Abstract
Genome sequencing efforts have revealed a strikingly large number of unannotated and uncharacterized genes that fall into metabolic enzymes classes, likely indicating that our current knowledge of biochemical pathways in normal physiology, let alone in disease states, remains largely incomplete. This realization presents a daunting challenge for post-genomic-era scientists in deciphering the biochemical and (patho)physiological roles of these enzymes and their metabolites and metabolic networks. This is further complicated by many recent studies showing a rewiring of normal metabolic networks in disease states to give rise to unique pathophysiological functions of enzymes, metabolites, and metabolic pathways. This review focuses on recent discoveries made using metabolic mapping technologies to uncover novel pathways and metabolite-mediated posttranslational modifications and epigenetic alterations and their impact on physiology and disease.
a major realization made from the human genome sequencing efforts was that a significant portion of the genome remained completely uncharacterized, including genes that encode enzymes that presumably metabolize small-molecule metabolites (48). This realization has led to the daunting task of deciphering the genetic blueprint by assigning biochemical and physiological functions to each of these encoded proteins in both normal and disease states. Metabolomics, an -omic technology that has arisen to profile the entirety of metabolites in a complex biological sample, has emerged as a powerful approach for mapping metabolic pathways and deciphering enzyme function in complex mammalian systems through the measurement of small-molecule metabolites or tracking isotopic incorporation of tracers into the metabolome(49). A variety of metabolomic platforms have arisen including nuclear magnetic resonance (NMR), gas chromatography-mass spectrometry (GC-MS), and liquid chromatography-MS (LC-MS), all of which possess their various strengths and weaknesses, in terms of ideal methods for characterizing certain physicochemical classes of metabolites, towards comprehensively mapping the metabolome. In this review, we specifically discuss 1) how targeted and untargeted metabolomic profiling approaches have been used to characterize the functions of previously uncharacterized enzymes, 2) how metabolomic profiling has uncovered unique functions for previously characterized enzymes, 3) how isotopic tracing-based approaches have been utilized to map pathway utilization in complex biological systems, and 4) how metabolic pathways influence posttranslational and epigenetic regulation of the proteome and genome.
Metabolomic Profiling Approaches Used to Characterize the Functions of Previously Uncharacterized Enzymes
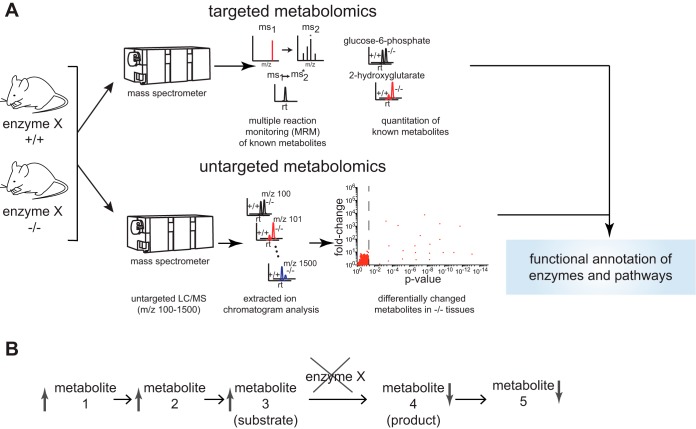
Targeted and untargeted metabolomic strategies have proved to be a fruitful strategy for deciphering functions of previously uncharacterized enzymes. Targeted metabolomics is most often used in metabolomic profiling experiments in which several hundred known metabolites are quantified in complex biological systems. Targeted metabolomics can be performed using GC- or LC-MS-based platforms and multiple-reaction monitoring (MRM)-based approaches. MRM quantitates fragment ions arising from specific parent ion mass-to-charge ratios (m/z), based on previous fragmentation analysis of metabolite standards (11). This MRM feature allows for accurate and quantitative analysis of even very low abundance metabolites. However, the metabolome is highly physicochemically diverse and likely consists of many yet-unknown metabolites or metabolites that may not be in standard targeted metabolomic methods. Thus, untargeted metabolomics has emerged as a complementary approach to broadly identify metabolites that are altered between comparison groups (49). These experiments are performed by setting the mass spectrometer to scan a broad m/z range. The mass spectra collected from these experiments are then analyzed by bioinformatics platforms that quantitate all detectable ions and then statistically sort for those ions that are altered between comparison groups (Fig. 1A). Subsequent database searching coupled with traditional analytical chemistry procedures are used to identify the ions of interest. These targeted and untargeted strategies have been used to uncover the biochemical functions of many uncharacterized enzymes, thus providing a foundation for understanding their (patho)physiological roles (Fig. 1B).
Fig. 1.
