Abstract
A role for acid-sensing ion channels (ASICs) to serve as epithelial channels for Na+ uptake by the gill of freshwater rainbow trout was investigated. We found that the ASIC inhibitors 4′,6-diamidino-2-phenylindole and diminazene decreased Na+ uptake in adult rainbow trout in a dose-dependent manner, with IC50 values of 0.12 and 0.96 μM, respectively. Furthermore, we cloned the trout ASIC1 and ASIC4 homologs and demonstrated that they are expressed differentially in the tissues of the rainbow trout, including gills and isolated mitochondrion-rich cells. Immunohistochemical analysis using custom-made anti-zASIC4.2 antibody and the Na+-K+-ATPase (α5-subunit) antibody demonstrated that the trout ASIC localizes to Na+/K+-ATPase-rich cells in the gill. Moreover, three-dimensional rendering of confocal micrographs demonstrated that ASIC is found in the apical region of mitochondrion-rich cells. We present a revised model whereby ASIC4 is proposed as one mechanism for Na+ uptake from dilute freshwater in the gill of rainbow trout.
Keywords: sodium uptake, gill, acid-sensing ion channels, mitochondrion-rich cells, fish, ionoregulation
fishes living in freshwater (FW) must actively take up Na+ against a steep concentration gradient. Na+ uptake occurs via specialized mitochondrion-rich cells (MRCs), located on the fish gill epithelium (26, 29), and was initially proposed to be linked to NH4+/H+ excretion (26). Subsequent studies have described two models of Na+ transport (for review see Refs. 8 and 18). In the first proposed model, Na+ is exchanged for H+ via an electroneutral Na+/H+ exchanger (NHE) located on the apical membrane of MRCs in fish gill epithelia. The identification of multiple NHEs in the gills of zebrafish (Danio rerio) (51), Osorezan dace (Tribolodon hakonensis) (15), rainbow trout (Oncorhynchus mykiss) (7, 19), and tilapia (Oreochromis mossambicus) (48) by immunocytochemistry, Western blot analysis, and RT-PCR, supports this model. However, significant thermodynamic constraints associated with the function of NHEs at very low ion concentrations (Na+ <0.1 mM) and low environmental pH (pH <5) (1, 34) suggest that fishes living in very soft and poorly buffered water would not be able to rely on a NHE-based mechanism for sufficient Na+ uptake. Recently, the NHE model was revised, whereby the ammonia transporter [rhesus (Rh) protein] present on the apical membrane of MRCs (30, 31) forms a functional metabolon with NHE2/3 (50). The revised model does alleviate the thermodynamic constraints associated with a low-pH environment, but not those imposed by low Na+ concentrations in the FW aquatic environment (6). Therefore, it is unlikely that this mechanism is the sole contributor to Na+ uptake by FW fishes living in low-ionic-strength and poorly buffered waters. The second proposed model for Na+ uptake is based on the Na+ uptake models in frog skin (for review see Ref. 14). In this model, an epithelial Na+ channel (ENaC) is electrochemically linked to an apical vacuolar-type H+-ATPase (VHA) (1). Several studies have provided functional evidence supporting the role of VHA in Na+ uptake in FW fish gills (9, 39) and apical localization of VHA in gill MRCs (44). However, a major drawback of this model has been the inability to identify the channel responsible for Na+ uptake, since no ENaC homologs have been found in the available teleost fish genomes. Therefore, we hypothesized that other channels related to ENaCs may perform this function.
ENaC is a member of the amiloride-sensitive ENaC/degenerin family of ion channels (23). In vertebrates, the closest relatives to ENaC are the acid-sensing ion channels (ASICs), with which they share ∼25% identity (24). Therefore, we investigated the possibility that ASICs may be responsible for Na+ uptake in FW fishes. ASICs are voltage-insensitive Na+ channels gated by extracellular H+ (47) and are associated with the nervous system of mammals and fishes (16, 35, 40, 42, 46). They have been implicated in mechanoreception and sensory transduction of taste and pain (for review see Ref. 23). In mammals, at least seven different ASIC subunits (ASIC1a, ASIC1b, ASIC2a, ASIC2b, ASIC3, ASIC4, and ASIC5) are encoded by five ASIC genes, while in zebrafish, six ASIC subunits (ASIC1.1, ASIC1.2, ASIC1.3, ASIC2, ASIC4.1, and ASIC4.2) are encoded by six different genes (13, 35, 40). No ASIC3 or ASIC5 has been demonstrated in zebrafish. Recently, Chen and colleagues (5) described a group of potent ASIC inhibitors, the diarylamidines. Diarylamidines are a non-amiloride-derived ASIC blockers class of compounds that inhibit ASIC currents at very low concentrations but do not have an effect on ENaC. This finding can be exploited to pharmacologically investigate a potential role for ASICs in Na+ uptake in FW fishes.
