Abstract
Microcystis aeruginosa is a planktonic unicellular cyanobacterium often responsible for seasonal mass occurrences at the surface of freshwater environments. An abundant production of intracellular structures, the gas vesicles, provides cells with buoyancy. A 8.7-kb gene cluster that comprises twelve genes involved in gas vesicle synthesis was identified. Ten of these are organized in two operons, gvpAIAIIAIIICNJX and gvpKFG, and two, gvpV and gvpW, are individually expressed. In an attempt to elucidate the basis for the frequent occurrence of nonbuoyant mutants in laboratory cultures, four gas vesicle-deficient mutants from two strains of M. aeruginosa, PCC 7806 and PCC 9354, were isolated and characterized. Their molecular analysis unveiled DNA rearrangements due to four different insertion elements that interrupted gvpN, gvpV, or gvpW or led to the deletion of the gvpAI-AIII region. While gvpA, encoding the major gas vesicle structural protein, was expressed in the gvpN, gvpV, and gvpW mutants, immunodetection revealed no corresponding GvpA protein. Moreover, the absence of a gas vesicle structure was confirmed by electron microscopy. This study brings out clues concerning the process driving loss of buoyancy in M. aeruginosa and reveals the requirement for gas vesicle synthesis of two newly described genes, gvpV and gvpW.
Many aquatic bacteria are provided with buoyancy due to subcellular structures, the gas vesicles (GV) (for a review, see reference 36). GV have long been known in aquatic anoxyphototrophic bacteria, cyanobacteria, and halophilic archaea and have been described more recently for a soil bacterium, Bacillus megaterium (20). These hollow, gas-filled structures allow the cells to float at the surface of water environments and can be used to position themselves under optimal light and oxygen conditions for growth. The structural shape, despite variations in width and length, is well conserved among different groups of organisms. Formed solely of proteins, the GV are spindle-shaped cylinders with conical end caps. They cluster into gas vacuoles (visible under phase-contrast microscopy as light-refractile bodies). The small hydrophobic protein GvpA is the major constituent and assembles to form the ribbed wall of the GV. The other less-abundant structural protein GvpC is located on the outside of the shell and has been shown to strengthen the resistance of GV to pressure (for a review, see reference 36).
Apart from the gvpA and gvpC genes encoding the structural proteins, several other genes are involved in GV formation in various microorganisms (see Fig. S1 in the supplemental material). In halophilic archaea, 8 to 10 of the 14 gvp genes identified are sufficient for functional synthesis of GV (7, 24). In B. megaterium, 11 genes represent the minimal set required for GV formation and function in Escherichia coli (20). Up to 10 genes have been identified in cyanobacteria, but the number of gvp genes necessary for GV formation remains obscure (1, 3, 6, 13, 16, 17).
The occurrence of spontaneous GV-deficient mutants (GV−) in laboratory cultures has been reported for both halophilic archaea (8) and cyanobacteria (4). In halobacteria, various types of genetic rearrangements in the vac region responsible for GV synthesis have been described. In H. halobium, GV mutants, which arise at a frequency of about 1% (8), were originally placed into three classes. Class I mutants were partially Vac− (that is, GV−) and highly unstable, and the phenotype was shown to be due to reduced copy numbers of gvpA. Class II mutants were also partially Vac− but were more stable, with insertion sequences (IS) either in the gvpA promoter or within gvpD and gvpE (15), thus suggesting (as later confirmed) a role for the two latter genes in the regulation of GV formation (10, 11, 25). Large deletions (59 to 67 kb) resulting in the strict GV− phenotype of highly stable class III mutants encompassed the entire gvp cluster, and the deletion junction mapped to the termini of insertion elements (23). For cyanobacteria, genetic characterization of nonbuoyant mutants has been carried out exclusively with strains of Planktothrix, and different gvp gene arrangements have been described for filament isolates from environmental samples and for laboratory cultures (2, 3, 4). Strains from Nordic lakes displayed variations in the number and order of the alternating copies of gvpA and gvpC and in the length of the gvpC gene (2). Nonbuoyant mutants isolated from a laboratory culture showed deletions and recombination events in the gvpA-gvpC region. They all retained the ability to produce GV, albeit less abundantly and with a decreased capacity to withstand pressure, resulting from the reduction in the number of gene copies (4).
Microcystis aeruginosa is a unicellular, planktonic freshwater cyanobacterium that often synthesizes hepatotoxins, the microcystins, which are responsible both for acute poisonings (29) and for promotion of liver cancer at sublethal concentrations (14, 34). Mass occurrences of M. aeruginosa are commonly found at the surface of water bodies in spring and summer. Buoyancy keeps Microcystis cells close to the surface, where the availability of light and oxygen greatly favors proliferation and hence the formation of scums. In laboratory cultures, the spontaneous occurrence of GV-deficient mutants is often observed. In this report, we present the identification of a 8,703-bp-long region of the M. aeruginosa genome that contains a gene cluster encoding GV (gvp genes) and the molecular characterization of nonbuoyant mutants due to DNA rearrangements involving insertion of endogenous mobile elements in 4 of the 12 genes of the gvp cluster.
MATERIALS AND METHODS
Strains and growth conditions.
This study was carried out on PCC 7806 and PCC 9354, two axenic strains of M. aeruginosa from the Pasteur Culture Collection. Experimental cultures were grown at 28°C in BG110 (30) supplemented with 2 mM NaNO3 and 10 mM NaHCO3 and exposed (unless otherwise specified) to continuous light provided by Osram Universal White fluorescent tubes under conditions of photosynthetic photon flux density of 30 μmol of photons m−2 s−1, as measured with a LICOR LI-185B quantum radiometer-photometer equipped with a LI-190SB quantum sensor. The cultures of 500 ml for DNA, RNA, and protein extractions were supplemented with 20 mM NaHCO3 and bubbled with 1% (vol/vol) CO2 in air. Cells in exponential-growth phase (optical density at 750 nm [OD750] = 0.5 to 0.6) were collected by centrifugation (10,000 × g, 10 min, 25°C).
Two of the mutants deficient in GV were obtained spontaneously during reisolation of axenic (PCC 9354 M4) or impure (PCC 7806 M1) cultures. Cells were diluted so as to obtain isolated colonies and plated on the standard growth medium solidified with 0.8% (wt/vol) Sigma washed agar (A 8678). After 4 weeks of incubation at 23°C with a photosynthetic photon flux density of 10 μmol of photons m−2 s−1, light-green colonies (indicative of GV− mutants) were picked and grown in liquid medium as described previously. The mutants PCC 7806 M2 and M3 were obtained after a cryogenic treatment of a culture derived from a freshly reisolated GV+ phenotype. Cryopreservation conditions were as follows: cells were exposed to ethylene glycol (5% [vol/vol] for M2 and 2.5% [vol/vol] for M3), cooled progressively (−1°C per minute until reaching −80°C) during 1 h 30 min, and subsequently stored in liquid nitrogen for 2 months. The cryovials were rapidly thawed at 37°C, and the cells were diluted and plated as described above. Mutant frequencies for a freshly isolated GV+ clone of an axenic strain PCC 7806 and percent survival (before and after cryoconservation in ethylene glycol) were estimated by counting the total number of GV+ and putative GV− colonies (light green) observed on individual plates and pooling the data of three independent experiments.
Physical organization and sequencing of the gvp region.
