Abstract
Chlamydia trachomatis is a major cause of ocular and sexually transmitted diseases worldwide. While much of our knowledge about its genetic diversity comes from serotyping or ompA genotyping, no quantitative assessment of genetic diversity within serotypes has been performed. To accomplish this, 507 urogenital samples from a multicenter U.S. study were analyzed by phylogenetic and statistical modeling. No B, Da, or I serotypes were represented. Based on our analyses, all but one previous urogenital B serotype was identified as Ba. This, coupled with the lack of B serotypes in our population, suggests that B has specific tropism for ocular mucosa. We identified a Ba/D recombinant (putative crossover nucleotide 477; P < 0.0001) similar to a B/D mosaic we described previously from an African trachoma patient. Computational analyses of the Ba/D recombinant indicated that upstream changes were less important for tissue tropism than downstream incorporation of the D sequence. Since most serotypes had nonsynonymous/synonymous ratios of <1.0, the major outer membrane protein, encoded by ompA, has many functional constraints and is under purifying selection. Surprisingly, all serotype groups except for J had a unimodal population structure indicating rapid clonal expansion. Of the groups with a unimodal structure, E and Ia and, to a lesser extent, G and K were prevalent, had infrequent incorporation of mutations, and, compared to other groups, had a relatively greater degree of diversifying selection, consistent with a selective sweep of mutations within these groups. Collectively, these data suggest a diverse evolutionary strategy for different serogroups of the organism.
Chlamydia trachomatis is the leading cause of bacterial sexually transmitted diseases (STDs) in the developed world, producing lower and upper genital tract infections (LGTIs and UGTIs, respectively) in males and females, including nongonococcal urethritis, epididymitis and proctitis in males; endocervicitis and pelvic inflammatory disease (PID) in females; and proctitis and reactive arthritis in both. PID and its sequelae—chronic pelvic pain, ectopic pregnancy, and infertility—account for approximately 80% of the estimated $2.5 billion annual cost of these infections in the United States (9, 23).
The major outer membrane protein (MOMP) of C. trachomatis has been a major focus of research, as it contains serological variant (serotype)-, subspecies-, and species-specific epitopes (2); has important neutralizing determinants (42); elicits T-cell help for antibody production (1, 51); and may play a role in the attachment and invasion of host cells and in tissue tropism (36). Not surprisingly, MOMP has been the focus of epidemiologic studies of the organism and has been the primary candidate for a chlamydial vaccine. Yet, little is known about the genetic variation of ompA, the gene that encodes MOMP, or the diversity and evolution of MOMP epitopes among STD populations in the United States and around the world.
Strain typing of C. trachomatis is based on monoclonal antibodies that recognize MOMP epitopes. Over 18 serotypes of the organism have been identified (17, 54). Serotypes A, B, Ba, and C are the primary cause of trachoma, a chronic ocular disease found predominantly in developing countries of the world. Serotypes D, Da, E, F, G, H, I, Ia, J, Ja, and K cause LGTIs and UGTIs (12). The lymphogranuloma venereum (LGV) serotypes, which include L1, L2, L2a, and L3, also cause STDs, but these infections tend to be associated with more severe invasive disease, such as suppurative proctitis and lymphadenitis (10).
While serotyping has been useful in epidemiologic studies by revealing, for example, an association of specific serotypes with proctitis in homosexual males (4, 7) and same-serotype recurrent infections with concurrent gonorrhea (5), ompA genotyping has provided additional epidemiologic insights. ompA comprises four variable segments (VSs) and five constant segments (CSs). ompA polymorphisms have been identified in both VSs and CSs among 39 to 66% of trachoma (11, 15, 24, 25) and STD (3, 8, 14, 31, 37, 41, 53, 56) strains worldwide. Based on these polymorphic differences, isolates of the same serotype have been differentiated as ompA variants of the parent serotype (11, 13, 33, 54), trachoma isolates have been distinguished from those that cause STDs (21), and phylogenetic relationships among different serotypes have been inferred (50).
With the advent of genomics and the explosion of bioinformatics tools available to analyze sequence data, novel insights into the population genetics of human pathogens and advances in infectious disease epidemiology have been made. For example, allelic variants of group A Streptococcus producing pyogenic exotoxin A have been linked with toxic shock syndrome (38, 43, 46). Resistance to penicillin has been ascribed to horizontal gene transfer between Neisseria species (48). Moreover, an overabundance of nonsynonymous changes per site in the surface-exposed loops of the major porin of Neisseria suggests that this protein is under diversifying selection (47).
For Chlamydia, our previous statistical analyses revealed recombination within ompA, including one instance of horizontal gene transfer between Chlamydia species (36). We found a high level of recombination relative to substitution processes in an area that contains T-cell epitopes which are important for eliciting protective immunity (28, 49). Consequently, the genetic variation of ompA reflects the cumulative effects of immune selection pressures, functional constraints, and other adaptive pressures that contribute to the ability of the organism to evade immune surveillance. The possibility of recombination in this immunogenic region strengthens the need for a quantitative assessment of these pressures and constraints as they vary by serotype. While previous studies have described ompA point mutations and recombination events, adequate assessment of ompA genetic variability and stability of MOMP, and thus the above factors, has been limited by small sample sizes, restricted geographic sampling regions, and the use of partial sequences that do not include all VSs (8, 21, 24-26, 37, 53, 56).
Thus, to investigate the population genetics and evolution of this organism, we conducted a population-based study utilizing over 500 clinical C. trachomatis STD samples from a multicenter nationwide study of STDs and included all VSs of ompA in the analyses. The use of clinical samples obviated many potential problems with in vitro-adapted serotypes. Our analyses provide a more comprehensive understanding of the population genetics and evolution of C. trachomatis strains that can then be used for robust epidemiologic studies and vaccine development.
MATERIALS AND METHODS
Study population and specimen collection.