Using targeted and untargeted metabolomics to decipher enzyme and pathway functions. A and B: functional targeted and untargeted metabolomic platforms (A) have arisen to assist in characterizing the functions of metabolic enzymes in complex biological systems (B). In a typical functional metabolomic experiment, one can comparatively profile alterations in the metabolome, comparing a control group to impairment or overexpression of a given metabolic enzyme in a complex biological or disease sample. This experiment can be comparing groups such as wild-type (+/+) vs. knockout (−/−) samples, vehicle- vs. enzyme inhibitor-treated samples, shControl vs. shEnzyme, or control vs. enzyme overexpression. Elevated or reduced metabolites from blocking an enzyme will assist in assigning substrate/product relationships but also metabolic pathway connectivities.
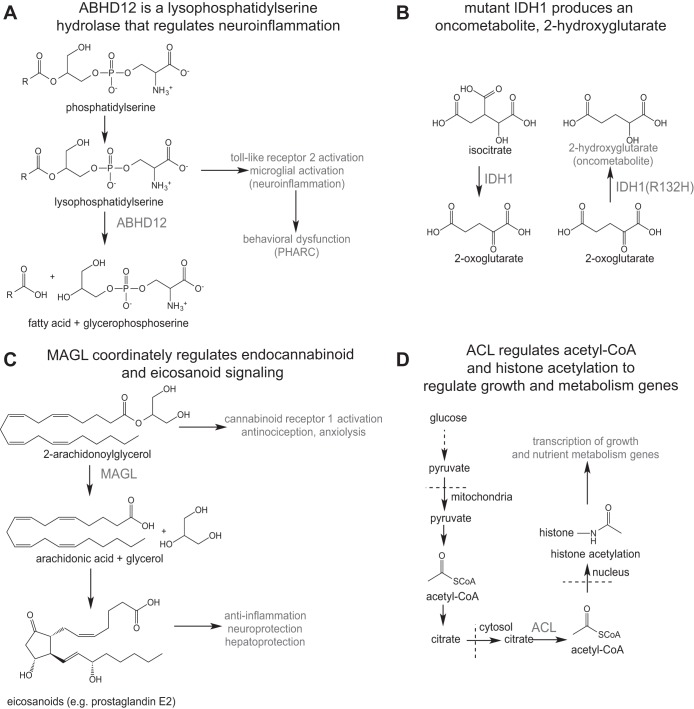
Blankman et al. (3) recently uncovered the function of the previously uncharacterized enzyme α/β-hydrolase domain-containing 12 (ABHD12), a serine hydrolase that was mutationally inactivated in patients with the neurodegenerative disorder PHARC (polyneuropathy, hearing loss, ataxia, retinosis, pigmentosa, and cataract). Blankman and colleagues generated ABHD12−/− mice, which present the PHARC phenotype, to use in metabolomic studies for mechanistic understanding of ABHD12 in PHARC. Untargeted metabolomics of ABHD12−/− mouse brain revealed elevated levels of many members of the lysophosphatidylserine (LPS) lipid class, and subsequent activity assays showed that ABHD12 is the principal LPS hydrolase in mouse brain (Fig. 2A). Blankman et al. went further to show that LPS accumulation in the brains of ABHD12-deficient mice led to neuroinflammation by activation of toll-like receptor 2, leading to microglial activation and motor and auditory defects reminiscent of PHARC (3). This work on ABHD12 shows that untargeted metabolomics can be used to implicate unexpected pathways in disease towards linking genotypes to phenotypes.
Fig. 2.
Examples of metabolic pathways that have been discovered by metabolomics. A: ABHD12 was recently characterized as a lysophosphatidylserine hydrolase. ABHD12 inactivation leads to accumulations in lysophosphatidylserine, leading to neuroinflammation, and behavioral dysfunction and PHARC. B: while wild-type IDH1 converts isocitrate to 2-oxoglutarate, mutant IDH1 found in gliomas was shown to convert 2-oxoglutarate to an oncometabolite, 2-hydroxyglutarate, shown to inhibit demethylases and alter the epigenetic landscape to drive cancer pathogenicity. C: MAGL was found to control endocannabinoid (2-AG) signaling and also control the major arachidonic acid precursor pool for generating proinflammatory eicosanoids. MAGL blockade leads to accumulation in 2-AG and cannabinoid-dependent antinocicpetion and anxiolysis while also lowering arachidonic acid and eicosanoids and reducing inflammation that subserves neurodegeneration and hepatic injury. D: recent studies have uncovered that acetyl-CoA levels and metabolism directly control histone acetylation and transcription of growth and nutrient metabolism genes. See text for definitions.
Long et al. (20) used an untargeted metabolomics screening approach wherein they overexpressed a series of uncharacterized serine hydrolases in HEK293T cells and mined for depletion or accumulation of endogenous metabolites. Using this approach, the authors identified ABHD3 as a hydrolase of medium-chain phospholipids. The authors then confirmed this in vitro finding in vivo by showing that ABHD3 knockout mice possessed elevated levels of myristoyl-phospholipids, including the bioactive lipid myristoyl-lysophosphatidylcholine.