The gvpA gene of M. aeruginosa PCC 7806 was initially amplified from total DNA through the use of the degenerate primers 635 and 636 (Table 1). The PCR product was cloned in the pGEM-T plasmid (Promega Corporation, Madison, Wis.) and sequenced on both strands. The resulting nucleotide sequence was used to detect the corresponding gvpA gene in the sequence of one of the contigs from a nonpublic database containing the partial genome sequence of M. aeruginosa PCC 7806 (three times coverage). This database sequence is the preliminary result of shotgun sequencing of 700-bp fragments of M. aeruginosa PCC 7806 genomic DNA cloned in the pcDNA-2.1 vector (Invitrogen Life Technologies, Rockville, Md.) and forms part of a current sequencing genome project at the Génopole-Ile de France, Institut Pasteur, Paris, France. A second contig from the same database was identified by sequence comparison to other known gvp genes. Gaps between contigs were filled by the sequencing of PCR products corresponding to the missing fragments (Genome Express, Meylan, France). Primers designed on the nucleotide sequence of the wild-type strain PCC 7806 were used for PCR amplification of the gvp cluster of the wild-type strain PCC 9354. A region encompassing the gvpF to gvpW genes in PCC 9354 was sequenced (Genome Express, Meylan, France).
TABLE 1.
Primers used for PCR, RT-PCR, and sequence determination
| Primer | Sequence (5′ to 3′) | Gene or spacera | Orientationb | Localization (bp)c |
|---|---|---|---|---|
| 635 | CGGCATGGCWGTKGARAARACKAAY | gvpA | F | 1014-1035; 1584-1605; 2130-2151 |
| 636 | CGGCCCKGTKGCYTCKGCRTAYTT | gvpA | R | 1194-1180; 1764-1750; 2310-2296 |
| 870 | TGCTTTGCGTCAGTCTTTCC | gvpC | F | 2751-2770 |
| 871 | TCCTTCACCTGTTTGGCTCT | gvpC | R | 3146-3127 |
| 893 | CTGAAGCGAAACAGCTAAAAG | gvpC | F | 3179-3199 |
| 949 | AATCGGGTTGATGGCTAGTG | gvpN | R | 4334-4315 |
| 950 | GAATCCGTCACCGAACAACT | gvpF | F | 6713-6732 |
| 964 | TAGAAGAAGAGGCGGCAGAA | gvpG | F | 7421-7440 |
| 965 | GCACGCTCCTGAATTTTCTC | gvpG | R | 7286-7267 |
| 972 | TGTTGTTACGCCTTCGATTG | gvpN | F | 3904-3923 |
| 973 | AGGCCATTGTTAACCGAGAA | gvpN | R | 4208-4189 |
| 974 | TGATAAAGCCAGGGAGATGG | gvpJ | F | 5112-5131 |
| 975 | TCAGTTTCCAGGGTTTGGAG | gvpJ | R | 5234-5215 |
| 976 | CGGACTGGTATCATGGCTCT | gvpJ | R | 5295-5276 |
| 977 | CTTGCTTGCACACCTTACGA | gvpK | F | 5972-5991 |
| 978 | GAGAACCCCTTTTTCCATCC | gvpK | R | 6130-6111 |
| 979 | TGCTTCCGGTAAAAGAGTGC | gvpK | R | 6271-6252 |
| 981 | AGCGAATTCTTAGGCGATCA | gvpF | R | 7145-7126 |
| 983 | CCGTAGTTGAACCCGAAGTT | gvpW | F | 8135-8154 |
| 993 | GTCCAGTTATCCCCCAAGGT | 5′ gvpAI | F | 683-702 |
| 994 | CATGGCAGTCGAAAAAAC | gvpA | F | 1014-1031; 1584-1601; 2130-2147 |
| 995 | CGAAGCGATAACTACACGAG | gvpA | R | 1164-1145; 1734-1715; 2280-2261 |
| 1006 | GGGGGAGAGACAGTAGAGTCC | 5′ gvpC | R | 2536-2516 |
| 1008 | GGATCGTTTATCGATTGCTGA | gvpC | F | 2643-2663 |
| 1017 | AATTTCTGGCCAGTCAATCCA | 5′ gvpAI | F | 1-21 |
| 1018 | AAGCGATCGATCAGCAGTTT | gvpF | F | 7104-7123 |
| 1020 | CGAGGCATGGTGCTAATTTT | gvpV | F | 7872-7891 |
| 1021 | GGGGGAGAAACTTGTCCTAA | gvpW | R | 8687-8668 |
| 1076 | TTCTTGGCCATGAGAACAAA | gvpW | R | 8325-8306 |
| 1077 | GCCCTTGCTTTCAGAAGAGA | gvpW | R | 8427-8408 |
| 1079 | CTGATCTGGATTGGGGAGAA | gvpG | F | 7252-7271 |
| 1080 | TCTGCCGCCTCTTCTTCTAA | gvpG | R | 7439-7420 |
| 1081 | GCCTCGGAAACAATCTGA | gvpV | R | 7877-7860 |
| 1082 | TTCTAGGTAAAAAGCTTGGAAA | gvpV | F | 7598-7619 |
| 1108 | AGCCTTCGACAGGAAATCAA | gvpN | F | 4809-4828 |
| 1109 | TGCGTGAAATTCCTCACTTG | gvpX | F | 5319-5338 |
| 1110 | GGACTGCCATCACGTCTCTC | gvpX | R | 5544-5525 |
| 1111 | TGGAAGGAAATGTTGATCTGG | gvpK | F | 6354-6374 |
| 1113 | AGTTGTTCGGTGACGGATTC | gvpF | R | 6732-6713 |
5′ gvpAI is the noncoding region located upstream of gvpAI; 5′ gvpC indicates the intergenic gvpAIII-gvpC region.
Orientation of primer with respect to direction of the ORF: forward (F) and reverse (R).
Localization is given with respect to the sequence deposited in EMBL under no. AJ57736.
Nucleic acid extraction.
Genomic DNA was extracted from a cell pellet (1-g wet weight) following Nucleobond AXG500 kit instructions (Macherey-Nagel, Düren, Germany) with the following modifications: a 2-h incubation at 80°C was performed with gentle shaking after the proteinase K step to improve cell disruption and inactivate DNases. The lysate was then loaded on the flowthrough cartridge and processed as described by the manufacturer.
For RNA isolation, the cell pellet (1-g wet weight) was frozen in liquid nitrogen immediately after centrifugation and resuspended in 2.5 ml of TRIzol reagent (Invitrogen Life Technologies). The cells were broken by eight pulses of vigorous vortexing for 15 s, and RNA was extracted according to the supplied protocol. The final RNA pellet was dissolved in 100 μl of diethyl pyrocarbonate-treated H2O and incubated for 2 h at 37°C with 5 U of RNase-free DNase (Roche Diagnostics, Mannheim, Germany) in 1× Superscript II buffer (Invitrogen Life Technologies).
PCR.
Amplification of the gvp genes was performed with the primers listed in Table 1. The rnpB amplicon used as a probe in RNA-DNA hybridization was obtained with the primers 5′-GGCTCCCGAAAGACCAAA-3′ and 5′-ACACCAAATGACGAAAAAGG-3′ through the use of (as a template) the plasmid pAV3002 containing a 0.7-kb fragment of the rnpB gene from Calothrix sp. strain PCC 7601 (35). DNA (100 ng), 10 pmol of each primer, 250 μM of each deoxynucleoside triphosphate, and 1 U of Taq polymerase in 1× buffer (Promega Corporation) were mixed and subjected to an initial step of 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min and a final elongation step of 72°C for 7 min in a 9700 Perkin-Elmer thermocycler (Applied Biosystems, Foster City, Calif.). A total of 10 μl of each sample was analyzed by gel electrophoresis on 1% (wt/vol) agarose in 1× Tris-borate-EDTA buffer and stained with ethidium bromide (31).
Transcription analysis.