The following study population participated in a large parent study designed by the Centers for Disease Control and Prevention: consenting men and women age 14 to 49 years who presented to family planning and STD clinics serving the Birmingham, Ala., Indianapolis, Ind., New Orleans, La., Seattle, Wash., and San Francisco, Calif., metropolitan areas and who met the inclusion criteria given below. The study was multicenter, primarily cross-sectional in design, and included enrollment into three components, designated A, B1, and C, from October 1995 to August 1997 (6, 29, 55). Specimens were collected as previously described (6).
Component A was designed to evaluate laboratory diagnostic methods for C. trachomatis. Consecutive patients seen at STD and family planning clinics were enrolled and consented unless they had taken antibiotics within the past 30 days, were pregnant, had a hysterectomy, were males with symptoms of urethritis, were females not receiving a pelvic examination, or refused participation. Component B1 was designed to evaluate risk factors for recurrent C. trachomatis infection. Women seen at family planning clinics, health center clinics, teen clinics, and STD clinics were consented and enrolled if they were under 34 years of age and had laboratory documentation of C. trachomatis infection. The resulting cohort had follow-up at 4 weeks and 6 months after the baseline visit. Component C was designed to determine prevalence and evaluate risk factors for C. trachomatis infection. Sentinel surveillance networks composed of adolescent, STD, family planning, and community health center clinics in the five metropolitan areas were established. Health care providers from individual sentinel clinics were selected from each group of clinics to universally enroll consenting women and men who provided medical histories and underwent routine lower and upper genital tract examination. In Birmingham, universal enrollment of consenting men and women was not done. At this site, enrollment was limited to 50 to 60 female patients sampled from the clinics 1 week per month.
Those participants enrolled in the parent study with C. trachomatis urethral (male) or cervical (female) infection documented by culture, commercial Amplicor PCR test (Amplicor test kit; Roche Diagnostics, Indianapolis, Ind.), and/or commercial ligase chain reaction (LCR) test (LCX; Abbott Diagnostics, Abbott Park, Ill.) were eligible for the present study. The diagnostic tests were performed according to standard culture techniques in the respective laboratory and according to manufacturer's instructions for the commercial assays, except that M4 transport medium was used. Of those patients who were eligible, approximately 50 men and 50 women from each study center were randomly selected for inclusion.
Determination of ompA genotypes.
Over 1,000 clinical samples were received from the five study centers. Of these, 532 were randomly selected for ompA genotyping, where approximately equal numbers of specimens for males and females (except for Birmingham, where no males were enrolled in component C) and for each center were randomly selected. The specimens were from the cervix (female) or urethra (male) and were positive by culture, LCR, or Amplicor PCR. The samples were original, nonpropagated culture remnant samples and remnant, nonamplified LCR or Amplicor PCR samples. DNA extraction, purification, amplification, and automated sequencing were performed as previously described (13, 14) with technicians blinded to all clinical, demographic, geographic, and microbiologic data. Samples with ambiguous nucleotides were resequenced using newly purified DNA from the respective remnant sample. Mixed infections were identified and confirmed as described previously (16).
Estimation of C. trachomatis serotypes and analysis of putative recombinants.
The sequence was first manually aligned against all prototype serotype sequences (57) using the multiple alignment sequence editor (MASE, version 3.1; Dana-Farber Cancer Institute, Harvard School of Public Health, [ftp.ebi.ac.uk/pub/software/unix/]). The similarity between the sequence and each prototype serotype sequence was computed as the number of like nucleotides divided by the total number of nucleotides under comparison. The serotype of the prototype sequence to which the sequence was most similar provided the serotype estimate. Statistical significance of putative recombinants and their breakpoints were assessed by Sawyer's runs test and the maximum chi-square test as previously described (36).
Statistical analyses.
Of the 532 samples that were ompA genotyped, 25 were excluded due to the presence of a mixed infection, leaving 507 sequences available for analysis. Differences in chlamydial serotype distribution by metropolitan area and by gender were determined by permutation analysis in the following manner (35): For each serotype, a 2 × 5 contingency table compared the number of sequences of that serotype in each metropolitan area or gender, respectively, to the number of sequences not of that serotype. A Pearson's chi-square homogeneity statistic was computed for the observed data. The test statistic was recomputed after 10,000 random reassignments of serotype, keeping the frequencies in the population constant. The P value was the proportion of randomizations in which the test statistic exceeded that for the observed data. The Bonferroni method was used to adjust for multiple serotypes.
Since there is extensive evidence for recombination in the ompA gene, phylogenetic inference and estimates of genetic distance for urogenital strains in this study were based on simple Hamming distances (denoted by p) without the inclusion of multiple substitution models. Tree topologies for all strains in a serotype class were generated by the neighbor-joining method and MEGA (molecular evolutionary genetics analysis), version 2.1 (32). The mean genetic distance within a serotype group was estimated as the average of all p distances between sequences of the same serotype for all possible sequence pairs obtained with MEGA, version 2.1 (32). The standard error of the estimate was generated using the bootstrap procedure with 500 replicates, and 95% confidence intervals (CIs) were calculated in the usual fashion. The distribution of genetic distances within a serotype group was determined by plotting a histogram of these distances. For each serotype group, the mean proportion of synonymous nucleotide differences per synonymous site (ps) and the mean proportion of nonsynonymous nucleotide differences per nonsynonymous site (pn) were computed between same serotype strains for all possible sequence pairs. This was accomplished by the method of Nei and Gojobori (39) using MEGA, version 2.1 (32). The standard error of the estimate and 95% CIs were determined similarly to the methods described above.
