localize predominantly in the nucleus,38 while shRNAs are exported to the cytoplasm through the exportin 5-dependent pathway.
For both SOFA-HDV-Rz1498 and shRNA1498, the inhibition of intracellular CA expression was more apparent than inhibition of Gag (Figure 5b) and a similar effect was observed for shRNAs targeting the 5′UTR and overlapping Tat/Rev reading frame (Supplementary Figure S3). It is possible that in these experiments both Rzs and shRNAs inhibit intracellular Gag processing to the CA protein, a mechanism that has been described as contributing to the inhibition of HIV-1 production provided by modified U1 snRNAs.43 Another possibility is that intracellular Gag is more stable compared to CA in these experiments, making the effect on CA expression more apparent at the sampling time evaluated. Further studies will be required to evaluate whether this effect is reproducible in different cell types and experimental conditions and determine whether it could be a contributing mechanism to the inhibition of viral production provided by Rzs and shRNAs targeting HIV-1 RNA.
An important consideration for the design of therapies targeting HIV-1 is that they target the majority of HIV-1 circulating strains. According to our estimates, the target site for both SOFA-HDV-Rz-1498 and shRNA1498 was highly conserved at the nt level (Figure 5a, Gag). The LANL dataset we used to generate our estimates was made up of 1850 complete HIV-1 sequences, compared to 170 and 495 in earlier studies that used this dataset to identify shRNA21 or siRNA20 target sites, respectively. A comparison of the subtype distribution between the LANL dataset we used and global estimates44 (Supplementary Figure S1) suggests that HIV-1 subtype C is under-represented in the LANL dataset while subtype B, unique recombinant forms (URFs) and CRFs other than AG or AE are all over-represented. While not exactly representative of the global distribution, the LANL dataset includes all published HIV-1 sequences with representatives from all major subtypes and circulating recombinant forms and can easily be used to generate conservation estimates at the nt level for the identification of antisense target sites.45 In addition to being highly conserved across the different sequences available in the LANL database, our results suggest that the 1498 target site may also have a conserved structure, as the effects of shRNA1498 were similar against diverse viral strains with sequence variation within and around the target site (Figure 6).
Another important consideration for the design of antisense molecules targeting HIV-1 RNA is their potential to target cellular RNAs with complementary target sites. While target sites can be compared to the human genome, it is not always possible to predict off-target effects, as many antisense technologies do not require perfectly matched target sites. In agreement with in vitro studies,9,10 a single nt mismatch in SOFA-HDV-Rz1498 (Figure 4) or its binding site (strain Indie-C1 and 94UG114, Figure 6) greatly affected its ability to inhibit HIV-1 production. In contrast, an imperfectly matched snoRNA linked hammerhead Rz targeting HIV-1 RNA was an effective inhibitor of HIV-1 production18 and several shRNAs have been shown to tolerate single and even double mismatches in HIV-1 RNA.25,46 Consistent with previous results,25 a single nt mismatch at position 17 did not affect the inhibition of viral production provided by shRNA1498 (strain Indie-C1 and 94UG114, Figure 6) suggesting that SOFA-HDV-Rzs are more sensitive to mismatches in their target sites compared to shRNAs. While this sensitivity would be beneficial in terms of limiting off-target effects, it is a liability for targeting a broader range of HIV-1 circulating strains and would render SOFA-HDV-Rzs more sensitive to the emergence of resistant virus in a therapeutic setting. At levels where both SOFA-HDV-Rz1498 and shRNA1498 inhibited HIV-1 production, only small changes in gene expression were observed (Figure 7, Supplementary Figure S4), suggesting that their target site may be sufficiently different from human RNA sequences to avoid major off-target effects on human mRNAs. Combined with its potency and effects on diverse HIV-1 strains, in addition to our predictions for sequence conservation, these results suggest that shRNA1498 has the potential to be used safely and effectively against a broad range of HIV-1 circulating strains in a therapeutic setting. Future studies to directly compare the safety and efficacy of shRNA1498 to other shRNA candidates in more relevant cell lines and HIV-1 infection models are planned.