In another example, studies had shown that the uncharacterized serine hydrolase KIAA1363 was highly upregulated across aggressive human cancer cells and malignant human breast tumors, but the function of this enzyme in cancer cells remained enigmatic. Using untargeted metabolomics, Chiang et al. (6) showed that KIAA1363 was a deacetylase of the ether lipid 2-acetyl monoalkylglycerol ether (2-acetyl MAGE). KIAA1363 inactivation led to lower levels of the product MAGE and the MAGE phosphorylation product alkyl-lysophosphatidic acid (alkyl-LPA). Alkyl-LPA was a known oncogenic signaling lipid that fueled cancer pathogenicity through LPA receptor signaling. Chiang et al. showed that KIAA1363 inactivation led to impaired cancer cell motility and tumorigenesis by lowering alkyl-LPA signaling in cancer cells.
Metabolomic Profiling Approaches Used to Uncover Unique Functions of Previously Characterized Enzymes
Targeted and untargeted metabolomic approaches have also been crucial in elucidating the oftentimes rewired, retasked, or novel functions of enzymes in tissue-specific or disease-specific contexts. In 2009, Dang et al. (8) used untargeted metabolomic profiling to uncover a neomorphic function of mutant isocitrate dehydrogenase (IDH) in cancer cells. IDH1 mutations at Arg132 have been found in many types of brain tumors. Wild-type IDH1 catalyzes the oxidative decarboxylation of isocitrate to 2-oxoglutarate as part of the tricarboxylic acid (TCA) cycle. Dang et al. used untargeted metabolomics to profile alterations in the metabolome in cells overexpressing IDH1 Arg132His compared with wild-type IDH1-overexpressing cells and found a dramatic accumulation in the novel metabolite, 2-hydroxyglutarate (2-HG) (Fig. 2B). The authors showed that mutant IDH1 performs a neomorphic function of converting 2-oxoglutarate to 2-HG. It is still unclear whether mutations in IDH1 are tumor promoting through a loss of function of IDH1 endogenous function or through its novel function of generating 2-HG. However, subsequent studies have shown that 2-HG acts as an oncometabolite by acting as a competitive inhibitor of α-ketoglutarate-dependent demethylases, including histone demethylases and the TET family of 5-methyl-cytosine hydroxylases, leading to epigenetic changes that drive cancer (52). Inhibitors of mutant IDH1 are now under consideration for cancer therapy (9).
Targeted and untargeted metabolomic profiling was successfully used to reveal novel biochemical functions for the lipolytic enzyme monoacylglycerol lipase (MAGL) in both normal and pathological states. It was already known that MAGL hydrolyzed monoacylglycerols to glycerol and fatty acids in vitro and that it was likely the primary enzyme responsible for the ultimate step of triacylglycerol hydrolysis (21, 47). MAGL was also postulated to be the primary hydrolase for the monoacylglycerol endocannabinoid signaling lipid 2-arachidonoylglycerol (2-AG) (10). A series of studies using targeted and untargeted metabolomic profiling revealed that MAGL not only controls monoacylglycerol metabolism, including 2-AG levels, but also arachidonic acid and arachidonic acid-derived eicosanoids levels (22–24, 33, 34, 37, 42) (Fig. 2C). MAGL blockade in the brain, for example, was shown to cause dramatic elevations in 2-AG levels and robust reductions in arachidonic acid and eicosanoid levels. In other tissues, MAGL blockade showed greater changes in other monoacylglycerols compared with modest increases in 2-AG levels, indicating differential monoacylglycerol metabolism in central vs. peripheral tissues (23). Nomura et al. (37) revealed that the connectivity between 2-AG hydrolysis and arachidonic acid and eicosanoids was also tissue specific in that MAGL linked these two metabolic pathways only in brain, liver, and lung, whereas other enzymes such as phospholipase 2g4a (Pla2g4a) mediated arachidonic acid release for eicosanoid production in spleen and the gut. More importantly, these insights showing that MAGL blockade leads to enhanced 2-AG endocannabinoid signaling and suppressed proinflammatory eicosanoid signaling in certain tissues led to key biological discoveries showing that MAGL inhibitors exhibit endocannabinoid-dependent antinociceptive, anxiolytic, and anti-inflammatory effects as well as eicosanoid suppression-dependent anti-inflammatory, neuroprotective, and hepatoprotective effects (5, 15, 16, 22, 37, 38, 53).