We compared the relative prevalence of shared mutations between strains of the same serotype group for different serotype groups. We postulated that a serotype group with high prevalence and infrequent incorporation of mutations within its strain pool might have undergone a selective sweep in which strains have exhibited rapid clonal expansion. To graphically depict these differences, we represented a mutation within a serotype group pool as a radial spoke on a wheel with spoke length inversely proportional to the frequency at which that mutation was present in the serotype group pool. The radial wheel, or starburst, represented the relative frequency of all mutations observed for that serotype group in this urogenital U.S. population. The location of the spoke within a 360° circle was determined by the nucleotide location of the mutation − 250 (nucleotide [nt] location of the beginning of the sequence)/810 (total base pair length of the sequence) multiplied by 360°. For example, serotype group Z has 100 sequences and a mutation at nt 250 is present in 50 of these sequences, such that the radial spoke length = 1/50/100 = 2 at 0°. A phylogenetic reconstruction was generated to establish tree topology between serotype groups based on Hamming distances between strains of dissimilar serotype groups for all possible sequence pairs using the neighbor-joining method and MEGA, version 2.1. Each starburst was superimposed onto the phylogenetic tree at the end of the branch that corresponded to the appropriate serotype group.
Nucleotide sequence accession number.
The sequences of the ompA genes determined in this study have been submitted to GenBank (accession no. AY535080 through AY535172).
RESULTS
Of the 507 study participants, 228 were men and 279 were women. Of these, 94 were seen in the Birmingham metropolitan area, 106 in Indianapolis, 112 in New Orleans, 88 in San Francisco, and 107 in Seattle. When comparing ompA nucleotide sequence data generated from this population, there were 329 polymorphic sites out of the possible 810 bp that were sequenced, aligned, and analyzed. The 810 bp (nt 244 to 1053) included VS1 through VS4 in their entirety. In the alignment of the urogenital strains from this study, locations of the VSs defined by Yuan et al. (57) were as follows: VS1 from nt 256 to 324, VS2 from nt 490 to 567, VS3 from nt 757 to 798, and VS4 from nt 949 to 1053.
Table S1 in the supplemental material describes each unique sequence in this population, including the serotype estimate for that sequence, changes with respect to its most similar prototype sequence, and the geographic region and gender for the strains represented by that sequence. The distribution of serotypes was similar to those in other studies. Serotypes D, E, F, and Ia were the most prevalent; G, J, and K were intermediate; and Ba, H, Ja, and a putative Ba/D recombinant were rare: Ba, 5 (1%) of 507; D, 71 (14%) of 507; Ba/D, 1 (0.2%) of 507; E, 150 (30%) of 507; F, 98 (19%) of 507; G, 21 (4%) of 507; H, 3 (1%) of 507; Ia, 73 (14%) of 507; J, 35 (7%) of 507; Ja, 24 (5%) of 507; and K, 26 (5%) of 507. Of note, there were no A, B, C, Da, or I serotypes. The putative Ba/D recombinant identified in this study had significant mosaicism (P < 0.0001) based on the Sawyer's runs test and the maximum chi-square test. The recombinant was markedly similar to serotype Ba before nt 477, the breakpoint predicted by the maximum chi-square test, and was markedly similar to serotype D after the breakpoint (see Fig S1 in the supplemental material).
Serotype distribution by metropolitan area was not homogeneous (Table 1). Permutation analysis was used to test whether a serotype was significantly associated with a metropolitan area. The rare serotypes were observed to a greater extent in San Francisco than in other metropolitan areas, with a significantly higher proportion of Ba sequences than in Birmingham, Indianapolis, and Seattle (P = 0.02) and a significantly higher proportion of G sequences than in any other city (P = 0.03) after adjustment for multiple comparisons. In contrast, there was no significant difference in the distribution of serotype by gender.
TABLE 1.
Distribution of Chlamydia trachomatis serovars by metropolitan area in the United States and by gender
| Parameter | No. of sequences of serovara:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ba | D | Ba/D | E | F | G | H | Ia | J | Ja | K | Total | |
| Metropolitan area | ||||||||||||
| Birmingham | 0 | 9 | 0 | 30 | 19 | 1 | 1 | 15 | 7 | 4 | 8 | 94 |
| Indianapolis | 1 | 21 | 0 | 32 | 19 | 2 | 0 | 16 | 8 | 6 | 1 | 106 |
| New Orleans | 0 | 19 | 0 | 27 | 19 | 4 | 0 | 20 | 3 | 10 | 10 | 112 |
| San Francisco | 4 | 7 | 1 | 25 | 12 | 10 | 0 | 12 | 11 | 2 | 4 | 88 |
| Seattle | 0 | 15 | 0 | 36 | 29 | 4 | 2 | 10 | 6 | 2 | 3 | 107 |
| Gender | ||||||||||||
| Male | 2 | 33 | 1 | 62 | 42 | 13 | 0 | 35 | 21 | 10 | 9 | 228 |
| Female | 3 | 38 | 0 | 88 | 56 | 8 | 3 | 38 | 14 | 14 | 17 | 279 |
| Total | 5 | 71 | 1 | 150 | 98 | 21 | 3 | 73 | 35 | 24 | 26 | 507 |
Significant differences in serovar distribution by metropolitan area were deduced by permutation testing. San Francisco had a significantly higher proportion of Ba sequences than Birmingham, Indianapolis, and Seattle (P = 0.02) and a significantly higher proportion of G sequences than any other city (P = 0.03) after adjustment for multiple comparisons.
Figure 1 shows the phylogenetic tree topologies based on Hamming distances for the urogenital strains in this study. For the B seroclass (Ba, D, Da, and E) sequences, the Ba/D recombinant clustered midway between the B and D serotype groups, and the D serotype sequences formed two main groups differentiated by the presence or absence of nucleotide changes at nt 246 and 249 (D/B120, D-CDC4, D-CDC6, D-CDC11, and D-CDC13 versus other D serotype sequences; Fig. 1A), which did not differentiate the D and Da subtypes. For the intermediate seroclass (F and G), F-CDC9 and G-CDC7 clustered apart from the rest of the sequences within their respective serotype groups (Fig. 1B). In 1994, Ossewaarde et al. (40) also reported phylogenetically distinct G sequences that they designated as Ga subtypes. These Ga strains had T547A and T1003G changes compared to G/UW-57. None of our G sequences contained the T547A change; however, 6 (29%) of 21 G sequences contained the T1003G change (T1036G in our alignment), confirming that these changes are prevalent population differences not restricted to the European continent. For the C seroclass (A, C, H, I, Ia, J, and Ja), the J serotype sequences formed two main groups consistent with the J and Ja subtypes (Fig. 1C).