For RNA-DNA hybridization analyses, RNA gel electrophoresis was performed as previously described (21). Blotting was achieved by capillary transfer (31) to nylon membranes (Hybond N+; Amersham Biosciences, Freiburg, Germany). Prehybridization (4 h) and hybridization (16 h) were carried out at 42°C in the presence of 50% (vol/vol) formamide as described previously (5). Probes were obtained by PCR amplification and labeled using [α-32P]ATP and a Megaprime random labeling kit (Amersham Biosciences).
The presence of transcripts was also determined by reverse transcriptase PCR (RT-PCR) with the primers described in Table 1. The reverse transcription reaction was carried out in 1× Superscript II buffer (Invitrogen Life Technologies)-0.5 to 1 μg of total RNA-10 pmol of R primer-each deoxynucleoside triphosphate at a concentration of 250 μM-1 mM dithiothreitol-H2O up to a final volume of 45 μl. A touchdown primer-annealing procedure (consisting of 70°C for 1 min, 65°C for 1 min, 60°C for 1 min, 55°C for 1 min, 50°C for 1 min, and 45°C for 1 min) was performed in a thermocycler prior to adding 5 μl of a fivefold dilution of Superscript II reverse transcripase (Invitrogen Life Technologies). The reaction was allowed to proceed for 30 min at 42°C followed by 5 cycles of 50°C for 1 min, 53°C for 1 min, and 56°C for 1 min. A total of 1 μl of the reverse transcription reaction-10 pmol of each primer-250 μM of each deoxynucleoside triphosphate-1 U of Taq polymerase (Promega Corporation) in 1× buffer was used for subsequent PCR. Genomic DNA (100 μg ml−1) (1 μl) and a 10× dilution of total RNA (1 μl) were used instead of cDNA as positive and negative controls, respectively. PCR was initiated by a 94°C step for 2 min followed by 35 cycles of 94°C for 10 s, 50°C for 20 s, and 72°C for 1 min and a final elongation step of 72°C for 7 min.
Immunodetection of GvpA.
GV fractions of M. aeruginosa strain PCC 7806 were enriched by collecting (with a Pasteur pipette) the white buoyant layer that accumulates at the top of nonagitated senescent cultures in which cells lyse spontaneously. The suspension containing GV was left standing until a new and more concentrated floating layer had accumulated, which was then collected as described above. For the preparation of cell extracts, the cell pellets (25 to 50 ml of culture at OD750 = 0.5 to 0.6) were resuspended in 0.5 ml of 20 mM Tris-HCl (pH 8.2) containing 5 mM dithiothreitol. After sonication on ice for eight pulses of 15 s, the protein concentration was estimated with Bradford reagent as described by the manufacturer (Bio-Rad, Hercules, Calif.). The crude cell extracts, as well as the enriched GV fraction, were treated with formic acid (0.1 ml for 20 μg of total protein, irrespective of the volume) for 2 h at 25°C to solubilize hydrophobic proteins, dried in a Speed-Vac, and resuspended in loading buffer. After being resolved by sodium dodecyl sulfate-16% polyacrylamide gel electrophoresis (19), proteins were transferred to a nitrocellulose membrane (ECL; Amersham Biosciences) using a Criterion Blotter (Bio-Rad). The GvpA protein was identified by use of polyclonal antibodies raised in rabbits by intradermal injection of the insoluble fraction of GV from strain Calothrix sp. strain PCC 7601 (gift from T. Damerval, Institut Pasteur, Paris, France). Detection was performed with an ECL Western blotting system (Amersham Biosciences) in Tris-buffered saline containing 0.1% (vol/vol) Tween 20 (TBS-T buffer) according to the manufacturer's instructions. Dilution of the primary antibody was 1/10,000.
Electron microscopy.
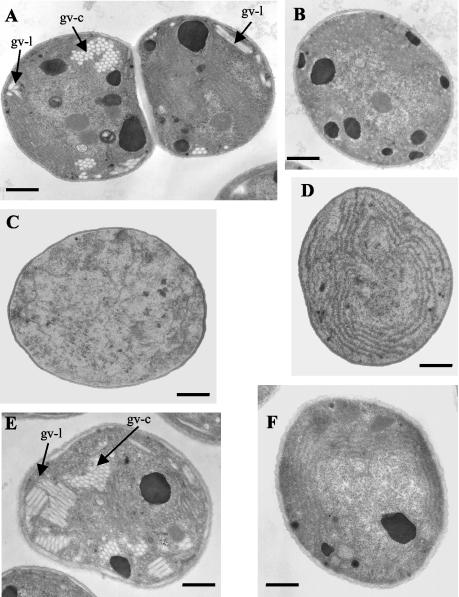
Since GV collapse by centrifugation, 0.5 ml of cells in exponential-growth phase (OD750 = 0.5 to 0.6) were used directly for fixation in 2% (vol/vol) glutaraldehyde prepared in 0.1 M cacodylate buffer. The cells were then collected by careful centrifugation at low speed (2,000 × g, 5 min, 25°C), and subsequent fixation, dehydration, and embedding in Spurr's resin were performed (as described previously) (12) at the Plate-Forme de Microscopie Electronique, Institut Pasteur, Paris, France (see Fig. 6).
FIG. 6.
Electron micrographs of the wild-type strains (A and E) and the GV-deficient mutant strains (B, C, D, and F) of M. aeruginosa. (A) PCC 7806 wild type; (B) PCC 7806 M1; (C) PCC 7806 M2; (D) PCC 7806 M3; (E) PCC 9354 wild type; (F) PCC 9354 M4. gv-l, GV in longitudinal section; gv-c, GV in cross-section. Bars, 500 nm.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper are available in the DDBJ-EMBL-GenBank database under the following accession numbers: AJ577136 for the gvp region; AJ577137 for ISMae1; AJ577138 for ISMae2; AJ577139 for ISMae3; and AJ577140 for ISMae4.
RESULTS
M. aeruginosa gvp gene cluster.
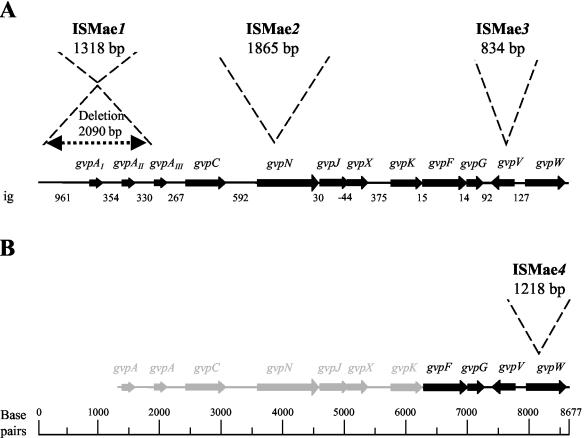
Sequence analysis of a 8,703-bp region of the genome of M. aeruginosa strain PCC 7806 involved in GV synthesis was performed as described in Materials and Methods. It revealed a cluster of twelve open reading frames (ORFs), all but one of which read in the same direction (Fig. 1A). PCR amplification analyses of M. aeruginosa PCC 9354 genomic DNA with the primers designed for M. aeruginosa PCC 7806 (see Table 1) produced fragments that were of similar sizes (data not shown). This allowed the construction of a map showing that a cluster of gvp genes with the same physical organization is present in M. aeruginosa PCC 9354. The gene organization in the region spanning gvpF to gvpW was confirmed by sequencing (Fig. 1B).
FIG. 1.