Using untargeted metabolomic platforms, Nomura et al. (37) also found that MAGL plays a unique and distinct role in regulating a fatty acid network enriched in protumorigenic signaling lipids in aggressive human cancer cells. Nomura et al. showed that MAGL was highly upregulated in aggressive human cancer cells and primary human tumors, but the role of MAGL in cancer was unclear. Untargeted metabolomics was used to show that MAGL inactivation lowered cancer cell fatty acid levels and fatty acid-derived oncogenic signaling lipids such as LPA and eicosanoids. LPA and eicosanoids have been shown to fuel aggressive and tumorigenic features of cancer cells by stimulating their respective G protein-coupled receptors. The authors showed that MAGL inactivation thwarted migratory, invasive, and tumorigenic properties of aggressive cancer cells by lowering fatty acids and protumorigenic signaling lipids (35, 36). This control of MAGL over lipolytic processes was a unique feature of this enzyme in cancer cells, as MAGL blockade does not result in global changes in fatty acid levels in most tissues.
Benjamin et al. (1) recently used targeted and untargeted metabolomic profiling to define the role of the ether-lipid generating enzyme alkylglycerone phosphate synthase (AGPS) in cancer cells. AGPS is a critical early step in ether lipid synthesis and catalyzes the reaction of acyl-glycerone phosphate to alkyl-glycerone phosphate, which is then converted to alkyl-LPA and other lipids in the ether lipid family. Ether lipids were known to be heightened in proliferative cells and primary tumors, but the biochemical and pathophysiological function of these lipids were unknown. Benjamin et al. showed that AGPS and ether lipid levels were heightened in aggressive human cancer cells and that AGPS knockdown led to impairments in aggressive and tumorigenic properties of cancer cells. Using a combination of targeted and untargeted metabolomics, Benjamin et al. found that AGPS knockdown led to not only a depletion of ether lipid levels but also alterations in fatty acids, acylglycerophospholipids, and eicosanoids. Using isotopic tracing methods, the authors showed that AGPS knockdown led to a diversion of arachidonic acid away from oncogenic signaling lipids such as prostaglandins and toward structural lipids such as phosphatidylcholines, through the upregulation of lysophatidylcholine acyltransferase-1 (1). Overall, the authors showed that AGPS drives cancer malignancy through heightening the levels of key oncogenic signaling lipids such as alkyl-LPA and prostaglandins and that AGPS modulates not only ether lipids but fatty acid utilization to promote an optimal landscape of signaling lipids that drives cancer pathogenicity.
In another example, Possemato et al. (39) and Locasale et al. (18) both established independently that phosphoglycerate dehydrogenase (PHGDH) and metabolic flux into serine and glycine metabolism was a critical node that drives cancer pathogenicity. Utilizing heteronuclear single quantum coherence spectroscopy NMR and isotopic tracing using targeted LC-MS-based metabolomics of [13C]glucose-labeled cells, Locasale et al. found significant 13C incorporation into 3-phosphoserine and serine pathways through PHGDH (18). Possemato et al. screened a set of metabolic genes associated with aggressive breast cancer by using negative-selection RNAi in a human breast cancer xenograft model to identify metabolic enzymes important for tumorigenesis, and they found PHGDH a critical enzyme in breast cancer pathogenicity (39). Both Locasale et al. and Possemato et al. used functional metabolomics to define the roles of PHGDH in cancer cells. Locasale et al. showed that PHGDH knockdown in Sk-Mel28 melanoma cancer cells lowers phosphoserine levels and an accumulation in glycolytic intermediates near the point of diversion into serine metabolism, confirming that the level of PHGDH expression alters glucose metabolism in cells with high PHGDH expression. Possemato et al. showed that PHGDH knockdown in MDA-MB-231 breast cancer cells results in decreases in TCA cycle metabolites, including citrate, isocitrate, α-ketoglutarate, succinate, fumarate, and malate, and quite provocatively showed that one-half of α-ketoglutarate was derived from the serine pathway. Both groups showed that inactivation of PHGDH led to substantial impairments in cancer pathogenicity (18, 39). Thus, metabolomic profiling by two independent groups identified the serine pathway through PHGDH as a critical metabolic node that drives cancer pathogenicity.
Metabolomics has also been useful in defining metabolic drivers of viral infection. Viral infection by herpes simplex virus 1 (HSV-1) results in widespread changes in host metabolism to provide energy and macromolecular precursors to fuel viral replication (46). To explore the role of individual enzymes in viral infection, Grady et al. (12) used a siRNA screen to show that argininosuccinate synthase-1 (AS1) knockdown increased virus yield. AS1 catalyzes the rate-limiting step of the synthesis of the essential amino acid arginine from aspartate and is reduced upon HSV-1 infection. Metabolomics revealed that AS1 knockdown resulted in a metabolic signature that closely mimicked that of HSV-1 infection. Specifically, AS1 knockdown led to reductions in aspartate levels and the levels of carbamoyl-aspartate, one of the first committed metabolites on the pathway to nucleotide synthesis, as well as the levels of many nucleotides and their precursors (12). Metabolomic strategies have also been used to survey altered host metabolism upon viral infection for many other viruses, including human cytomegalovirus (hCMV), influenza, Dengue, and hepatitis C, which alter host metabolism during infection (2, 31, 32, 41). Thus, metabolomic profiling has been essential toward identifying key metabolic nodes that may be targeted for fighting viral infection.