FIG. 1.
Phylogenetic reconstructions for the unique ompA gene sequences in the B seroclass (A), the intermediate seroclass (B), and the C seroclass (C) using the neighbor-joining method. The values at the nodes are the bootstrap confidence levels (BCL) for the interior branch. The BCL represents the percentage of 500 bootstrap resamplings for which the strain to the right was separated from the other strains.
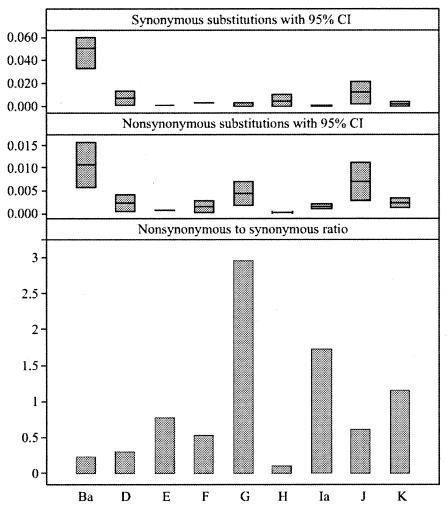
As there is extensive evidence for recombination in the ompA gene (36), we used simple Hamming distances to compare genetic distance estimates within serotype groups. When comparing strains within a serotype group, serotypes E, Ba, H, Ia (0.001; 95% CI, 0.0006 to 0.002) and F (in descending order) were the most conserved, while serotypes J, G, D, and K (0.002; 95% CI, 0.0009 to 0.003) were the most divergent (Fig. 2). Serovar J (0.008; 95% CI, 0.005 to 0.01) had a significantly higher mean genetic distance than all serovars other than G (0.004; 95% CI, 0.002 to 0.006) and D (0.003; 95% CI, 0.001 to 0.006); serovar E (0.0004; 95% CI, 0.0002 to 0.0006) had a significantly lower mean genetic distance than all serovars other than Ba (0.0005; 95% CI, 0 to 0.002), F (0.002; 95% CI, 0.0005 to 0.003), and H (0.0009; 95% CI, 0 to 0.003); and serovar Ba had a significantly lower mean genetic distance than serovars G and J. Using the methods of Nei and Gojobori, serotype Ba had the highest mean proportion of nonsynonymous nucleotide differences per nonsynonymous site (pn; Fig. 3) among serotypes, followed by J. These two serotypes also had the highest mean proportion of synonymous nucleotide differences per synonymous site (ps; Fig. 3). Serotypes G, Ia, and K had pn/ps ratios greater than 1.0; all other serotypes had ratios less than 1.0 (Fig. 3).
FIG. 2.
Mean Hamming distance within each serotype group based on average Hamming distance between all possible pairs of sequences of similar serotypes. Minimum and maximum values represent lower and upper limits of the 95% CI of the estimate, while the value plotted at the horizontal bar level represents the mean estimate.
FIG. 3.
Mean synonymous (top panel) and nonsynonymous (middle panel) mutation rates, respectively, for serotype groups based on the method of Nei and Gojobori. Minimum and maximum values represent lower and upper limits of the 95% CI of the estimate, while a value plotted at horizontal bar level represents the mean estimate. The bottom panel shows the nonsynonymous/synonymous mutation rate ratio for serotype groups based on the mean estimates shown in the prior two graphs.
The distribution of the pairwise distances within a serotype group was primarily unimodal (Fig. 4C; shown for serotype E). Only serotype J had a bimodal population structure, with two prevalent sequence groups representing the J and Ja subtypes roughly 98.5% similar (Fig. 4B). Serotype D had a unimodal population structure with a substantial shoulder (Fig. 4A); 47 sequences had two mutations, one at nt 246 (T→C) and the other at nt 249 (G→C), compared to the prototype D/B-120 sequence; 24 sequences did not have these two mutations. Sequences with these two mutations would be 99.7% similar to the D/B120 sequence. Thus, the mutations at nt 246 and 249 probably represent the differences in these populations. It is possible that we might have detected a bimodal population structure for serotype D if we had included additional sequence data upstream of where our sequences began in CS1. Other studies have reported six prevalent nucleotide changes in this region. However, even if serotype D has a bimodal structure, the two population groups are not likely to represent the D and Da subtypes, because none of the previously reported nucleotide changes in CS1 or the prevalent changes present in our data differentiate between the D and Da subtypes.
FIG. 4.
Population structure of serotype groups in this study based on histogram of Hamming distances between all possible pairs of sequences of similar serotypes. (A) Unimodal structure with shoulder was seen for serotype D, with the main group and shoulder group roughly 99.7% similar. (B) The only serotype group to have a bimodal structure was serotype J, with the two main groups roughly 98.5% similar. (C) Unimodal structure was typical of most serotypes exemplified by serotype E.
Figure 5 shows the degree to which ompA nucleotide changes have been incorporated into the C. trachomatis population for different serotypes. Larger and more uniform spoke lengths within the starburst represent both infrequent incorporation of mutations within the serotype group and a prevalent serotype group. Serotypes in which we observed nucleotide changes compared to the prototype that were at least 25% prevalent in the population (Ba, C246T, A249G, A532G, G607T, and T781G; D, C246T, A249G, A660T, and G1000A; F, G269; H, A272G and C865T; and J, T625C and G627C) had the least uniform radial spoke lengths, while those without prevalent changes had more uniform spoke lengths (E, Ia, G, and K). Because of their high prevalence and a low frequency of shared mutations, serotype groups E and Ia had both uniform spoke lengths and large-diameter circles.