The gvp gene cluster region in M. aeruginosa and IS found in the GV-deficient mutants. (A) Strain PCC 7806; (B) strain PCC 9354. From top to bottom, names and sizes of the IS, localizations of the insertions, names of the putative genes, maps of the ORFs and their orientations, and intergenic regions (ig) in base pairs (bp) are shown. Arrows, ORFs; black arrows and lines, sequenced regions; grey-shaded arrows and lines, nonsequenced regions. Precise locations (with respect to the sequence submitted to DDBJ/EMBL/GenBank under no AJ577136) of the insertion elements are as follows: between nucleotides 115 and 2206 for ISMae1, between nucleotides 4033 and 4034 for ISMae2, between nucleotides 7633 and 7634 for ISMae3, and between nucleotides 8301 and 8302 for ISMae4.
Table 2 presents the characteristics of the predicted amino acid sequences of the ORFs from strain PCC 7806 and the results of BLAST searches performed to determine their similarity to known or putative gene products of other microorganisms. Three copies (named gvpAI, gvpAII, and gvpAIII) of the gene encoding the major structural protein GvpA are repeated in tandem with 354- and 330-bp-long intergenic sequences. Levels of identity between gvpAI and gvpAII, between gvpAI and gvpAIII, and between gvpAII and gvpAIII are 96, 97, and 97%, respectively. The deduced amino acid sequences of GvpAII and GvpAIII are identical, while GvpAI differs by one amino acid at the C-terminal end. This difference arises from a single base change (G to T) in the nucleotide sequence, leading to the replacement of Ala68 by Ser68 in the predicted protein. These low-molecular-weight hydrophobic proteins are homologs of other cyanobacterial GvpA sequences. The M. aeruginosa PCC 7806 GvpA sequences also display 43% similarity to the hydrophobic N-terminal sequence of GvpJ and 28% similarity to the C-terminal sequence of GvpK but no similarity to the GvpX predicted protein sequence (Table 2). The gvpJ gene has no ATG but has two putative GTG start codons, neither of which is preceded by a Shine-Dalgarno sequence. However, the second GTG is more likely to be the translation initiation codon with respect to the preferred surrounding sequence described for Synechocystis sp. strain PCC 6803 genes (32). Of the three ATG codons in the 5′ nucleotide sequence of the gvpX gene, only the first one is preceded by a putative 5′-GGAG-3′ Shine-Dalgarno sequence. The gvpX gene might therefore overlap the end of gvpJ by 44 nucleotides. The predicted protein GvpJ is more similar to other cyanobacterial GvpJs than to the M. aeruginosa PCC 7806 GvpX (even though the C terminus of GvpJ shares 54% similarity with the C terminus of GvpX). The N terminus of GvpJ also shows 30% similarity to the 40 C-terminal amino acids of GvpK (Table 2).
TABLE 2.
Sequence analysis results
| Gene(s) | No. of amino acids | Mol mass (Da) | Isolectric point | Best homology to known and putative proteins (accession no.)a | E value | Identity (%) to other ORFs in the Microcystis clusterb |
|---|---|---|---|---|---|---|
| gvpAI | 71 | 7,487 | 4.37 | GvpA A. flos-aquae (P10397) | 3e−19 | GvpAII (98), Nt of GvpJ (43), Ct of GvpK (28) |
| gvpAII, gvpAIII | 71 | 7,487 | 4.37 | GvpA A. flos-aquae (P10397) | 3e−19 | GvpAI, GvpJ, GvpK (as indicated above) |
| gvpC | 212 | 25,153 | 10.22 | GvpC A. flos-aquae (P09413) | 2e−06 | None |
| gvpN | 346 | 38,469 | 5.24 | GvpN Nostoc sp. strain PCC 7120 (AD2087) | 1e−135 | None |
| gvpJ | 136 | 14,743 | 4.71 | GvpJ Nostoc sp. strain PCC 7120 (Q9AKS3) | 2e−27 | Ct:Ct of GvpX (54), Nt:Ct of GvpK (30) |
| gvpX | 102 | 11,752 | 10.01 | GvpJ Nostoc sp. strain PCC 7120 (Q8YUT1) | 0.016 | GvpJ (as indicated above) |
| gvpK | 157 | 17,234 | 4.20 | GvpK A. flos-aquae (P55148) | 4e−50 | GvpJ (as indicated above), GvpAI (as indicated above) |
| gvpF | 244 | 28,402 | 4.72 | GvpF Nostoc sp. strain PCC 7120 (AI2086) | 5e−90 | None |
| gvpG | 86 | 9,819 | 3.68 | GvpG Nostoc sp. strain PCC 7120 (AH2086) | 3e−24 | None |
| gvpV | 111 | 12,991 | 10.29 | Alr2246 Nostoc sp. strain PCC 7120 (AG2086) | 1e−09 | None |
| gvpW | 227 | 26,256 | 4.86 | All2245 Nostoc sp. strain PCC 7120 (AF2086) | 7e−49 | None |
Accession numbers are SwissProt when starting with P or Q and Protein Information Resource when starting with A.
None, no discernible similarity to known sequences and Protein Information Resource; Ct, ORF carboxy terminal; Nt, ORF amino terminal.
The predicted proteins of six other ORFs of this gene cluster share no obvious similarities to each other, and four of them (namely, GvpC, GvpF, GvpG, and GvpN) can be identified by their high degree of similarity to other cyanobacterial Gvp proteins present in the protein databases (Table 2). The GvpC amino acid sequence contains four conserved repeats of 33 residues (data not shown). GvpN contains the amino acid sequence LCGPAGTGKT that represents a putative nucleotide binding site. The predicted products of two ORFs downstream of gvpG (hereafter designated GvpV and GvpW) share similarities to two putative proteins of unknown function (Alr2246 and All2245, respectively) of Nostoc (also called Anabaena) sp. strain PCC 7120 (Table 2).
Most of the ORFs in the M. aeruginosa PCC 7806 gvp gene region display acidic isoelectric points, with the exceptions of GvpC, GvpX, and GvpV, for which the pI values are more than 10 (Table 2). Hydropathy profiles revealed that (like those of GvpA) the N-terminal parts of GvpJ, GvpG, and GvpK are hydrophobic. The amino acid sequences of GvpC, GvpX, and GvpV are hydrophilic, and those of GvpF, GvpN, and GvpW are amphiphilic (data not shown).
Transcription analysis.