Isotopic Tracing-Based Metabolomic Approaches Used to Map Metabolic Pathway Utilization in Disease
Isotopic labeling strategies to measure metabolic fluxes and pathway utilization have been used for many decades. In cell-based isotopic labeling experiments, cells are fed with isotopically labeled metabolites, and the isotope is traced through downstream metabolites by use of mass spectrometry. We will focus on some of the recent discoveries using isotopic tracing-based metabolomic approaches.
Isotope tracing has been recently used to show that Ras-transformed cancer cells utilize macropinocytosis, the uptake of extracellular fluid and its contents, to replenish amino acid stores (7). By use of isotopic tracing, cancer cells were shown to uptake and catabolize yeast protein to utilize the building blocks for glutamine synthesis and many other metabolic pathways required for cellular biochemistry. Inhibition of macropinocytosis inhibited the growth of pancreatic tumors and represents another branch of metabolism that can be exploited to fight cancer (7).
In another example, Kamphorst et al. (14) used [13C]glucose and [13C]glutamine to trace de novo lipogenesis as a function of oncogene expression and oxygen availability. [13C]glucose and [13C]glutamine can be traced into isotopically labeled citrate and then into acetyl-CoA, which can be used by cells to make palmitate in the de novo lipogenesis pathway. In normoxia, glucose was found to be the primary carbon source for lipogenic acetyl-CoA, whereas glutamine-derived carbon had a larger contribution in hypoxia (14). Interestingly, the authors showed that Ras-transformed cells and hypoxic cells increase fatty acid import through scavenging media sources of fatty acids primarily in the form of lysophospholipids, thus relying less on de novo lipogenesis (14). These results were quite striking, since cancer cells were thought to rely almost solely on de novo lipogenesis rather than exogenous uptake of lipids for generating cellular lipid pools (28, 29). Indeed, Nomura et al. (36) had shown previously that addition of exogenous fatty acids rescued the pathogenic defects conferred by knocking down MAGL, the lipolytic supplier of fatty acids, in aggressive cancer cells and showed that isotopically labeled fatty acids could be taken up and metabolically incorporated into more complex lipids in cancer cells. Louie et al. (25) recently also used isotopic fatty acid tracing-based targeted and untargeted metabolomic profiling to show that cancer cells robustly incorporate exogenous palmitic acid into complex phospholipids, sphingolipids, acylcarnitines, and ether lipids, including several key oncogenic signaling lipids such as LPA and ceramide 1-phosphate. Those authors also showed that aggressive cancer cells divert fatty acid incorporation away from oxidative pathways and toward structural and oncogenic signaling lipids.
Metabolic Control of Posttranslational and Epigenetic Regulation of the Proteome and Genome
Small-molecule metabolites have been historically known to confer posttranslational and epigenetic modifications onto the proteome and genome, respectively, such as with acetyl-CoA and acetylation, S-adenosylmethionine (SAM) and methylation, or ATP and phosphorylation (51). However, most of the attention around this metabolite-mediated regulation on protein and gene function has focused on the enzymes that append or remove these modifications rather than the pathways that generate the metabolites themselves. Recent studies have revealed the exciting and provocative realization that metabolic pathways and fluxes play an important role in dictating posttranslational and epigenetic regulation. This intersection between metabolism and protein/gene regulation is likely to be greatly expanded upon in the coming years as we uncover novel metabolites, metabolic pathways, and protein/gene modifications that influence overall (patho)physiology. We provide some recent examples of these discoveries.
In addition to the aforementioned well-known posttranslational protein modifications, there are likely to be additional yet-uncharacterized modifications that are important in protein regulation. Moellering et al. (30) recently discovered a novel nonenzymatic 3-phosphoglyceryl-lysine (pgK) posttranslational modification generated by covalent modification of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) product 1,3-bisphosphoglycerate (1,3-BPG) to lysine residues. Many glycolytic enzymes were found to have pgK modifications, including GAPDH itself and enolase-1 (ENO1), leading to inhibition of their activities. The authors showed that cells exposed to high glucose accumulated pgK modifications on glycolytic enzymes to create a potential feedback mechanism that contributes to the build-up and redirection of glycolytic intermediates to biosynthetic pathways (30).