FIG. 5.
Phylogenetic reconstructions with a starburst superimposed at the end of the branch corresponding to the appropriate serotype group. Starbursts represent the relative frequency of shared mutations within a serotype group pool as described in Materials and Methods. The radial spoke length is inversely proportional to the frequency of mutations, so that longer spoke lengths correspond to less frequent incorporation of mutations among similar serotype group strains. The location of the radial spoke within a 360° circle was determined by the nucleotide location of the mutation as described in Materials and Methods. The starbursts were each magnified to ×250 in order to discern the radial spoke length and star-like quality of the radial wheel for each serotype group.
DISCUSSION
Previous studies of chlamydial STDs have attempted to describe the genetic variability of ompA by inferring the phylogeny of their population with laboratory-maintained strains and published sequences (50). However, these studies used small population sizes which do not reasonably approximate random ones, and the combined populations represent strains from vastly different time periods, since the laboratory-maintained strains were generally isolated decades before the other strains. Phylogenetic reconstructions of ompA should only be used to describe the short-term evolution of Chlamydia, since intra- and intergenic recombination have previously been documented for this gene (8, 19, 26, 36) and recombination over time disrupts the bifurcating tree-like phylogeny and clonal structure of the organism (27, 36).
We assessed the genetic variability of ompA within the context of a large population-based study and analyzed the data using methods that were appropriate for recombination in the gene. Consequently, the data were analyzed without multiple substitution models so as to not introduce an inaccurate evolutionary model. Further, we presented tree topologies composed entirely of sequences from this study, which we used to infer the short-term evolution of C. trachomatis. This was reasonable, as these strains were isolated over a 2-year period.
The overall serotype distribution in our data was similar to other studies of genital C. trachomatis infections with minor differences (20, 44, 56). In our study, there were no A, B, C, Da, or I serotypes represented. The lack of A and C serotypes is consistent with the universal and near universal absence, respectively, of these serotypes among urogenital infections (21, 34, 41). Of the three urogenital studies that reported C serotypes, two used restriction fragment length polymorphisms (RFLP) to assign the serotype. In one study (41), all strains typed as C by RFLP were found to be either F or G by sequencing, suggesting that RFLP is less reliable for strain typing.
While Ba serotypes have regularly been found at both urogenital and ocular sites, B serotypes have only been found twice in urogenital samples—B/Alpha-95 (18) and B/IU1226 (50). When comparing these two urogenital B sequences to prototype sequences, B/Alpha-95 was most similar to B/TW-5, justifying its assignment as serotype B. However, B/IU1226 was most similar to Ba/Apache2, as were the four sequences in this study. Thus, we feel that B/IU1226 should be reassigned as a Ba isolate and believe that the occurrence of B sequences in urogenital infections is indeed a rare event.
The lack of Da strains in our population contradicts a 1995 study by Sayada et al. in which 9 (12%) of 73 urogenital strains were found to be Da by RFLP (45). Based on sequence comparison, the D (D/B120) and Da (Da/TW-448) subtypes can be distinguished by 3 nt differences, one of which codes for an amino acid change in VS4 (G1036A for Da/TW-448 in our alignment). None of our D sequences had this amino acid change, and all of them were most similar to D/B120. In GenBank, three urogenital sequences have previously been assigned to Da: Da/IU-1554 (AF063209), Da/TW-488 (X62921), and Da/Ev-293 (X77365). However, based on our analyses, Da/IU-1554 and Da/TW-488 sequences were most similar to D/IC-Cal8 and Da/Ev-293 was most similar to D/B120. Only Da/Ev-293 had the G1036A nonsynonymous change noted above. These findings suggest that Da urogenital infections are also rare. However, the importance of Da as a urogenital subtype should be evaluated in additional studies that include other global regions where chlamydial STDs are prevalent.
Finally, there were no I sequences: only Ia sequences were represented. Recently, in other STD populations, serotype I has also been less frequently identified (52). This may reflect an adaptive response of serotype I to host or antimicrobial pressure to evolve as an Ia. Alternatively, there might have been an evolutionary disadvantage ascribed to serotype I that has driven this process. Thus, these cumulative data suggest that occurrences of A, B, C, and Da serotypes in urogenital infections are, at best, rare events and that serotype I is being replaced by Ia.
The Ba/D mosaic identified in this study is similar to a B/D recombinant we previously isolated from a Tunisian trachoma patient in the 1970s (15). While both strains had similar crossover points, three nonsynonymous mutations upstream of VS1 that were identical to those described for B from other trachoma populations (21) were present in the Tunisian recombinant while absent in the STD mosaic. Thus, recombination between Ba or B and D seems to confer an evolutionary advantage for both STD and trachoma populations. Interestingly, the Ba/D recombinant did not have two changes that were present in all the Ba sequences in our population (C246T and A249G at amino acids 60 and 61, respectively, in the Frost alignment) compared to B/TW-5, a trachoma strain. These changes were also specific for the urogenital specimens in the population described by Frost et al. (21). Consequently, the Ba/D mosaic was substantially more similar to B/TW-5 than the other urogenital Ba sequences in this study, suggesting that changes upstream of nt 477 are less important for tissue specificity than the incorporation of the D sequence downstream of 477. Moreover, there is only one change between the urogenital Ba sequences in this study and B/TW-5 downstream of nt 477. This indicates that for serotype B, MOMP functional constraints are high in this region, and thus there is little opportunity for tissue-specific change.