In this study we used a variety of whole-animal, cellular, and molecular approaches to investigate a potential role for ASICs in Na+ uptake in FW fishes. We used FW rainbow trout as our model organism, since their natural habitat includes very-low-ionic-strength waters with Na+ concentrations and pH levels below the theoretical limits of the NHE/Rh metabolon model. We identified two trout subunits of ASICs (ASIC1 and ASIC4) that are expressed in gill MRCs and demonstrated an apical MRC localization for trout ASIC4. This is the first study to identify a role for ASICs in Na+ uptake in FW fishes and is also the first study to demonstrate a nonneuronal function for ASICs in an organism.
MATERIALS AND METHODS
Animal holding.
Rainbow trout embryos were obtained from the Raven Brood Trout Station (Caroline, AB, Canada) and grown to the appropriate size for experimentation (∼2 g for flux experiments and 200–500 g as adults for other experiments). Fish were maintained indoors in flow-through 450-liter fiberglass tanks supplied with aerated and dechlorinated Edmonton tap water (hardness as CaCO3: 1.6 mmol/l, alkalinity: 120 mg/l, NaCl: 0.5 mmol/l, pH 8.2, 15°C). Fish were fed ground dry commercial trout pellets (Purina trout chow) once daily and kept on a 14:10-h light-dark photoperiod. All animal use was approved by the University of Alberta Animal Care Committee under Protocol AUP00000072.
Pharmacological inhibition of Na+ uptake.
For flux studies, juvenile trout were first acclimated to low-ionic-strength pH 6.0 water for 4 days prior to Na+ flux experiments. The duration of acclimation was deemed sufficient on the basis of a previous soft water exposure study in rainbow trout (Salmo gairdneri) (43). Fish were transferred to 60-liter plastic tubs supplied with aerated recirculating low-ionic-strength water (Na+: 30.0 ± 3.0 μmol/l, Ca2+: 24 μmol/l, Cl−: 78.0 ± 2.0 μmol/l) and kept at a constant temperature of 15°C. After acclimation, two series of flux experiments were completed. The first experiment investigated concentration-dependent effects of two ASIC-inhibiting diarylamidines, 4′,6-diamidino-2-phenylindole (DAPI; 0.001, 0.01, 0.1, 1.0, and 10 μmol/l; Sigma) and diminazene (0.0003, 0.003, 0.03, 0.3, and 3.0 μmol/l; Sigma), on Na+ uptake in juvenile trout. This experiment established the 50% inhibitory concentrations (IC50; Hill function) and optimal concentrations (>80% inhibition) for DAPI and diminazene. The second flux experiment compared the effects of two well-known pharmacological Na+ uptake inhibitors [amiloride (500 μmol/l) and phenamil (50 μmol/l); Sigma] and the two ASIC inhibitors [DAPI (1 μmol/l) and diminazene (3 μmol/l)] on Na+ uptake in juvenile trout.
For all flux experiments, fish were transferred to individual darkened 60-ml flux chambers, supplied with a constant flow of aerated low-ionic-strength, low-pH water, and acclimated to the chambers for 24 h prior to experimentation. Unidirectional Na+ influx was measured using radiolabeled 22Na, as described previously (12). Briefly, flow of water to chambers was turned off, 0.1 μCi/l 22Na was added to each chamber and allowed to mix for 5 min, and either the DMSO control (0.05% DMSO) or pharmacological agents dissolved in DMSO were added to the chambers. After a 5-min equilibration period, 6-ml water samples were collected at time 0 and 90 and 180 min as appropriate. At the completion of the experiment, fish were terminally anesthetized (MS-222, 1 g/l) and weighed. Water samples (3 ml) were analyzed for 22Na radioactivity using a gamma counter (Packard Cobra II, Auto Gamma, model 5010, Perkin Elmer, Waltham, MA), and total concentration of Na+ was measured using atomic absorption spectrophotometry (model 3300, Perkin Elmer, Shelton, CT). Unidirectional 22Na+ influx (μmol·kg−1·h−1) was calculated for each flux period according to the following equation: JNain = Δcpm × V/(SA × t × M), where Δcpm is the difference between the initial and final radioactivities in the water (cpm/ml), V is the water flux volume (ml), SA is the average specific activity (cpm/μmol Na+) as measured at the beginning and end of the flux period, t is the time elapsed (h), and M is the mass of the fish (kg).