Acetyl-CoA is a critical metabolite that influences acetylation, especially of histones, which in turn influences epigenetic regulation of gene transcription. Histone acetylation by histone acetyltransferases (HATs) and removal of acetylation by histone deacetylases (HDACs) were thought to be the main regulators of histone acetylation. However, recent studies have highlighted the importance of acetyl-CoA levels and the metabolic pathways that regulate acetyl-CoA levels in histone acetylation and gene transcription. Using two-dimensional GC-MS-based metabolomic profiling, Tu et al. (43) intriguingly showed that intracellular metabolites, particularly acetyl-CoA levels, periodically changed during different phases of the yeast cell cycle, indicating that the abundance of key metabolites might help control the temporal regulation of cellular processes and establishment of cell cycle. Subsequently, Cai et al. (4) provocatively showed that acetyl-CoA levels increase substantially upon entry into growth in yeast and consequently induce the Gcn5p/SAGA-catalyzed acetylation of histones at genes important for growth, thereby promoting rapid transcription and commitment to growth. Cai et al. showed that acetyl-CoA functions as a carbon source rheostat that signals the initiation of cell growth programming through specific histone acetylation (4). Wellen et al. (50) also showed that acetyl-CoA levels, generated by ATP-citrate lyase (ACL) in mammalian cells influences cell growth and differentiation and that ACL-mediated acetyl-CoA production drove histone acetylation and gene expression, prompting growth factor-induced increases in nutrient metabolism (Fig. 2D). Many metabolic genes were shown to be regulated by ACL-dependent histone acetylation, including glucose transporter Glut4 and glycolytic regulators hexokinase II (HKII), phosphofructokinase-1 (PFK-1), and lactate dehydrogenase A (LDH-A), showing a powerful link between metabolism and histone acetylation, even in mammalian cells (50).
In another example, Ulanovskaya et al. (45) showed an exciting link between nicotinamide N-methyltransferase (NNMT), histone methylation, and epigenetic regulation that drives cancer aggressiveness. NNMT methylates nicotinamide, a precursor to NAD+, using S-adenosylmethionine (SAM), modulating two important energy metabolites. SAM is the principle biological methyl donor utilized in transmethylation, transsulfuration, and polyamine synthesis (27). NNMT is overexpressed in a variety of tumors. Ulanovskaya et al. used metabolomics to uncover that overexpression of NNMT led to reduced SAM levels and increased levels of products S-adenosylhomocysteine (SAH) and 1-methylnicotinamide (1MNA) (45). This resulted in a decrease in cellular methylation potential by diverting SAM methylation toward 1MNA and away from histone methylation. Subsequent DNA microarray experiments showed that NNMT overexpression upregulates many protumorigenic gene products, including SNAI2, TGFB2, and CNTN1 (45). This work showed yet another example of how a metabolic pathway, namely NNMT and generation of 1MNA, could divert SAM-mediated methylation away from histone methylation enough to exert control over the epigenetic and aggressive features of cancer cells.
NNMT has also been recently implicated in obesity and type 2 diabetes. Kraus et al. (17) discovered using DNA array analysis that NNMT expression is inversely correlated with Glut4 glucose transporter expression in white adipose tissue (WAT), Glut4 expression is decreased in adipocytes in both obesity and type 2 diabetes, and Glut4 expression alters insulin sensitivity. NNMT knockdown protects against diet-induced obesity by raising levels of NAD+ and SAM, increasing flux of SAM into polyamine biosynthesis, and augmenting energy expenditure. Additionally, using ChIP-qPCR, Kraus et al. showed that NNMT inhibition modifies histone methylation leading to activation of rate-controlling enzymes involved in polyamine flux, resulting in increased energy expenditure. Thus, by use of genomics and epigenomics, the metabolic node NNMT was identified as a potential target for the treatment of obesity and type 2 diabetes (17).
Many recent studies have also shown how metabolic pathways can generate metabolites that inhibit enzymes that confer epigenetic modifications. For example, IDH1 mutations are correlated with extensive, coordinated hypermethylation at specific loci known as the CpG island methylator phenotype (CIMP) in glioma. Turcan et al. (44) analyzed the genomic DNA of mutant or wild-type IDH1-overexpressing cells, using the Ilumina Infinium Human Methylation 450 Platform, to uncover that mutant IDH1 caused increased hypermethylation at a number of genes. The structural similarity of α-KG and 2-HG led to the hypothesis that accumulation of 2-HG could bind and function as a competitive inhibitor to α-KG-dependent dioxygenases. Over 60 mammalian deoxygenases utilize α-KG as a substrate, including histone demethylases and 5-methylcytosine hydroxylases (19). Xu et al. (52) showed that 2-HG inhibits multiple α-KG-dependent dioxygenases at the high concentrations observed in human glioma tumors. Lu et al. (26) also showed that IDH mutations impair histone demethylation, resulting in a block-to-cell differentiation. IDH1 mutations thus confer unique epigenetic control over the expression of a large number of genes, linking central carbon metabolism to gene regulation and cancer pathogenicity.