We found significant differences in serotype distribution by geographic region. While the chlamydial reservoir for the majority of strains seems to be relatively homogeneous in the United States, the rare serotypes, Ba and G, were significantly overrepresented in San Francisco. Perhaps the abundance of Ba and G serotypes in San Francisco reflects the population dynamics of this international city, which is constantly undergoing turnover with a large immigrant population from Asia and a large homosexual/bisexual community. In contrast, there was no significant difference in serotype by gender, suggesting that specific serotypes do not preferentially infect one gender.
We have provided the first quantitative assessment of the genetic diversity within each serotype group for ompA. We found that serotypes E, Ba, H, Ia, and F (in descending order) were the most conserved, while J, G, D, and K were the most divergent. By comparing genetic variability, pn/ps substitution ratios, and the relative degrees of incorporation of substitutions in the population, the data suggest that C. trachomatis has had occasional selective sweeps of mutations through a serotype group or groups similar to those described for human immunodeficiency virus type 1 (HIV-1) (22). HIV-1 is comprised of M, N, and O groups, and the majority of infections are caused by the M group, which is made up of subtypes A to J. The effective population size of subtypes A (heterosexually transmitted) and B (transmitted via needle sharing or male homosexual sex) based on both gag and env gene sequence alignments were recently analyzed and found to be surprisingly smaller than expected, similar to what we found for chlamydiae. The authors suggested that this could be explained by genetic drift or a selective sweep of mutations through the respective subtype populations.
For Chlamydia, we found a predominantly unimodal population structure for ompA (except for serotype J), consistent with a rapid expansion of strains with little time for propagation of mutations. Under this model, we expect C. trachomatis to have prevalent serotype groups with nucleotide and amino acid changes that are infrequently shared among strains within the same serotype. This can be seen for serotypes E and Ia and less so for G and K. The former serotypes are among the most prevalent and least variable and are without prevalent population changes. Moreover, there is laboratory evidence that serotype E outcompetes other strains for nutrients and growth factors; this is objective evidence for rapid expansion of this serotype group (30). The high pn/ps ratios for serotypes Ia, G, and K, which also had rapid expansion, indicate that, compared to most other serotypes, they may be under a greater degree of diversifying selection even though, for the most part, ompA has many functional constraints and is under purifying selection. Rapid expansion of selected serotypes may in part explain the relative replacement of I with Ia and the overabundance of serotype G in San Francisco if rapid expansion of G has occurred with preference for geographical boundaries.
Serotypes Ba, D, F, H, and J appear to have had less rapid population growth exemplified by the presence of prevalent population changes and greater genetic variability overall. For these serotypes, mutations have had time to propagate throughout the population, and a lesser degree of expansion helps to explain the lower prevalence for Ba, H, and J. Moreover, they appear to be under a greater degree of purifying selection than those with rapid expansion consistent with the lower observed pn/ps.
For serotypes D and J, the prevalent population changes observed in this population agree with those previously reported, suggesting they have been present for at least a decade (17, 21). We found that serotype J had a bimodal population structure, which likely represents the J and Ja subtypes with mutations that occurred before the two groups diverged. The prevalent population changes observed for serotype D are also present in our Ba sequences and have previously been ascribed to differentiate urogenital from trachoma B sequences (21), although these changes were not universally found in our population and thus appear to be nonspecific. Since the same changes for serotype D and urogenital serotype Ba have propagated through many populations, the evolution of these serotypes appears to be linked, either by the early incorporation of a mutation occurring before their divergence or by recombination events between the two.
In summary, by taking a population-based approach to the genetic and evolutionary analysis of ompA for C. trachomatis urogenital infections in the United States, we were able to quantitatively and qualitatively assess the variation of this gene and determine how immune selection pressures, functional constraints, and other adaptive pressures vary among serotypes. Importantly, our analyses provide the first evidence that the evolution of the organism likely includes selective sweeps. This is encouraging for vaccine design in that mutations that occur during selective sweeps are unlikely to be shared, and immune protection against recent clinical strains will be similar to the response mounted against prototype strains. Finally, this research provides precise genetic strain differentiation that can be applied to analyses of phenotypic disease markers to determine if MOMP is a virulence factor for the organism.
Supplementary Material
Acknowledgments
We would like especially to thank Sally Liska and the following individuals for their excellent technical assistance: Mitchell Pate, Barbara van der Pol, Robert Suchland, and Cathy Cammarata.
This work was supported by a grant from the Centers for Disease Control and Prevention (POHC96000762 to D. Dean) and Public Health Service grants AI39499 and EY/AI12219 from the National Institutes of Health (to D. Dean).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Allen, J. E., R. M. Locksley, and R. S. Stephens. 1991. A single peptide from the major outer membrane protein of Chlamydia trachomatis elicits T cell help for the production of antibodies to protective determinants. J. Immunol. 147:674-679. [PubMed] [Google Scholar]
- 2.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes, R. C., A. M. Rompalo, and W. E. Stamm. 1987. Comparison of Chlamydia trachomatis serovars causing rectal and cervical infections. J. Infect. Dis. 156:953-958. [DOI] [PubMed] [Google Scholar]
- 5.Batteiger, B. E., J. Fraiz, W. J. Newhall, B. P. Katz, and R. B. Jones. 1989. Association of recurrent chlamydial infection with gonorrhea. J. Infect. Dis. 159:661-669. [DOI] [PubMed] [Google Scholar]
- 6.Black, C. M., J. Marrazzo, R. E. Johnson, E. W. Hook III, R. B. Jones, T. A. Green, J. Schachter, W. E. Stamm, G. Bolan, M. E. St. Louis, and D. H. Martin. 2002. Head-to-head multicenter comparison of DNA probe and nucleic acid amplification tests for Chlamydia trachomatis infection in women performed with an improved reference standard. J. Clin. Microbiol. 40:3757-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boisvert, J. F., L. A. Koutsky, R. J. Suchland, and W. E. Stamm. 1999. Clinical features of Chlamydia trachomatis rectal infection by serovar among homosexually active men. Sex. Transm. Dis. 26:392-398. [DOI] [PubMed] [Google Scholar]
- 8.Brunham, R., C. Yang, I. Maclean, J. Kimani, G. Maitha, and F. Plummer. 1994. Chlamydia trachomatis from individuals in a sexually transmitted diseases core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Investig. 94:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 1993. Recommendations for the prevention and management of Chlamydia trachomatis infections. Morb. Mortal. Wkly. Rep. 42:1-39. [PubMed] [Google Scholar]
- 10.Dean, D. 1997. Chlamydia trachomatis sexually transmitted diseases, p. 473-490. In D. H. Connor, D. A. Schwartz, and F. W. Chandler (ed.), Pathology of infectious diseases. Appleton and Lange Publishers, Stamford, Conn.