Tissue collection and preparation.
RNA isolation for expression analysis was performed on adult fish. Briefly, fish were euthanized as described above, a blood sample was withdrawn from the caudal arch, and the brain, head kidney, and trunk kidney were dissected out and immediately freeze-clamped in liquid N2 for later processing. For gill tissue, the animal was first perfused with ice-cold, heparinized (15 mg) phosphate-buffered saline (PBS; in mM: 137 NaCl, 2.7 KCl, 4.3 Na2HPO4, and 1.4 NaH2PO4, pH 7.8), and gill tissue or MRCs (as appropriate) were obtained according to the protocols described elsewhere (10). After perfusion, gill arches were processed for MRC isolation, freeze-clamped in liquid N2 for RNA isolation, or placed in fixative for immunohistochemistry or scanning electron microscopy (SEM) (see below).
MRC isolation and cellular imaging.
Adult rainbow trout (∼300–500 g) gills were perfused with PBS to remove blood according to original protocols (10, 33). Subsequently, gill filaments were removed from the rakers, cut into sections (2–5 mm, 3–6 filaments), rinsed in PBS (in mM: 137 NaCl, 2.7 KCl, 4.3 Na2HPO4, and 1.4 NaH2PO4, pH 7.8), and subjected to three (20-min) incubations in 5 ml of 0.05% trypsin-EDTA with shaking (200 rpm) at room temperature. The subsequent cellular suspensions following each incubation were passed through a 64-μm nylon mesh filter into 10 ml of ice-cold fetal bovine serum and rinsed through with PBS to halt trypsin activity. The cells were then centrifuged (5 min, 1,500 g, 4°C) and washed three times with 25 ml of PBS. The final pellet was resuspended in 2 ml of ice-cold PBS and applied to a three-step Percoll density gradient (2 ml, 1.09 g/ml; 2 ml, 1.05 g/ml; and 3 ml, 1.03 g/ml) and centrifuged (45 min, 2,000 g, 4°C, 0-brake). After centrifugation, the cell layer at the 1.09-1.05 interface was removed, washed three times in ice-cold PBS and resuspended in 800 μl of ice-cold PBS.
Isolated MRCs (∼500,000) in PBS with an additional 2 μl of MgCl2 (1 M) and CaCl2 (1 M) to aid in cell attachment were placed on pretreated (1 M HCl acid-washed, 0.1% poly-l-lysine-coated, and rinsed with double-distilled H2O and 70% ethanol) 15-mm round glass coverslips (catalog no. CS-15R, Warner Instruments) and allowed to attach for 2 h at 4°C. After attachment, coverslips were placed at room temperature, and, after careful removal of PBS, the cells were briefly rinsed in Na+-containing buffer (in mM: 142.5 NaCl, 5.0 CaCl2, 1.0 MgCl2, 4.0 KCl, 15 HEPES, and 2.5 NaHCO3, pH 7.8). The cells were then incubated in 200 μl of Na+-containing buffer, which contained 2 μl of 5 mM pH-sensitive BCECF-AM (50 μg in 16 μl of DMSO and 20% pluronic acid), for 30 min at 18°C. Coverslips were placed into a 70-μl imaging chamber (model RC-20H, Warner Instruments) for the perfusion experiments. The chamber was fixed to an inverted fluorescence microscope (Nikon Eclipse TE300), and the cells were subjected to differential interference contrast microscopy and fluorescent imaging. To allow for BCECF-AM excitation at 495 and 440 nm, the microscope was fitted with a xenon arc lamp (Lambda DG-4, Sutter Instruments, Novato, CA). Images were digitally obtained at 440 and 495 nm on a mono 12-bit charge-couple device camera (Retiga EXi, QImaging, Burnaby, BC, Canada) every 1.7 s during perfusion experiments. Ratios of fluorescence at 495 nm to fluorescence at 440 nm were digitally compiled using Northern Eclipse software (Mississauga, ON, Canada).