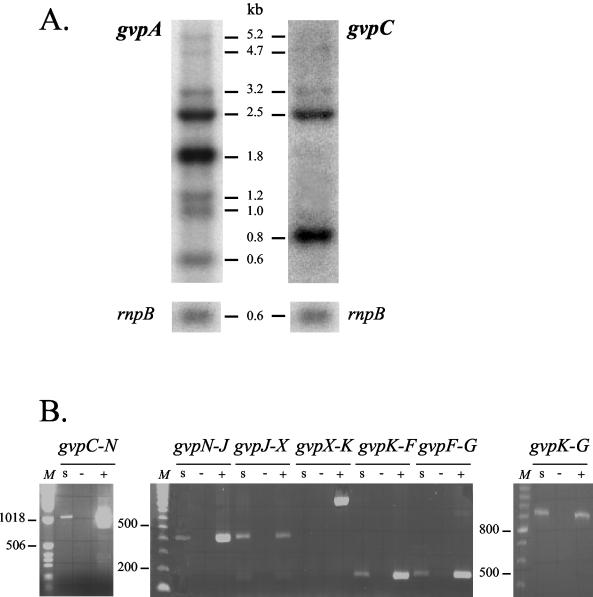
Expression of the genes encoding GvpA and GvpC, the two known structural proteins of the cyanobacterial GV, was examined by RNA-DNA hybridization in the wild-type strain M. aeruginosa PCC 7806 (Fig. 2A). This analysis revealed two major mRNA species of 1.8 and 2.5 kb when a probe internal to the gvpA gene was used. The size of the 1.8-kb transcript species is consistent with cotranscription of the three gvpA genes, while the 2.5-kb transcript may span the region starting with gvpAI and ending after gvpC. Indeed, hybridization of the same blot with a gvpC probe revealed the same 2.5-kb mRNA species but not the 1.8-kb-long one (Fig. 2A). An additional transcript, 0.8 kb long, was also detected that may correspond to an mRNA species containing only gvpC. Hybridization with the gvpA probe also revealed three less-intense mRNA species, which may correspond to the transcription of one (0.6 kb) and two (1.0 and 1.2 kb) copies of the gvpA gene. Since the hybridization bands of the 1.0- and 1.2-kb mRNA species are generally more diffuse than those of the 0.6-kb-long species, they may also correspond to the degradation products of the longer transcripts. Finally, three additional minor mRNA species (3.2, 4.7, and 5.2 kb long) could be detected using both the gvpA and gvpC probes (Fig. 2A). To ascertain the existence of these large transcripts, RT-PCR analyses were conducted. Amplification products corresponding to cotranscription of gvpC and gvpN, of gvpN and gvpJ, and of gvpJ and gvpX were obtained (Fig. 2B). Negative and positive control experiments were performed using RNA and genomic DNA, respectively, as templates. Together, the results of the RNA-DNA hybridization and the RT-PCR analyses indicate that the gvpA to gvpX genes (Fig. 1A) may be expressed as an operon. No transcript longer than 5 kb could be detected by RNA-DNA hybridization; accordingly, no cotranscription between the gvpX and gvpK genes was observed (Fig. 2B). In contrast, RT-PCR products were generated between gvpK and gvpF, between gvpF and gvpG, and between gvpK and gvpG (Fig. 2B). A second operon, gvpKFG, should therefore be transcribed independently from the other gvp genes. The gvpV gene that reads in the reverse orientation must be expressed individually. This is also the case for gvpW, which is not cotranscribed with the downstream ORF in the genome (data not shown).
FIG. 2.
Transcription analysis of the gvp cluster. (A) RNA blot analysis of the gvpA, gvpC, and rnpB transcripts from M. aeruginosa PCC 7806 grown under a day-night cycle (16 h-8 h). The cells were harvested for RNA extraction after 1 h of light when the culture reached OD750 = 0.4. The same blot (15 μg of total RNA per lane) was successively hybridized with different probes: a 180-bp gvpA fragment (obtained by PCR amplification with primers 635 and 636), a 400-bp gvpC fragment (amplified with primers 870 and 871), and (as a control for RNA loading and transfer) a 0.6-kb rnpB fragment (obtained by PCR with the primers mentioned in Materials and Methods). The sizes of the different transcripts are indicated in kilobases (kb). (B) RT-PCR analysis showing cotranscription between the genes of the gvp cluster. The gvpC-N fragment was amplified with primers 893 and 973, the gvpN-J fragment was amplified with primers 975 and 1108, the gvpJ-X fragment was amplified with primers 974 and 1110, the gvpX-K fragment was amplified with primers 978 and 1109, the gvpK-F fragment was amplified with primers 1111 and 1113, the gvpF-G fragment was amplified with primers 965 and 1018, and the gvpK-G fragment was amplified with primers 965 and 1111. s, analyzed sample (with cDNA as a template); −, negative control (with RNA as a template); +, positive control (with genomic DNA as a template). A DNA ladder (100 bp [Amersham Biosciences] or 1 kb [Invitrogen Life Technologies]) was used as a size marker (lanes M).
Mutants carry IS in the gvp genes.
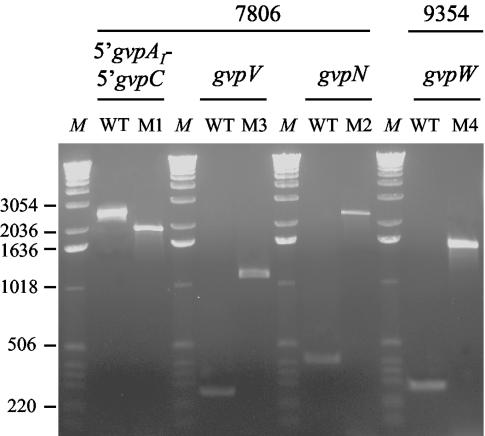
GV-deficient (GV−) mutants appear spontaneously, particularly following stress, at high frequencies in cultures of the M. aeruginosa strains (R. Rippka, unpublished observations). As determined more thoroughly for strain PCC 7806 (see Materials and Methods), the average frequency of GV− mutants in a culture originating from a newly reisolated GV+ phenotype was 8 × 10−4. Significantly higher frequencies (about 7 × 10−2) were observed after the same culture had been cryopreserved in ethylene glycol (2 and 5% [vol/vol]), conditions under which survival of the cells was very low (<10%). In an attempt to elucidate the basis for this phenomenon and to possibly localize the mutations in the gvp region, four mutants deficient in GV were isolated (see Materials and Methods for details): three of strain PCC 7806 and one of strain PCC 9354. With these, a PCR-based molecular analysis was conducted using the primers listed in Table 1. Mutants M2 and M3 of strain PCC 7806 and M4 of strain PCC 9354 revealed PCR products for gvpN, gvpV, and gvpW, respectively, that migrated more slowly on agarose gels than those of the wild-type strains (Fig. 3). Sequencing and analysis of these PCR products revealed the presence of typical IS structures including terminal inverted repeats, direct target repeats, and encoded ORFs with homology to known transposases of the IS4, IS1, and IS5 families (A. Mlouka et al., unpublished data). They were therefore assigned the following names (in accordance with recommended nomenclature guidelines): ISMae2, ISMae3, and ISMae4 for the 1,865-bp insertion in gvpN (mutant PCC 7806 M2), the 834-bp insertion in gvpV (mutant PCC 7806 M3), and the 1,218-bp insertion in gvpW (mutant PCC 9354 M4), respectively (Fig. 1).
FIG. 3.
Detection of the IS by PCR analysis of the gvp region of the wild-type (WT) and the GV-deficient mutant (M1, M2, M3, and M4) strains of M. aeruginosa. Primers used for amplification (see Table 1) were as follows: primers 1006 and 1017 for the 5′ gvpAI-5′ gvpC region, primers 1081 and 1082 for gvpV, primers 949 and 972 for gvpN, and primers 983 and 1077 for gvpW. A 1-kb DNA ladder (Invitrogen Life Technologies) was used as a size marker (lanes M).
No gvpA gene could be amplified with primers 994 and 995 (see Table 1) in mutant PCC 7806 M1 (data not shown). To understand what kind of DNA rearrangement had resulted in the loss of gvpA, primer 1017 (designed 1 kb upstream of gvpAI) was chosen and used together with primer 1006 (located in the gvpAIII-gvpC intergenic region) for PCR amplification (see Table 1). The DNA fragment amplified for the mutant PCC 7806 M1 was approximately 1 kb smaller than that in the wild-type strain (Fig. 3). Sequencing of the PCR products revealed that a complex DNA rearrangement had occurred, since a sequence of 1,318 bp replaced a 2,089-bp fragment starting in the gvpAI upstream region and ending within the coding region of gvpAIII. Therefore, no intact copy of any of the three genes encoding GvpA in the mutant PCC 7806 M1 remained. The inserted sequence, which encodes ORFs displaying some homology to known transposases, was named ISMae1 (Mlouka et al., unpublished). The locations and lengths of each of the four insertion elements are indicated in Fig. 1.
Mutants express the structural gvpA gene but not the GvpA protein.