- 11.Dean, D. 1994. Molecular characterization of new Chlamydia trachomatis serological variants from a trachoma endemic region of Africa, p. 259-262. In J. Orfila, G. I. Byrne, M. A. Chernesky, J. T. Grayston, R. B. Jones, G. L. Ridgway, R. Saikku, J. Schachter, W. E. Stamm, and R. S. Stephens (ed.), Chlamydial infections. Societa Editrice Esculapio, Bologna, Italy
- 12.Dean, D. 2002. Pathogenesis of chlamydial ocular infections, p. 1-20. In W. Tasman and E. A. Jaeger (ed.), Duane's foundations of clinical ophthalmology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 13.Dean, D., and K. Millman. 1997. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J. Clin. Investig. 99:475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 15.Dean, D., J. Schachter, C. R. Dawson, and R. S. Stephens. 1992. Comparison of the major outer membrane protein variant sequence regions of B/Ba isolates: a molecular epidemiologic approach to Chlamydia trachomatis infections. J. Infect. Dis. 166:383-392. [DOI] [PubMed] [Google Scholar]
- 16.Dean, D., and R. S. Stephens. 1994. Identification of individual genotypes of Chlamydia trachomatis from experimentally mixed serovars and mixed infections among trachoma patients. J. Clin. Microbiol. 32:1506-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean, D., R. Suchland, and W. Stamm. 2000. Evidence for long-term cervical persistence of Chlamydia trachomatis by omp1 genotyping. J. Infect. Dis. 182:909-916. [DOI] [PubMed] [Google Scholar]
- 18.Farencena, A., M. Comanducci, M. Donati, G. Ratti, and R. Cevenini. 1997. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect. Immun. 65:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitch, W. M., E. M. Peterson, and L. M. de la Maza. 1993. Phylogenetic analysis of the outer-membrane-protein genes of chlamydiae, and its implication for vaccine development. Mol. Biol. Evol. 10:892-913. [DOI] [PubMed] [Google Scholar]
- 20.Frost, E. H., S. Deslandes, and D. Bourgaux-Ramoisy. 1993. Chlamydia trachomatis serovars in 435 urogenital specimens typed by restriction endonuclease analysis of amplified DNA. J. Infect. Dis. 168:497-501. [DOI] [PubMed] [Google Scholar]
- 21.Frost, E. H., S. Deslandes, D. Gendron, D. Bourgaux-Ramoisy, and P. Bourgaux. 1995. Variation outside variable segments of the major outer membrane protein distinguishes trachoma from urogenital isolates of the same serovar of Chlamydia trachomatis. Genitourin. Med. 71:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grassly, N. C., P. H. Harvey, and E. C. Holmes. 1999. Population dynamics of HIV-1 inferred from gene sequences. Genetics 151:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groseclose, S. L., A. A. Zaidi, S. J. DeLisle, W. C. Levine, and M. E. St. Louis. 1999. Estimated incidence and prevalence of genital Chlamydia trachomatis infections in the United States, 1996. Sex. Transm. Dis. 26:339-344. [DOI] [PubMed] [Google Scholar]
- 24.Hayes, L. J., R. L. Bailey, D. C. Mabey, I. N. Clarke, M. A. Pickett, P. J. Watt, and M. E. Ward. 1992. Genotyping of Chlamydia trachomatis from a trachoma-endemic village in the Gambia by a nested polymerase chain reaction: identification of strain variants. J. Infect. Dis. 166:1173-1177. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, L. J., S. Pecharatana, R. L. Bailey, T. J. Hampton, M. A. Pickett, D. C. Mabey, P. J. Watt, and M. E. Ward. 1995. Extent and kinetics of genetic change in the omp1 gene of Chlamydia trachomatis in two villages with endemic trachoma. J. Infect. Dis. 172:268-272. [DOI] [PubMed] [Google Scholar]
- 26.Hayes, L. J., P. Yearsley, J. D. Treharne, R. A. Ballard, G. H. Fehler, and M. E. Ward. 1994. Evidence for naturally occurring recombination in the gene encoding the major outer membrane protein of lymphogranuloma venereum isolates of Chlamydia trachomatis. Infect. Immun. 62:5659-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes, E. C., R. Urwin, and M. C. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 28.Ishizaki, M., J. E. Allen, P. R. Beatty, and R. S. Stephens. 1992. Immune specificity of murine T-cell lines to the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 60:3714-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson, R. E., T. A. Green, J. Schachter, R. B. Jones, E. W. Hook III, C. M. Black, D. H. Martin, M. E. St. Louis, and W. E. Stamm. 2000. Evaluation of nucleic acid amplification tests as reference tests for Chlamydia trachomatis infections in asymptomatic men. J. Clin. Microbiol. 38:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, R. B., J. A. Williams, and B. van der Pol. 1998. Competitive growth of serovars E and F combined in mixed tissue culture infections, p. 523-526. In R. S. Stephens, G. I. Byrne, G. Christiansen, I. N. Clarke, J. T. Grayston, R. G. Rank, G. L. Ridgway, P. Saikku, J. Schachter, and W. E. Stamm (ed.), Chlamydial infections. Proceedings of the Ninth International Symposium on Human Chlamydial Infection. International Chlamydia Symposium, Napa, Calif.