Cell perfusion.
Solutions were perfused across the attached MRCs in the holding chamber by gravity feed. A six-input manifold (model Mp-6, Warner Instruments) attached to 60-ml syringe holder blocks controlled by pinch valves (model VE-6, Warner Instruments) monitored manually with VC-6 valve controllers (Warner Instruments) was used to perfuse the solutions at a rate of ∼0.5 ml/min. Cells were monitored from original resting state for changes in intracellular pH (pHi) when exposed to Na+-free and Na+-containing solutions. Starting in Na+-free conditions (in mM: 142.5 N-methyl-d-glucamine-Cl, 2.5 C5H14NO·HCO3−, 5 CaCl2, 1 MgCl2, 4 KCl, and 15 HEPES, pH 7.8) followed by replacement with a Na+-containing solution, we monitored the activity of the NHE in trout MRCs following a pHi disturbance. An identical transition between Na+-free and Na+-containing solutions was made, but this time in the presence of 1 μM DAPI or 100 μM ethylisopropylamiloride (EIPA). EIPA was used in this experiment, since it potently inhibits NHE but has a low affinity for Na+ channels (25). Perfusion solutions were bubbled with 0.3% CO2 balanced with O2 throughout the experiments. At the end of each experimental perfusion, cells were subjected to a final high-K+ solution calibration protocol (in mM: 120 potassium gluconate, 20 KCl, 2 MgCl2, and 20 HEPES) in which four solutions were adjusted to pH levels (∼8.40, 7.80, 7.20, and 6.60) by the addition of the ionophore nigericin (5 μM) to equilibrate pHi and extracellular pH. The ratios of fluorescence at 495 nm to fluorescence at 440 nm at each step of the pH calibration were used to form a regression equation for each individual cell trace. This equation was extrapolated to the raw ratio data obtained over the entire time course, yielding a calibrated pHi for each individual cell monitored during the perfusion experiment. Data were collected and analyzed for each calibrated individual cell by comparing the rate of alkalinization (ΔpHi/Δt) under the control parameters with that in the presence of the drug to determine percent inhibition.
Preparation of total RNA.
Total RNA was extracted from frozen tissues using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. RNA was treated with DNase I (Ambion, Austin, TX) according to the manufacturer's protocol; then an on-column cleanup was carried out using an RNeasy Mini Kit (Qiagen, Mississauga, ON, Canada). The quality of the extracted RNA was assessed by a bioanalyzer (model 2100, Agilent Technologies, Palo Alto, CA) or by visualization on a formaldehyde gel. RNA concentration was measured with a spectrophotometer (ND-1000 UV-Vis, NanoDrop Technologies, Rockland, DE).
Molecular cloning and phylogenetic analysis.
Rapid amplification of cDNA ends (RACE)-ready cDNA was synthesized from gill RNA using a SMARTer RACE cDNA amplification kit (Clontech, Mountain View, CA) according to the manufacturer's instructions. cDNA solutions were diluted 1:10 with tricine-EDTA buffer provided in the kit and stored at −20°C.
Oligonucleotides for cloning of ASIC1 and ASIC4 genes were designed with PrimerQuest (Integrated DNA Technology) (Table 1) on the basis of conserved regions in available ASIC mRNA sequences from zebrafish, tilapia, pufferfish, stickleback, and cod (National Center for Biotechnology Information, Ensembl). Sequences were aligned with GeneDoc (version 2.6.0.2, http:/www.psc.edu/biomed/genedoc) and ClustalX (version 1.81) software. DNA sequences encoding the 5′- and 3′-termini of the ASIC1 gene and the 5′-terminus of the ASIC4 gene were obtained using the Advantage 2 PCR kit (Clonetech) with RACE-ready cDNA templates generated from gill RNA. The ASIC4 3′-terminus was obtained using degenerate primer (Table 1). Resulting PCR products were excised from the gel and purified with a QIAquick gel extraction kit (Qiagen). Subsequently, amplicons of interest were sequenced directly or ligated into pJET1.2 vector using a CloneJET kit (Thermo Scientific) for verification. Recombinant plasmids were transformed into competent cells (One Shot TOP10 chemically competent Escherichia coli, Invitrogen). Colonies were screened for plasmids containing inserts of the correct size by agarose gel electrophoresis and sequenced with pJET1.2-specific primers provided in the CloneJET kit.