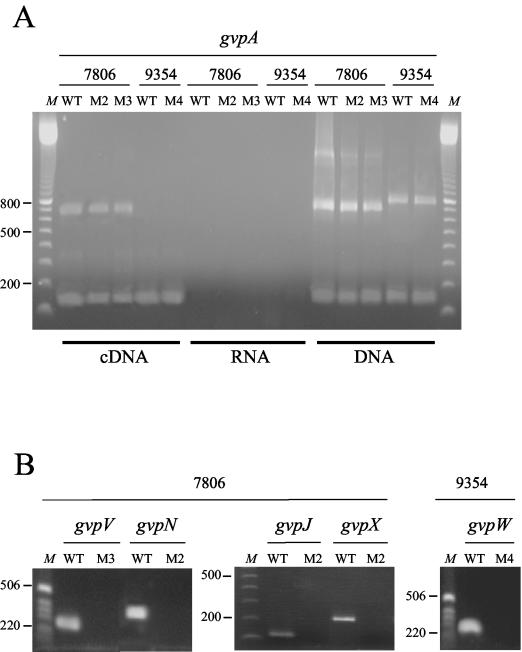
Expression of the genes encoding GvpA was examined by RT-PCR in PCC 7806 M2, PCC 7806 M3, and PCC 9354 M4, three mutant strains of M. aeruginosa. In strain PCC 7806, two fragments were amplified with primers 994 and 995 (Fig. 4A): one of approximately 150 bp, corresponding to one gvpA gene copy (gvpAI, gvpAII, or gvpAIII), and one of 700 bp, corresponding to an amplification between two gvpA gene copies (gvpAI-gvpAII or gvpAII-gvpAIII). Although at least two copies of gvpA are present in strain PCC 9354, only the shorter transcript covering a single gvpA gene copy could be detected when cDNA was used as a template (Fig. 4A). Since the larger fragment amplified with genomic DNA of strain PCC 9354 is slightly longer than that obtained for strain PCC 7806 (Fig. 4A), there might exist some divergence in the gvpA intergenic regions of these two strains. The amplicons obtained from the cDNA were the same size as those obtained with genomic DNA as a positive control. The negative controls confirmed that the RNA samples did not contain any contaminating genomic DNA (Fig. 4A).
FIG. 4.
Transcription analysis of gvp genes in the wild-type and the GV-deficient mutant strains of M. aeruginosa. (A) RT-PCR with the gvpA-specific primers 994 and 995 (see Table 1). The results for PCC 7806 wild-type (WT) and mutant (M2 and M3) strains and PCC 9354 wild-type and mutant (M4) strains are shown. (B) RT-PCR results for the three genes carrying an IS (gvpN, gvpV, and gvpW) and for gvpJ and gvpX. The results for PCC 7806 wild-type (WT) and mutant (M3 and M2) strains and PCC 9354 wild-type and mutant (M4) strains are shown. The gvpV gene was amplified with primers 1081 and 1082, gvpN was amplified with primers 949 and 972, gvpJ was amplified with primers 974 and 975, gvpX was amplified with primers 1109 and 1110, and gvpW was amplified with primers 983 and 1077. A DNA ladder (100 bp [Amersham Biosciences] or 1 kb [Invitrogen Life Technologies]) was used as a size marker (lanes M).
The presence of transcripts for gvpN, gvpV, and gvpW, the genes containing IS, was tested in the mutant strains PCC 7806 M2, PCC 7806 M3, and PCC 9354 M4, respectively (Fig. 4B). While all three genes were expressed in the wild-type strains under the growth conditions tested, the respective genes of the mutants carrying an insertion were not transcribed (Fig. 4B). The gvpN gene was cotranscribed with the downstream gvpJ and gvpX genes in the wild-type strain PCC 7806, but no transcription of gvpJ and gvpX could be detected by RT-PCR in the mutant strain (Fig. 4B), confirming that (as previously mentioned) these genes are part of a large operon.
Immunodetection of the major structural protein GvpA was performed with cell extracts of the wild-type and mutant strains (Fig. 5). Both wild-type strains revealed a single 7.5-kDa band equivalent in mass to that obtained with a fraction enriched in GV and corresponding to the small hydrophobic protein GvpA. No such band was detected for any of the mutants.
FIG. 5.
Immunodetection of GvpA on isolated GV and cell extracts from the wild-type and the GV-deficient mutant strains of M. aeruginosa. GV, enriched GV fraction (20 μg) from PCC 7806 wild-type strain. Other lanes show the results for crude extracts (200 μg) from PCC 7806 wild-type (WT) and mutant (M1, M2, and M3) strains and from PCC 9354 wild-type and mutant (M4) strains.
Mutants lack GV structures.
No gas vacuoles were detectable using phase-contrast microscopy with the M. aeruginosa mutant strains PCC 7806 M1, M2, and M3 and PCC 9354 M4 (data not shown). Analysis of thin sections of cells by electron microscopy confirmed the absence of GV structures in all the mutants (Fig. 6).
DISCUSSION
In this study, we identified a gene cluster involved in the formation of GV in the bloom-forming, hepatotoxic cyanobacteria M. aeruginosa PCC 7806 and PCC 9354. All the previously identified gvp genes in cyanobacteria are present in M. aeruginosa. The overall physical organization of the gvp region of M. aeruginosa PCC 7806 and PCC 9354 is equivalent to that of the Anabaena/Nostoc sp. strain PCC 7120, including the two newly described gvp genes gvpW (a counterpart of alr2245 in the genome of Anabaena/Nostoc sp. strain PCC 7120) and gvpV (a counterpart of alr2246 in the genome of Anabaena/Nostoc sp. strain PCC 7120), the latter being the only gene of this region that reads in an opposite orientation. The main difference is the absence of gvpX from the Anabaena/Nostoc sp. strain PCC 7120 gvp cluster. Genes encoding counterparts of GvpW and GvpV are also found in close proximity to gvp genes in the unannotated genomes of Trichodesmium erythraeum (Tery1205 and Tery1213, respectively) and Nostoc punctiforme ATCC 29133 (Npun5344 and Npun5335, respectively). Available data concerning the gvp cyanobacterial sequences are in line with a common set of genes among members of this phylum, whose physical organization, as well as gene composition and number, differs somewhat from that observed in halobacteria or B. megaterium (Fig. 6).
Spontaneous GV-deficient mutants often appear after prolonged maintenance of M. aeruginosa clones even under standard growth conditions, but their frequency is particularly high following a physiological stress such as cryopreservation. The mutants examined here resulted from independent DNA rearrangements caused by insertion events. There was apparently no preference for a given insertion element, since four different IS, ISMae1-4, were found to interrupt the gvp genes. Although it cannot be excluded that the gvp region of the genome is a hot spot for transposition events, the high frequency of mutations observed may result from a biased selection towards GV-deficient mutants if their survival or growth is favored compared that of to the wild-type cells. Indeed, GV in M. aeruginosa account for up to 10% of the total protein content (13); therefore, their synthesis represents a very costly energetic demand for the cells. In contrast to field conditions, in which M. aeruginosa cells need to regularly rise up to the water surface to increase their photosynthetic activities (36), laboratory culture conditions may provide cells with an environment in which ample nutrient availability (in particular, that of CO2 and light irradiance) render GV production no longer competitive. It is interesting that for a strain of the genus Planktothrix, the only other planktonic cyanobacterium for which GV mutants have been studied, the mutations resulted from DNA rearrangements that did not implicate insertion elements (4).
The GV-deficient mutant PCC 7806 M1 retains no complete copy of any of the three gvpA genes, which explains the absence of the GvpA protein and of a GV structure. As seen with other cyanobacteria that produce abundant GV, multiple copies of the gene encoding the major structural GvpA protein are found in M. aeruginosa PCC 7806 and PCC 9354 (6, 13). The presence of multiple copies of gvpA permits fast synthesis of large amounts of GvpA, which represents 90% in mass of the GV content (36), with minimal participation of the transcriptional machinery. As has been suggested for Anabaena flos-aquae (13), moreover, an operon producing multiple gvpA and gvpC transcripts may represent an efficient means to control the relative abundances of mRNA species of each gene and hence to obtain the correct GvpA/GvpC ratio incorporated in the GV.