- 31.Jurstrand, M., L. Falk, H. Fredlund, M. Lindberg, P. Olcén, S. Andersson, K. Persson, J. Albert, and A. Bäckman. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 33.Lampe, M. F., R. J. Suchland, and W. E. Stamm. 1993. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect. Immun. 61:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lan, J., I. Melgers, C. J. L. M. Meijer, J. M. M. Walboomers, R. Roosendaal, C. Burger, O. P. Bleker, and A. J. C. van den Brule. 1995. Prevalence and serovar distribution of asymptomatic cervical Chlamydia trachomatis infections as determined by highly sensitive PCR. J. Clin. Microbiol. 33:3194-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manly, B. F. J. 1991. Randomization, bootstrap and Monte Carlo methods in biology, 2nd ed. Chapman and Hall, London, United Kingdom.
- 36.Millman, K. L., S. Tavare, and D. Dean. 2001. Recombination in the ompA gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J. Bacteriol. 183:5997-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morré, S. A., R. Moes, I. Van Valkengoed, J. P. Boeke, J. T. M. van Eijk, C. J. L. M. Meijer, and A. J. C. Van den Brule. 1998. Genotyping of Chlamydia trachomatis in urine specimens will facilitate large epidemiological studies. J. Clin. Microbiol. 36:3077-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 40.Ossewaarde, J. M., M. Rieffe, A. de Vries, R. P. Derksen-Nawrocki, H. J. Hooft, G. J. van Doornum, and A. M. van Loon. 1994. Comparison of two panels of monoclonal antibodies for determination of Chlamydia trachomatis serovars. J. Clin. Microbiol. 32:2968-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen, L. N., H. O. Kjæaer, J. K. Møller, T. F. Ørntoft, and L. Ostergaard. 2000. High-resolution genotyping of Chlamydia trachomatis from recurrent urogenital infections. J. Clin. Microbiol. 38:3068-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peeling, R. W., and R. C. Brunham. 1991. Neutralization of Chlamydia trachomatis: kinetics and stoichiometry. Infect. Immun. 59:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piffaretti, J. C., H. Kressebuch, M. Aeschbacher, J. Bille, E. Bannerman, J. M. Musser, R. K. Selander, and J. Rocourt. 1989. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 86:3818-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole, E., and I. Lamont. 1992. Chlamydia trachomatis serovar differentiation by direct sequence analysis of the variable segment 4 region of the major outer membrane protein gene. Infect. Immun. 60:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sayada, C., E. Vretou, J. Orfila, J. Elion, and E. Denamur. 1995. Heterogeneity within the first constant segment of the major outer membrane protein gene in Chlamydia trachomatis serovar D/Da distinguishes 2 lineages. C. R. Acad. Sci. Ser. III 318:943-949. [PubMed] [Google Scholar]
- 46.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, N. H., J. Maynard Smith, and B. G. Spratt. 1995. Sequence evolution of the porB gene of Neisseria gonorrhoeae and Neisseria meningitidis: evidence of positive Darwinian selection. Mol. Biol. Evol. 12:363-370. [DOI] [PubMed] [Google Scholar]
- 48.Spratt, B. G., L. D. Bowler, Q. Y. Zhang, J. Zhou, and J. M. Smith. 1992. Role of interspecies transfer of chromosomal genes in the evolution of penicillin resistance in pathogenic and commensal Neisseria species. J. Mol. Evol. 34:115-125. [DOI] [PubMed] [Google Scholar]
- 49.Stagg, A. J., W. A. Elsley, M. A. Pickett, M. E. Ward, and S. C. Knight. 1993. Primary human T-cell responses to the major outer membrane protein of Chlamydia trachomatis. Immunology 79:1-9. [PMC free article] [PubMed] [Google Scholar]
- 50.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su, H., R. P. Morrison, N. G. Watkins, and H. D. Caldwell. 1990. Identification and characterization of T helper cell epitopes of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 172:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suchland, R. J., L. O. Eckert, S. E. Hawes, and W. E. Stamm. 2003. Longitudinal assessment of infecting serovars of Chlamydia trachomatis in Seattle public health clinics: 1988-1996. Sex. Transm. Dis. 30:357-361. [DOI] [PubMed] [Google Scholar]
- 53.Takourt, B., B. de Barbeyrac, M. Khyatti, F. Radouani, C. Bebear, S. Dessus-Babus, and A. Benslimane. 2001. Direct genotyping and nucleotide sequence analysis of VS1 and VS2 of the Omp1 gene of Chlamydia trachomatis from Moroccan trachomatous specimens. Microbes Infect. 3:459-466. [DOI] [PubMed] [Google Scholar]
- 54.Wang, S. P., and J. T. Grayston. 1991. Three new serovars of Chlamydia trachomatis: Da, Ia, and L2a. J. Infect. Dis. 163:403-405. [DOI] [PubMed] [Google Scholar]
- 55.Whittington, W. L., C. Kent, P. Kissinger, M. K. Oh, J. D. Fortenberry, S. E. Hillis, B. Litchfield, G. A. Bolan, M. E. St. Louis, T. A. Farley, and H. H. Handsfield. 2001. Determinants of persistent and recurrent Chlamydia trachomatis infection in young women: results of a multicenter cohort study. Sex. Transm. Dis. 28:117-123. [DOI] [PubMed] [Google Scholar]
- 56.Yang, C. L., I. Maclean, and R. C. Brunham. 1993. DNA sequence polymorphism of the Chlamydia trachomatis omp1 gene. J. Infect. Dis. 168:1225-1230. [DOI] [PubMed] [Google Scholar]
- 57.Yuan, Y., Y.-X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.