Although SOFA-HDV-Rz1498 was more effective than our previously designed candidates targeting the Tat/Rev region of HIV-1 RNA it reached only a 50% inhibition of viral production in cotransfection assays compared to an almost complete inhibition at much lower levels by shRNA1498 (Figure 5b,c). In contrast, it showed a similar potential to support a sustained inhibition of viral replication in Jurkat T cells (Figure 8), suggesting a long-term persistence of its effects in an infection model. Improvements in the design or expression of SOFA-HDV-Rz1498 may further increase its efficacy and modifications to render it less sensitive to mutations in its target site are being considered. Overall, our results suggest that both SOFA-HDV-Rz-1498 and shRNA1498 could be further developed for expression in a gene therapy setting or by direct delivery with newly developed methods4 as RNA drugs.
Materials and methods
In silico identification of SOFA-HDV-Rz target sites. The use of the LANL dataset to estimate sequence conservation at the nt level has been previously described.45 Briefly, a multiple sequence alignment of all complete HIV-1 sequences were downloaded from the LANL database using the QuickAlign tool and a consensus sequence with % conservation at each position was generated using Jalview sequence editor47 and exported to Microsoft Excel (Supplementary File S1, raw data tab). Several positions in the consensus sequence were represented in only a small number of sequences and positions that occurred in less than 10% of the sequences were removed from the raw data to facilitate target site selection (Supplementary File S1, conservation estimates tab). Highly conserved target sites were selected based on criteria illustrated in Figure 2a. Nucleotide BLAST48 was then used to align target sites to HIV-1 NL4-3 (M19921) and Ribosubstrates software (http://www.riboclub.org/ribosubstrates) was used to evaluate the potential for the corresponding SOFA-HDV-Rzs to target human RNAs as previously described.26 Briefly, the software identifies potential target sites for SOFA-HDV-Rzs in a cDNA database allowing for variations in the length of the spacer sequence and biosensor (Figure 1a). Perfectly matched target sites are assigned a value of 0, and the score increases by 10 for each wobble base pair and by 100 for each mismatch. We set a value of 20 as the cut-off for potential off-target effects, representing at least two wobble base pairs between a SOFA-HDV-Rz targeting HIV-1 RNA and a potential target site in any human RNA.
Plasmid construction. All SOFA-HDV Rzs and shRNAs were expressed from the human RNaseP H1 promoter in the vector psiRNA-H1GFP::Zeo (InvivoGen, San Diego, CA). SOFA-HDV-Rz inserts were generated using an overlapping PCR strategy11,14,28 and shRNA inserts were generated by annealing complementary oligonucleotides. Sequences for shRNA522 and shRNA553 inserts were obtained from a previous study22 and using an identical design, shRNAns (nonsense, adapted from siControl20), shRNA5983 (adapted from sh149) and shRNA1498 inserts were designed. The cloning strategies and sequences of all oligonucleotides used for the generation of plasmid inserts are provided in the Supplementary Materials and Methods, variable SOFA-HDV-Rz DNA sequences are illustrated in Supplementary Table S1.
Transfections. Cotransfections of HIV-1 molecular clones with Rz or shRNA expressing plasmids were performed in either 24- or 12-well plate formats as indicated in the figure legends. Cell culture conditions are described in the Supplementary Materials and Methods. Twenty-four hours prior to transfection, HEK293T cells were plated at 2 × 105 cells/ml and transfections were carried out using TransIT reagent (Mirus, Madison, WI) according to the manufacturer's instructions. Viral production was estimated 48 hours following transfection by measuring HIV-1 RT activity in the culture supernatant. To account for differences in viral production between experiments, replicates for each construct evaluated were performed in parallel with the empty vector psiRNA-H1GFP::Zeo and all data are expressed as a percentage of viral production in the empty vector cotransfected cells (Relative RT activity). For each Rz or shRNA construct evaluated, we also included the irrelevant control SOFA-HDV-Rz-HBV or the nonsense control shRNAns, respectively.
Rz expression in HEK293T cells. Total RNA extracts were harvested from transfected cells using Trizol reagent (Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Ten microgram of total RNA was resolved on an 8% denaturing polyacrylamide gel, transferred to a nylon membrane (AmershamHybond-N+, GE Healthcare, Little Chalfont, UK) and UV cross-linked. Membranes were incubated with ProbeSOFA followed by Probe5S and visualized using a Phosphor screen. Probe sequences, labeling and details on the northern blot conditions are provided in the Supplementary Materials and Methods.