Table 1.
Primer sets for cloning and tissue distribution of ASICs
| Gene | Primer Sequence |
|---|---|
| Cloning | |
| ASIC1 | |
| 5′ | 5′-TCACCCAGCAACCCTGCGAACTCGT-3′ |
| 3′ | 5′-ACACCCTGCAACATGACGCGCT-3′ |
| ASIC4 | |
| 5′ | 5′-GATGTCCCGTCCGGTTGAAGATGTC-3′ |
| Forward | 5′-CCCAGTAACATCAAGTGTGTCG-3′ |
| Reverse | 5′-YYARCANGCRAARTCYTC-3′ |
| RT-PCR | |
| ASIC1 | |
| Forward | 5′-AAGTCCACTCCCATAGA-3′ |
| Reverse | 5′-CAGCCAGGTTATTCCTT-3′ |
| ASIC4 | |
| Forward | 5′-CTTTCGTTTCTCTGCTCTCACC-3′ |
| Reverse | 5′-CAAGACCAGGAAGTTGTCTCTG-3′ |
| GAPDH | |
| Forward | 5′-AAGGGTGAGGTGAGCATGGA-3′ |
| Reverse | 5′-GCTTTACCCCATGGGATCTCAT-3′ |
ASIC, acid-sensing ion channel; 5′, 5′-rapid amplification of cDNA ends (RACE) primer; 3′, 3′−RACE primer; Y = C + T; R = A + G; N = A + G + T + C.
The amino acid sequences derived from our cloned trout ASIC4 and ASIC1 genes (accession numbers KF964646 and KF964645, respectively), together with the previously annotated amino acid sequences of ASIC1, ASIC2, ASIC3, and ASIC4 from various fish species and other taxa, were aligned and analyzed using Seaview software (version 4; http://pbil.univ-lyon1.fr/software/seaview). A rooted phylogenetic tree showing evolutionary relationships between the different ASIC proteins was generated using the maximum-likelihood method and LG model and 700 bootstrap resamplings.
RT-PCR.
Tissue-specific cDNA templates utilized for RT-PCR were synthesized from 3 μg of RNA from gill, MRCs, kidney, brain, or blood using SuperScript III reverse transcriptase (Invitrogen) according to protocols provided by the manufacturer. ASIC4 and ASIC1 amplicons were obtained using primers designed with PrimerQuest (Table 1). PCR conditions were as follows: 98°C for 1 min of initial denaturation followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 62°C for 30 s, and elongation at 72°C for 40 s, with the final elongation at 72°C for 10 min. PCR products were visualized by 1% agarose gel electrophoresis followed by ethidium bromide staining.
Immunoprecipitation of ASIC4 and Western blot analysis.