Five repeats of 33 amino acids occur in GvpC of A. flos-aquae (13), whereas there are four repeats in M. aeruginosa PCC 7806 and Calothrix sp. strain PCC 7601 (6) and three in Anabaena/Nostoc sp. strain PCC 7120 (16). The variable number of repeats (thought to allow periodic interaction of GvpC with the ribs formed by GvpA) generally correlates with the width and strength of the GV (18, 36), as has been shown for natural variants of Planktothrix spp. (2, 3, 4). Beard and coworkers (2) proposed that Planktothrix strains with shorter GvpC proteins (hence, stronger GV) were favored in deep lakes and that those possessing a longer GvpC variant were better adapted to life close to the surface. The number of repeats found in M. aeruginosa suggests that its GV are stronger than those of A. flos-aquae. Indeed, this observation is in good agreement with the mean critical pressure values calculated for GV isolated from these two planktonic strains (critical pressure = 0.76 and 0.6 Mpa, respectively) (36). These relatively low values imply that such GV are unable to withstand very high pressures, which correlates with the natural habitat of these microorganisms most commonly found at the surface of lakes.
While the roles of GvpA and GvpC are well known (36), the functions of other GV proteins remain unclear. Three of the GV-deficient mutants described in this study retained the presence and transcription of gvpA. The lack of the corresponding GvpA protein might therefore result either from decreased transcriptional efficiency or from a defect of the expression machinery (presumably due to malfunctioning of other Gvp proteins) at a posttranscriptional level. In strains PCC 7806 M3 and PCC 9354 M4, IS interrupt two genes of unknown function (gvpV and gvpW, respectively); in strain PCC 7806 M2, an insertion element is located within the coding sequence of the previously identified gvpN. The lack of expression of these three genes is correlated with a defect in GV formation, thus implying their essential role in this phenomenon.
According to DasSarma and coworkers (7), GvpN plays a role at a late stage of GV formation. In contrast, Offner and coworkers (26) propose that GvpN, while being nonessential, may enhance GV production. Indeed, a conserved nucleotide-binding site (consistent with a regulatory function) is found in all GvpN proteins (reference 27 and this study). GvpN in M. aeruginosa PCC 7806 also shares on approximately half its length significant similarities with MoxR-like ATPases of the AAA+ superfamily, which can play a role in transcriptional regulation or protein quality control (9). Unlike the halobacterial gvpN mutants, strain PCC 7806 M2 does not synthesize any GV structure (see Fig. S1 in the supplemental material). This deficiency would be in favor of a regulatory role for GvpN in M. aeruginosa; no straight conclusion can be drawn, however, since the downstream gvpJ and gvpX genes are no longer expressed in this mutant strain.
Deletion studies carried out in halobacteria demonstrated the absolute requirement for GvpJ, GvpK, GvpF, and GvpL, which have all been suggested to play a role in GV prestructures or assembly (28). The gvp cluster of M. aeruginosa PCC 7806 and PCC 9354, like those of Anabaena/Nostoc sp. strain PCC 7120 and A. flos-aquae, contains three genes encoding GvpJ, GvpK, and GvpF (also named GvpF/L in A. flos-aquae) (17). The putative GvpJ protein displays all the typical characteristics of previously identified GvpJs in terms of length, acidic pI, and similarities to GvpA in its N-terminal part but has no ATG start codon. The functional meaning of a putative GTG initiation codon that would correspond to a low transcriptional level of the gene (reference 33 and references therein) would be consistent with the suggested role of GvpJ in an early stage of GV assembly, requiring only minor amounts of the protein (28). In contrast to GvpJ, GvpX has a very basic pI and shows no homology to GvpA but can be partly aligned with GvpJ. Although GvpX shows limited but detectable resemblance to the C-terminal ends of other cyanobacterial GvpJ proteins, it is far more similar to that of M. aeruginosa GvpJ. This observation leads us to predict that the gvpX gene might have arisen from a duplication of part of gvpJ.
A potential candidate for the function of GvpL in Microcystis spp. is GvpW, whose length and acidic pI are consistent with those calculated for the corresponding halobacterial proteins. The counterpart of GvpW in Anabaena/Nostoc sp. strain PCC 7120 (All2245) has been assigned no function, most likely because a BLAST search produces only low levels of similarity to proteins in the public databases. Nevertheless, the best match corresponds to a Gvp protein, namely, GvpL of B. megaterium. The absence of GV in the mutant strain PCC 9354 M4 (Fig. 6) reinforces the hypothesis that GvpW, like GvpL of halobacteria and B. megaterium, is essential for GV formation in M. aeruginosa. Moreover, although gvpA is expressed in this strain, no GvpA protein could be detected in crude extracts. If, as has been suggested for gvpL, gvpW encodes a minor structural protein, the absence of such a protein may impede the assembly or stabilization of the structures and the unassembled GvpA may be subsequently degraded. The gvp-associated location of this gene in four cyanobacterial genomes, the significant similarities of its product to GvpL proteins, and the phenotype observed when its coding sequence is interrupted by an IS prompt us to propose gvpW as a nonorthologous equivalent of the halobacterial gvpL in cyanobacteria. As for gvpV, the other newly identified gene of unknown function, the difficulty of predicting its roles arises from the lack of homology to proteins of the public databases, as well as from the various mutations resulting in similar phenotypes in the gvp gene cluster of M. aeruginosa.
The gvp gene cluster presented in this work is one of the most complete set of gvp genes described for cyanobacteria. All mutations in this region involved gene rearrangements due to insertion elements, with or without an associated deletion. Molecular characterization of GV-deficient spontaneous mutants led to the discovery of gvpV and gvpW, two new gvp genes shown here to be essential to GV formation. The fortuitously high mutation frequency, particularly after recovery from cryopreservation, can now be exploited further to gain additional new insights into the function of gvp genes in M. aeruginosa, a cyanobacterium so far relatively refractory to genetic manipulation.
Supplementary Material
Acknowledgments
We are very grateful to M.C. Prévost for electron microscopy and photography and to S. Ferris, C. Pichon, L. Frangeul, A. Marcel, P. Glaser, and S. Cole for the sequencing of the Microcystis genome. We express our gratitude to Rosmarie Rippka for valuable discussions. We also thank M. Herdman and A. Singer for helpful support and advice.