In vitro SOFA-HDV-Rz cleavage assay. Single-turnover conditions (Rz>>substrate) were used to evaluate the catalytic activity of SOFA-HDV-Rzs as previously described.28 Briefly, a trace amount of 5′-end-labeled substrate (<1 nmol/l) was incubated at 37 °C with a final concentration of 100 nmol/l of the selected SOFA-HDV-Rz. The cleavage reactions were initiated by the addition of MgCl2 and samples were taken at different time intervals and stopped with loading buffer. Recovered samples were resolved on a 20% denaturing polyacrylamide gel, visualized using a Phosphor Screen and quantified using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The control reaction was performed in the absence of Rzs (replaced by water) and its last time interval sample was used to subtract the background. For each time point, the percentage of cleavage was calculated (cleaved product counts over cleaved + uncleaved products counts). Details on the DNA templates used for in vitro transcription, RNA synthesis and labeling are provided in the Supplementary Materials and Methods. The kobs and Fmax were then calculated using GraphPad Prism 5 for each Rz. The rate of cleavage (kobs) was obtained by fitting the data to the equation Ft = Fmax (1 − e−kt), where Ft is the percentage of cleavage at time t, Fmax is the maximum percent cleavage and k is the rate constant (kobs).
HIV-1 protein expression in HEK293T cells. The detection of HIV-1 protein expression using an HIV-1 p24 antibody in HEK293T cells has been previously described.50 Briefly, 100 µg of total protein was resolved on a 10% denaturing poly-acrylamide gel and transferred to a Hybond ECL nitrocellulose membrane (GE Healthcare, Little Chalfont, UK). Membranes were incubated first with anti-HIVp24 (183-H12-5C) followed by anti-GAPDH (sc-32233, Santa Cruz Biotechnology, Dallas, TX), bands were visualized using ECL (GE Healthcare). The relative intensity of bands were calculated using Image J densitometry software (Version 1.48, National Institutes of Health, Bethesda, MD). Data are expressed as Gag or CA band intensities relative to the intensity of the Gag band in the control SOFA-HDV-RzHBV(Figure 5b) or shRNAns (Supplementary Figure S3) lanes.
Gene expression profiling in HEK293T cells. Gene expression levels relative to control transfections were analyzed by human gene expression microarrays (ACRI proprietary slides). The triplicate dye-swap experiments are described in the Supplementary Materials and Methods.
HIV-1 infection assay. Stable Jurkat T lymphocytes were generated by electroporation of psiRNA constructs followed by selection with Zeocin (InvivoGen, San Diego, CA). The relative expression of psiRNA constructs in the stable cell populations was estimated by measuring GFP expression from the integrated vector with a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA), and proliferation was determined by counting live cells by Trypan blue (Wisent, St Bruno, Canada) exclusion using a hemocytometer. Prior to infection, SOFA-HDV-Rz or shRNA expressing cells were plated in six-well plates at 2 × 105 cells per well. Viral replication was monitored by measuring RT activity in culture supernatants at various days after infection. Cell culture and selection conditions are described in the Supplementary Materials and Methods.
HIV-1 RT assay. The HIV-1 RT assay used in this study was performed as previously described.45 Briefly, 5 µl of supernatant was incubated with a polyadenylic acid template (Roche, Basel, Switzerland), an oligodT primer (Life Technologies) and [32P]-dTTP (3,000 Ci/mmol, Perkin Elmer, Waltham, MA) for 2 hours at 37 °C in 50 µl total reaction mixture. Five microliter of the reaction mixture was then spotted onto Diethylaminoethyl (DEAE) filter mat (Perkin Elmer, Waltham, MA) and washed five times in 2X SSC buffer, followed by two washes in 95% ethanol to remove [32P]-dTTP not incorporated into the poly dT RT product. Counts per minute (cpm) were calculated for each sample using a microplate scintillation counter (MicrobetaTriLux, Perkin Elmer) and are proportional to the amount of HIV-1 RT enzyme present in the reaction mixture.