Concluding Remarks
A major challenge faced by scientists in the postgenomic era is deciphering the functions of uncharacterized metabolic enzymes and pathways in both normal physiology and the oftentimes dysregulated functions of enzymes and pathways in disease. In this review, we show many examples of valiant efforts using advanced functional metabolomic platforms not only to uncover the metabolic basis of diseases but also to uncover unique treatment strategies that target these nodal metabolic drivers of diseases such as cancer, neurodegenerative diseases, and diabetes. We also demonstrate how recent studies have uncovered how metabolic pathways, enzymes, and metabolites can directly influence posttranslational and epigenetic regulation of the proteome and genome, respectively. These complex intersections between the metabolome, proteome, and genome present an even grander challenge for scientists but also represent exciting avenues for attenuating disease by manipulating metabolism and simultaneously controlling cellular genetic, metabolic, and signaling pathways.
Although the various examples shown in this review provide a general workflow for functional metabolomic profiling to characterize metabolic pathways, the majority of metabolic enzymes still remain uncharacterized. Thus, a more concerted and higher-throughput approach is necessary combined with the integration of functional metabolomic, proteomic, and genomic strategies to effectively annotate the entirety of the metabolic genome en masse and in parallel instead of our current approach of characterizing each enzyme one by one. With the advent of higher-throughput mass spectrometry and sequencing platforms, such large-scale endeavors may be attainable in the near future.
GRANTS
This work was supported by grants from the National Institutes of Health (R21 CA-170317, R01 CA-172667, R00 DA-030908) and the Searle Scholar Foundation.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: M.M.M. and D.K.N. drafted manuscript; M.M.M. and D.K.N. edited and revised manuscript; M.M.M. and D.K.N. approved final version of manuscript; D.K.N. prepared figures.
ACKNOWLEDGMENTS
We thank the members of the Nomura Research Group for critical reading of the manuscript.
REFERENCES
- 1.Benjamin DI, Cozzo A, Ji X, Roberts LS, Louie SM, Mulvihill MM, Luo K, Nomura DK. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc Natl Acad Sci USA 110: 14912–14917, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birungi G, Chen SM, Loy BP, Ng ML, Li SFY. Metabolomics approach for investigation of effects of dengue virus infection using the EA hy926 cell line. J Proteome Res 9: 6523–6534, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Blankman JL, Long JZ, Trauger SA, Siuzdak G, Cravatt BF. ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. Proc Natl Acad Sci USA 110: 1500–1505, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell 42: 426–437, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Z, Mulvihill MM, Mukhopadhyay P, Xu H, Erdelyi K, Hao E, Holovac E, Hasko G, Cravatt BF, Nomura DK, Pacher P. Monoacylglycerol lipase controls endocannabinoid and eicosanoid signaling and hepatic injury in mice. Gastroenterology 144: 808–817 or e815, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol 13: 1041–1050, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497: 633-+, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis M, Pragani R, Popovici-Muller J, Gross S, Thorne N, Salituro F, Fantin V, Straley K, Su M, Dang L, Simeonov A, Shen M, Boxer MB. ML309: A potent inhibitor of R132H mutant IDH1 capable of reducing 2-hydroxyglutarate production in U87 MG glioblastoma cells. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD: NIH, 2010 [PubMed] [Google Scholar]
- 10.Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, Kathuria S, Piomelli D. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA 99: 10819–10824, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudley E, Yousef M, Wang Y, Griffiths WJ. Targeted metabolomics and mass spectrometry. Adv Protein Chem Str 80: 45–83, 2010 [DOI] [PubMed] [Google Scholar]
- 12.Grady SL, Purdy JG, Rabinowitz JD, Shenk T. Argininosuccinate synthetase 1 depletion produces a metabolic state conducive to herpes simplex virus 1 infection. Proc Natl Acad Sci USA 110: E5006–E5015, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellerstein MK, Murphy E. Stable isotope-mass spectrometric measurements of molecular fluxes in vivo: emerging applications in drug development. Curr Opin Mol Therap 6: 249–264, 2004 [PubMed] [Google Scholar]