This work was supported by the Institut Pasteur, the Centre National de la Recherche Scientifique (URA 2172), and the Programme “Génopole” from the Ministère de l'Education Nationale, de la Recherche et de la Technologie (MENRT). A. Mlouka was a recipient of a Ph.D. fellowship from the MENRT. K. Comte was the recipient of a fellowship from the European project “COBRA” (QLRT-2000-01645).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Albouy, D., A. M. Castets, and N. Tandeau de Marsac. 2001. The gas vesicle gene (gvp) cluster of the cyanobacterium Pseudanabaena sp. strain PCC 6901. DNA Sequence 12:337-344. [DOI] [PubMed] [Google Scholar]
- 2.Beard, S. J., P. A. Davis, D. Iglesias-Rodríguez, O. M. Skulberg, and A. E. Walsby. 2000. Gas vesicle genes in Planktothrix spp. from Nordic lakes: strains with weak gas vesicles possess a longer variant of gvpC. Microbiology 146:2009-2018. [DOI] [PubMed] [Google Scholar]
- 3.Beard, S. J., B. A. Handley, P. K. Hayes, and A. E. Walsby. 1999. The diversity of gas vesicle genes in Planktothrix rubescens from Lake Zürich. Microbiology 145:2757-2768. [DOI] [PubMed] [Google Scholar]
- 4.Beard, S. J., B. A. Handley, and A. E. Walsby. 2002. Spontaneous mutations in gas vesicle genes of Planktothrix spp. affect gas vesicle production and critical pressure. FEMS Microbiol. Lett. 215:189-195. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, D., J. Houmard, and N. Tandeau de Marsac. 1993. Electron transport regulates cellular differentiation in the filamentous cyanobacterium Calothrix. Plant Cell 5:451-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damerval, T., J. Houmard, G. Guglielmi, K. Csiszàr, and N. Tandeau de Marsac. 1987. A developmentally regulated gvpABC operon is involved in the formation of gas vesicles in the cyanobacterium Calothrix 7601. Gene 54:83-92. [DOI] [PubMed] [Google Scholar]
- 7.DasSarma, S., P. Arora, F. Lin, E. Molinari, and L. R. S. Yin. 1994. Wild-type gas vesicle formation requires at least ten genes in the gvp gene cluster of Halobacterium halobium plasmid pNRC100. J. Bacteriol. 176:7646-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DasSarma, S., J. T. Halladay, J. G. Jones, J. W. Donovan, P. J. Giannasca, and N. Tandeau de Marsac. 1988. High-frequency mutations in a plasmid-encoded gas vesicle gene in Halobacterium halobium. Proc. Natl. Acad. Sci. USA 85:6861-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougan, D. A., A. Mogk, K. Zeth, K. Turgay, and B. Bukau. 2002. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 529:6-10. [DOI] [PubMed] [Google Scholar]
- 10.Englert, C., K. Krüger, S. Offner, and F. Pfeifer. 1992. Three different but related gene clusters encoding gas vesicles in halophilic archaea. J. Mol. Biol. 227:586-592. [DOI] [PubMed] [Google Scholar]
- 11.Englert, C., G. Wanner, and F. Pfeifer. 1992. Functional analysis of the gas vesicle gene cluster of the halophilic archaeon Haloferax mediterranei defines the vac-region boundary and suggests a regulatory role for the gvpD gene or its product. Mol. Microbiol. 6:3543-3550. [DOI] [PubMed] [Google Scholar]
- 12.Guglielmi, G., and G. Cohen-Bazire. 1982. Structure et distribution des pores et des perforations de l'enveloppe de peptidoglycane chez quelques cyanobactéries. Protistologica 18:151-165. [Google Scholar]
- 13.Hayes, P. K., and R. S. Powell. 1995. The gvpA/C cluster of Anabaena flos-aquae has multiple copies of a gene encoding GvpA. Arch. Microbiol. 164:50-57. [DOI] [PubMed] [Google Scholar]
- 14.Harada, K. I., M. Oshikata, H. Uchida, M. Suzuki, F. Kondo, K. Sato, Y. Ueno, S. Z. Yu, G. Chen, and G. C. Chen. 1996. Detection and identification of microcystins in the drinking water of Haimen city, China. Nat. Toxins 4:277-283. [DOI] [PubMed] [Google Scholar]
- 15.Jones, J. G., N. R. Hackett, J. T. Halladay, D. J. Scothorn, C. F. Yang, W. L. Ng, and S. DasSarma. 1989. Analysis of insertion mutants reveals two new genes in the pNRC100 gas vesicle gene cluster of Halobacterium halobium. Nucleic Acids Res. 17:7785-7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 17.Kinsman, R., and P. K. Hayes. 1997. Genes encoding proteins homologous to halobacterial Gvps N, J, K, F & L are located downstream of gvpC in the cyanobacterium Anabaena flos-aquae. DNA Sequence 7:97-106. [DOI] [PubMed] [Google Scholar]
- 18.Kinsman, R., A. E. Walsby, and P. K. Hayes. 1995. GvpCs with reduced numbers of repeating sequence elements bind to and strengthen cyanobacterial gas vesicles. Mol. Microbiol. 17:147-154. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Li, N., and M. C. Cannon. 1998. Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J. Bacteriol. 180:2450-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liotenberg, S., D. Campbell, R. Rippka, J. Houmard, and N. Tandeau de Marsac. 1996. Effect of the nitrogen source on phycobiliprotein synthesis and cell reserves in a chromatically adapting filamentous cyanobacterium. Microbiology 142:611-622. [DOI] [PubMed] [Google Scholar]
- 22.Mayr, A., and F. Pfeifer. 1997. The characterization of the nv-gvpACNOFGH gene cluster involved in gas vesicle formation in Natronobacterium vacuolatum. Arch. Microbiol. 168:24-32. [DOI] [PubMed] [Google Scholar]
- 23.Ng, W. L., P. Arora, and S. Dassarma. 1994. Large deletions in class III gas vesicle-deficient mutants of Halobacterium halobium. Syst. Appl. Microbiol. 16:560-568. [Google Scholar]
- 24.Offner, S., A. Hofacker, G. Wanner, and F. Pfeifer. 2000. Eight of fourteen gvp genes are sufficient for formation of gas vesicles in halophilic archaea. J. Bacteriol. 182:4328-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Offner, S., and F. Pfeifer. 1995. Complementation studies with the gas vesicle-encoding p-vac region of Halobacterium salinarium PHH1 reveal a regulatory role for the p-gvpDE genes. Mol. Microbiol. 16:9-19. [DOI] [PubMed] [Google Scholar]
- 26.Offner, S., G. Wanner, and F. Pfeifer. 1996. Functional studies of the gvpACNO operon of Halobacterium salinarium reveal that the GvpC protein shapes gas vesicles. J. Bacteriol. 178:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeifer, F., and C. Englert. 1992. Function and biosynthesis of gas vesicles in halophilic Archaea. J. Bioenerg. Biomembr. 24:577-585. [DOI] [PubMed] [Google Scholar]
- 28.Pfeifer, F., K. Krüger, R. Röder, A. Mayr, S. Ziesche, and S. Offner. 1997. Gas vesicle formation in halophilic Archaea. Arch. Microbiol. 167:259-268. [DOI] [PubMed] [Google Scholar]
- 29.Pouria, S., A. de Andrade, J. Barbosa, R. L. Cavalcanti, V. T. S. Barreto, C. J. Ward, W. Preiser, G. K. Poon, G. H. Neild, and G. A. Codd. 1998. Fatal microcystin intoxication in haemodialysis unit in Caruaru, Brazil. Lancet 352:21-26. [DOI] [PubMed] [Google Scholar]
- 30.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sazuka, T., and O. Ohara. 1996. Sequence features surrounding the translation initiation sites assigned on the genome sequence of Synechocystis sp. strain PCC6803 by amino-terminal protein sequencing. DNA Res. 31:225-232. [DOI] [PubMed] [Google Scholar]
- 33.Sussman, J. K., L. Simons, and R. W. Simons. 1996. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 21:347-360. [DOI] [PubMed] [Google Scholar]
- 34.Ueno, Y., S. Nagata, T. Tsutsumi, A. Hasegawa, M. F. Watanabe, H. D. Park, G. C. Chen, G. Chen, and S. Z. Yu. 1996. Detection of microcystins, a blue-green algal hepatotoxin, in drinking water sampled in Haimen and Fusui, endemic areas of primary liver cancer in China, by highly sensitive immunoassay. Carcinogenesis 17:1317-1321. [DOI] [PubMed] [Google Scholar]
- 35.Vioque, A. 1992. Analysis of the gene encoding the RNA subunit of ribonuclease P from cyanobacteria. Nucleic Acids Res. 20:6331-6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walsby, A. E. 1994. Gas vesicles. Microbiol. Rev. 58:94-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.