SUPPLEMENTARY MATERIAL Figure S1. Subtype distribution of HIV-1 sequences used to calculate conservation estimates in comparison to global distribution estimates. Figure S2. Effect of SOFA-HDV-Rz1498 on the quality of virions produced from cotransfected HEK293T cells. Figure S3. Gag and Capsid protein expression in cells cotransfected with HIV-1 pNL4-3 and different shRNAs targeting HIV-1 RNA. Figure S4. Change in mRNA expression ratios compared to a control vector for SOFA-HDV-Rz1498 and shRNA1498 transfected HEK293T cells. Figure S5. Stable Jurkat cell lines analysis. Table S1. SOFA-HDV-Rz target sites and DNA coding sequences. File S1. Data for sequence conservation estimates. File S2. Significantly up- and downregulated gene identities and log2 ratios in transfected HEK293T cells for SOFA-HDV-Rz1498 or shRNA1498, compared to the empty Rz expression plasmid (psiRNA). Materials and Methods
Acknowledgments
The authors thank Daniel Leger (Atlantic Cancer Research Institute) for gridding and analysis of the microarray data, Diane Singhroy and Thibault Mesplede from Mark Wainberg's lab (McGill University) for help in the selection and provision of HIV-1 molecular clones and Samantha Burugu (McGill University) for review of the manuscript. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: p94UG114.1.6 (Full-Length) from Beatrice Hahn and FengGao, and the UNAIDS Network for HIV Isolation and Characterization. This study was supported by the Canadian Institutes of Health Research (CIHR) (CIHR: HOP-93434 and DCB-120266 to A.G. and MOP-44022 to J.-P.P.): R.J.S, M.V.L., and S.M.D. were recipients of a Banting and Best Canada Graduate Scholarship, a doctoral fellowship from the Fonds de Recherche du Québec-Santé (FRQ-S) and a Vanier Canada Graduate Scholarship, respectively. J.-P.P. held the Canada Research Chair in Genomics and Catalytic RNA and now holds the Chaire de l'Université de Sherbrooke en structure et génomique de l'ARN. J.-P.P. is member of the Centre de Recherche Clinique Etienne-Le Bel and A.G. is a member of the Réseau SIDA-maladies infectieuses from the FRQ-S.
Supplementary Material
Subtype distribution of HIV-1 sequences used to calculate conservation estimates in comparison to global distribution estimates.
Effect of SOFA-HDV-Rz1498 on the quality of virions produced from cotransfected HEK293T cells.
Gag and Capsid protein expression in cells cotransfected with HIV-1 pNL4-3 and different shRNAs targeting HIV-1 RNA.
Change in mRNA expression ratios compared to a control vector for SOFA-HDV-Rz1498 and shRNA1498 transfected HEK293T cells.
Stable Jurkat cell lines analysis.
SOFA-HDV-Rz target sites and DNA coding sequences.
Data for sequence conservation estimates.
Significantly up- and downregulated gene identities and log2 ratios in transfected HEK293T cells for SOFA-HDV-Rz1498 or shRNA1498, compared to the empty Rz expression plasmid (psiRNA).
References
- De Clercq E. Antiretroviral drugs. Curr Opin Pharmacol. 2010;10:507–515. doi: 10.1016/j.coph.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Le Douce V, Janossy A, Hallay H, Ali S, Riclet R, Rohr O, et al. Achieving a cure for HIV infection: do we have reasons to be optimistic. J Antimicrob Chemother. 2012;67:1063–1074. doi: 10.1093/jac/dkr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer LJ, Rossi JJ. Ex vivo gene therapy for HIV-1 treatment. Hum Mol Genet. 2011;20 R1:R100–R107. doi: 10.1093/hmg/ddr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot J, Berkhout B. Nucleic acids-based therapeutics in the battle against pathogenic viruses. Handb Exp Pharmacol. 2009;189:243–263. doi: 10.1007/978-3-540-79086-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JJ. Ribozyme therapy for HIV infection. Adv Drug Deliv Rev. 2000;44:71–78. doi: 10.1016/s0169-409x(00)00085-5. [DOI] [PubMed] [Google Scholar]
- Zeng W, Chen YC, Bai Y, Trang P, Vu GP, Lu S, et al. Effective inhibition of human immunodeficiency virus 1 replication by engineered RNase P ribozyme. PLoS ONE. 2012;7:e51855. doi: 10.1371/journal.pone.0051855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif-Ullah M, Lévesque M, Robichaud G, Perreault JP. Development of ribozyme-based gene-inactivations; the example of the hepatitis delta virus ribozyme. Curr Gene Ther. 2007;7:205–216. doi: 10.2174/156652307780859008. [DOI] [PubMed] [Google Scholar]