- 14.Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA 110: 8882–8887, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinsey SG, Long JZ, O'Neal ST, Abdullah RA, Poklis JL, Boger DL, Cravatt BF, Lichtman AH. Blockade of endocannabinoid-degrading enzymes attenuates neuropathic pain. J Pharmacol Exp Therap 330: 902–910, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinsey SG, O'Neal ST, Long JZ, Cravatt BF, Lichtman AH. Inhibition of endocannabinoid catabolic enzymes elicits anxiolytic-like effects in the marble burying assay. Pharmacol Biochem Behav 98: 21–27, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraus D, Yang Q, Kong D, Banks AS, Zhang L, Rodgers JT, Pirinen E, Pulinilkunnil TC, Gong F, Wang YC, Cen Y, Sauve AA, Asara JM, Peroni OD, Monia BP, Bhanot S, Alhonen L, Puigserver P, Kahn BB. Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508: 258–262, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43: 869–U879, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 4: 152–156, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Long JZ, Cisar JS, Milliken D, Niessen S, Wang C, Trauger SA, Siuzdak G, Cravatt BF. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat Chem Biol 7: 763–765, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long JZ, Cravatt BF. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem Rev 111: 6022–6063, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nat Chem Biol 5: 37–44, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long JZ, Nomura DK, Cravatt BF. Characterization of monoacylglycerol lipase inhibition reveals differences in central and peripheral endocannabinoid metabolism. Chem Biol 16: 744–753, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long JZ, Nomura DK, Vann RE, Walentiny DM, Booker L, Jin X, Burston JJ, Sim-Selley LJ, Lichtman AH, Wiley JL, Cravatt BF. Dual blockade of FAAH and MAGL identifies behavioral processes regulated by endocannabinoid crosstalk in vivo. Proc Natl Acad Sci USA 106: 20270–20275, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louie SM, Roberts LS, Mulvihill MM, Luo K, Nomura DK. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochim Biophys Acta 1831: 1566–1572, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O'Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483: 474–478, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu SCS. Adenosylmethionine. Int J Biochem Cell Biol 32: 391–395, 2000 [DOI] [PubMed] [Google Scholar]
- 28.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 7: 763–777, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Menendez JA, Lupu R. Fatty acid synthase-catalyzed de novo fatty acid biosynthesis: from anabolic-energy-storage pathway in normal tissues to jack-of-all-trades in cancer cells. Arch Immunol Ther Exp 52: 414–426, 2004 [PubMed] [Google Scholar]
- 30.Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 341: 549–553, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathogens 2: e132, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26: 1179–1186, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nomura DK, Blankman JL, Simon GM, Fujioka K, Issa RS, Ward AM, Cravatt BF, Casida JE. Activation of the endocannabinoid system by organophosphorus nerve agents. Nat Chem Biol 4: 373–378, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura DK, Hudak CS, Ward AM, Burston JJ, Issa RS, Fisher KJ, Abood ME, Wiley JL, Lichtman AH, Casida JE. Monoacylglycerol lipase regulates 2-arachidonoylglycerol action and arachidonic acid levels. Bioorg Med Chem Lett 18: 5875–5878, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chem Biol 18: 846–856, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140: 49–61, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MC, Ward AM, Hahn YK, Lichtman AH, Conti B, Cravatt BF. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science 334: 809–813, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piro JR, Benjamin DI, Duerr JM, Pi Y, Gonzales C, Wood KM, Schwartz JW, Nomura DK, Samad TA. A dysregulated endocannabinoid-eicosanoid network supports pathogenesis in a mouse model of Alzheimer's disease. Cell Reports 1: 617–623, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476: 346–350, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabinowitz JD, Purdy JG, Vastag L, Shenk T, Koyuncu E. Metabolomics in drug target discovery. Cold Spring Harbor Symp Quant Biol 76: 235–246, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roe B, Kensicki E, Mohney R, Hall WW. Metabolomic profile of hepatitis C virus-infected hepatocytes. PloS One 6: e23641, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlosburg JE, Blankman JL, Long JZ, Nomura DK, Pan B, Kinsey SG, Nguyen PT, Ramesh D, Booker L, Burston JJ, Thomas EA, Selley DE, Sim-Selley LJ, Liu QS, Lichtman AH, Cravatt BF. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat Neurosci 13: 1113–1119, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci USA 104: 16886–16891, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LGT, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483: 479–U137, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol 9: 300–306, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathogens 7: e1002124, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaughan M, Berger JE, Steinberg D. Hormone-sensitive lipase and monoglyceride lipase activities in adipose tissue. J Biol Chem 239: 401–409, 1964 [PubMed] [Google Scholar]
- 48.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science 291: 1304–1351, 2001 [DOI] [PubMed] [Google Scholar]
- 49.Vinayavekhin N, Homan EA, Saghatelian A. Exploring disease through metabolomics. ACS Chem Biol 5: 91–103, 2010 [DOI] [PubMed] [Google Scholar]
- 50.Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol 13: 270–276, 2012 [DOI] [PubMed] [Google Scholar]
- 52.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong P, Wang W, Pan B, Liu X, Zhang Z, Long JZ, Zhang HT, Cravatt BF, Liu QS. Monoacylglycerol lipase inhibition blocks chronic stress-induced depressive-like behaviors via activation of mTOR signaling. Neuropsychopharmacology 39: 1763–1776, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]