- Bergeron LJ, Perreault JP. Target-dependent on/off switch increases ribozyme fidelity. Nucleic Acids Res. 2005;33:1240–1248. doi: 10.1093/nar/gki278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron LJ, Reymond C, Perreault JP. Functional characterization of the SOFA delta ribozyme. RNA. 2005;11:1858–1868. doi: 10.1261/rna.2112705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robichaud GA, Perreault JP, Ouellette RJ. Development of an isoform-specific gene suppression system: the study of the human Pax-5B transcriptional element. Nucleic Acids Res. 2008;36:4609–4620. doi: 10.1093/nar/gkn432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Anjou F, Routhier S, Perreault JP, Latil A, Bonnel D, Fournier I, et al. Molecular Validation of PACE4 as a Target in Prostate Cancer. Transl Oncol. 2011;4:157–172. doi: 10.1593/tlo.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque MV, Lévesque D, Brière FP, Perreault JP. Investigating a new generation of ribozymes in order to target HCV. PLoS ONE. 2010;5:e9627. doi: 10.1371/journal.pone.0009627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motard J, Rouxel R, Paun A, von Messling V, Bisaillon M, Perreault JP. A novel ribozyme-based prophylaxis inhibits influenza A virus replication and protects from severe disease. PLoS ONE. 2011;6:e27327. doi: 10.1371/journal.pone.0027327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiola K, Perreault JP, Cousineau B. Gene targeting in the Gram-Positive bacterium Lactococcus lactis, using various delta ribozymes. Appl Environ Microbiol. 2006;72:869–879. doi: 10.1128/AEM.72.1.869-879.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainé S, Scarborough RJ, Lévesque D, Didierlaurent L, Soye KJ, Mougel M, et al. In vitro and in vivo cleavage of HIV-1 RNA by new SOFA-HDV ribozymes and their potential to inhibit viral replication. RNA Biol. 2011;8:343–353. doi: 10.4161/rna.8.2.15200. [DOI] [PubMed] [Google Scholar]
- Bramlage B, Luzi E, Eckstein F. HIV-1 LTR as a target for synthetic ribozyme-mediated inhibition of gene expression: site selection and inhibition in cell culture. Nucleic Acids Res. 2000;28:4059–4067. doi: 10.1093/nar/28.21.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwalla HJ, Li H, Li SY, Abad D, Rossi JJ. Use of a U16 snoRNA-containing ribozyme library to identify ribozyme targets in HIV-1. Mol Ther. 2008;16:1113–1119. doi: 10.1038/mt.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Kuller T, Capalbo G, Klebba C, Engels JW, Klein SA. Identification and characterization of a highly efficient anti-HIV pol hammerhead ribozyme. Oligonucleotides. 2009;19:265–272. doi: 10.1089/oli.2008.0150. [DOI] [PubMed] [Google Scholar]
- Naito Y, Nohtomi K, Onogi T, Uenishi R, Ui-Tei K, Saigo K, et al. Optimal design and validation of antiviral siRNA for targeting HIV-1. Retrovirology. 2007;4:80. doi: 10.1186/1742-4690-4-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- McIntyre GJ, Groneman JL, Yu YH, Jaramillo A, Shen S, Applegate TL. 96 shRNAs designed for maximal coverage of HIV-1 variants. Retrovirology. 2009;6:55. doi: 10.1186/1742-4690-6-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung MB, Hampel A. Computer analysis of the conservation and uniqueness of ribozyme-targeted HIV sequences. Methods Mol Biol. 1997;74:27–36. doi: 10.1385/0-89603-389-9:27. [DOI] [PubMed] [Google Scholar]
- Sajic R, Lee K, Asai K, Sakac D, Branch DR, Upton C, et al. Use of modified U1 snRNAs to inhibit HIV-1 replication. Nucleic Acids Res. 2007;35:247–255. doi: 10.1093/nar/gkl1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Dykxhoorn DM, Kumar P, Ranjbar S, Song E, Maliszewski LE, et al. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood. 2005;106:818–826. doi: 10.1182/blood-2004-10-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucier JF, Bergeron LJ, Brière FP, Ouellette R, Elela SA, Perreault JP. RiboSubstrates: a web application addressing the cleavage specificities of ribozymes in designated genomes. BMC Bioinformatics. 2006;7:480. doi: 10.1186/1471-2105-7-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Luque FJ, Reyes-Darias JA, Puerta-Fernández E, Berzal-Herranz A. Inhibition of HIV-1 replication and dimerization interference by dual inhibitory RNAs. Molecules. 2010;15:4757–4772. doi: 10.3390/molecules15074757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévesque MV, Perreault JP. Target-induced SOFA-HDV ribozyme. Methods Mol Biol. 2012;848:369–384. doi: 10.1007/978-1-61779-545-9_23. [DOI] [PubMed] [Google Scholar]
- DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, Peden KW. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses. 1996;12:191–194. doi: 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Otsuka N, Matsuo K, Shiino T, Kojima A, Kurata T, et al. An infectious DNA clone of HIV type 1 subtype C. AIDS Res Hum Retroviruses. 1999;15:1321–1324. doi: 10.1089/088922299310223. [DOI] [PubMed] [Google Scholar]
- Ndung'u T, Renjifo B, Essex M. Construction and analysis of an infectious human Immunodeficiency virus type 1 subtype C molecular clone. J Virol. 2001;75:4964–4972. doi: 10.1128/JVI.75.11.4964-4972.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Robertson DL, Carruthers CD, Morrison SG, Jian B, Chen Y, et al. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusagawa S, Takebe Y, Yang R, Motomura K, Ampofo W, Brandful J, et al. Isolation and characterization of a full-length molecular DNA clone of Ghanaian HIV type 1 intersubtype A/G recombinant CRF02_AG, which is replication competent in a restricted host range. AIDS Res Hum Retroviruses. 2001;17:649–655. doi: 10.1089/088922201300119761. [DOI] [PubMed] [Google Scholar]
- Rossi JJ, June CH, Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good PD, Krikos AJ, Li SX, Bertrand E, Lee NS, Giver L, et al. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- Puerta-Fernández E, Barroso-del Jesus A, Romero-López C, Tapia N, Martínez MA, Berzal-Herranz A. Inhibition of HIV-1 replication by RNA targeted against the LTR region. AIDS. 2005;19:863–870. doi: 10.1097/01.aids.0000171399.77500.e0. [DOI] [PubMed] [Google Scholar]
- Chang Z, Westaway S, Li S, Zaia JA, Rossi JJ, Scherer LJ. Enhanced expression and HIV-1 inhibition of chimeric tRNA(Lys3)-ribozymes under dual U6 snRNA and tRNA promoters. Mol Ther. 2002;6:481–489. doi: 10.1006/mthe.2002.0696. [DOI] [PubMed] [Google Scholar]
- Michienzi A, Cagnon L, Bahner I, Rossi JJ. Ribozyme-mediated inhibition of HIV 1 suggests nucleolar trafficking of HIV-1 RNA. Proc Natl Acad Sci USA. 2000;97:8955–8960. doi: 10.1073/pnas.97.16.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal D, Feng Z, Stoltzfus CM. Excessive RNA splicing and inhibition of HIV-1 replication induced by modified U1 small nuclear RNAs. J Virol. 2010;84:12790–12800. doi: 10.1128/JVI.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. WHO-UNAIDS Network for HIV Isolation and Characterisation Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS. 2011;25:679–689. doi: 10.1097/QAD.0b013e328342ff93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough RJ, Lévesque MV, Perreault JP, Gatignol A. Design and evaluation of clinically relevant SOFA-HDV ribozymes targeting HIV RNA. Methods Mol Biol. 2014;1103:31–43. doi: 10.1007/978-1-62703-730-3_3. [DOI] [PubMed] [Google Scholar]
- Sabariegos R, Giménez-Barcons M, Tàpia N, Clotet B, Martínez MA. Sequence homology required by human immunodeficiency virus type 1 to escape from short interfering RNAs. J Virol. 2006;80:571–577. doi: 10.1128/JVI.80.2.571-577.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- Clerzius G, Shaw E, Daher A, Burugu S, Gélinas JF, Ear T, et al. The PKR activator, PACT, becomes a PKR inhibitor during HIV-1 replication. Retrovirology. 2013;10:96. doi: 10.1186/1742-4690-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Subtype distribution of HIV-1 sequences used to calculate conservation estimates in comparison to global distribution estimates.
Effect of SOFA-HDV-Rz1498 on the quality of virions produced from cotransfected HEK293T cells.
Gag and Capsid protein expression in cells cotransfected with HIV-1 pNL4-3 and different shRNAs targeting HIV-1 RNA.
Change in mRNA expression ratios compared to a control vector for SOFA-HDV-Rz1498 and shRNA1498 transfected HEK293T cells.
Stable Jurkat cell lines analysis.
SOFA-HDV-Rz target sites and DNA coding sequences.
Data for sequence conservation estimates.
Significantly up- and downregulated gene identities and log2 ratios in transfected HEK293T cells for SOFA-HDV-Rz1498 or shRNA1498, compared to the empty Rz expression plasmid (psiRNA).