An anti-zebrafish ASIC4b (zASIC4.2) polyclonal antibody against two peptides corresponding to regions near the NH2 terminus (aa 146–160; PKSRKGHRPSELQYP) and the COOH terminus (aa 519–533; CFEEVKVKAANDVAQP) of zASIC4.2 protein (accession no. Q708S3.1) was custom-made by 21st Century Biochemicals (Marlboro, MA). The polyserum was affinity-purified against each of these peptides independently, and the NH2-terminal peptide-purified antibody was used exclusively in this study. Immunoprecipitation was performed according to the technique described by Goss and colleagues (11). Briefly, trout gills were perfused with heparinized ice-cold PBS (Ca2+-free), and the second and third gill arches were dissected out and washed three times in ice-cold PBS. Gill rakers were removed, and the remaining gill arches were cut into 200- to 300-mg sections. Each section was lysed for 30 min in 2–3 ml of lysis buffer (100 mM NaCl, 4 mM KCl, 50 mM HEPES, 5 mM EDTA, pH 7.4, and 1% Triton-X) containing protease inhibitors (cOmplete Mini, EDTA-free protease inhibitor tablets; Roche Diagnostics, Mannheim, Germany). After centrifugation to remove the debris (5,000 g, 5 min, 4°C), the lysates were incubated with 2–4 μl of anti-zASIC4.2 antibody at 4°C overnight on an end-over-end rotator. On the next day, 60 μl of protein A-Sepharose CL4b beads (Sigma Chemical, St. Louis, MO) preswelled in immunoprecipitation (IP) buffer (in mM: 100 NaCl, 4 KCl, 50 HEPES, and 5 EDTA, pH 7.4) and preblocked with 1% bovine serum albumin (fraction V; Sigma) were added to the lysates, which were incubated at 4°C for 4–6 h on the rotator. After the incubation, the supernatant was removed and the beads were washed three times with 1 ml of IP buffer. After the washes, the beads were incubated in Laemmli buffer for 15 min at 65°C and centrifuged, and the supernatant was retained for Western blot analysis, as described previously (45). Briefly, the samples were separated on a 7.5% polyacrylamide minigel and transferred to a nitrocellulose (NC) membrane using a wet transfer system (Bio-Rad Laboratories, Hercules, CA). The NC membrane was blocked in 5% bovine serum albumin in Tris-buffered saline with 0.2% Triton X-100 (TBST) for 30 min and incubated with rabbit anti-zASIC4.2 antibody (1:1,000 dilution) on a rocker at 4°C overnight. The membrane was washed four times for 15 min each with TBST, blocked again with 5% bovine serum albumin for 15 min, and then incubated with a secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Santa Cruz Biotechnology, Dallas, TX) at 1:50,000 dilution at room temperature for 1 h. The NC membrane was washed four times in TBST, and the immunoreactive bands were visualized using a SuperSignal West Pico chemiluminescent substrate kit (Thermo Scientific) following the manufacturer's instructions.
SEM and immunohistochemistry.
For examination of the surface of the gill by SEM, filaments were fixed in 3% glutaraldehyde and 2% paraformaldehyde in 0.15 mM sodium cacodylate buffer (pH 7.4, 290 mosM) for 1 h at 4°C, dehydrated in a graded series of ethanol, and critical point dried. Samples were then mounted on an SEM stub, sputter-coated with gold, and examined with a scanning electron microscope (model JSM-6301FXV, Jeol).
For immunohistochemistry, gills were excised from adult rainbow trout, fixed in 4% paraformaldehyde in PBS (pH 7.4) overnight at 4°C, dehydrated in a graded series of ethanol, and embedded into paraffin blocks. Serial sections (5 μm) were cut and rehydrated in a decreasing ethanol series followed by double-distilled H2O. Rehydrated sections were incubated in 10 mM citrate buffer at 70°C for 1 h for epitope retrieval. After incubation, slides were washed in PBS and then blocked with 6% milk powder in a humidified chamber for 1 h. Sections were then incubated overnight at 4°C simultaneously with the anti-zASIC4.2 antibody (1:400 dilution) and anti-Na+-K+-ATPase (NKA) monoclonal antibody (1:400 dilution, anti-chicken α5-subunit; Developmental Studies Hybridoma Bank, University of Iowa) followed by secondary FITC-conjugated anti-mouse antibody (1:500 dilution; Invitrogen, Oregon) and tetramethylrhodamine-conjugated anti-rabbit antibody (1:500 dilution; Invitrogen, Oregon). Slides incubated with preimmune serum from the anti-zASIC4.2 antibody production in place of primary antibody were used as a negative control. Slides were viewed with a laser scanning confocal microscope (model LSM 710, Zeiss, Germany) at the Cross Cancer Institute Cell Imaging Facility (Edmonton, AB, Canada). Images were processed with LSM Image Browser (version 4.2.0.121; Carl Zeiss). Surface rendering and three-dimensional reconstruction of z-stack images were performed using Imaris software (version 6.2.2, Bitplane, Zurich, Switzerland).
Statistical analysis.
Na+ flux data were subjected to statistical analysis and are reported as means ± SE. All pharmacological experiment data sets were tested for homogeneity of variance and compared using one-way ANOVA (SigmaPlot version 11, Systat, Chicago, IL). If significant differences (P ≤ 0.05) were found, a post hoc multiple-comparisons Tukey's test was applied to determine these differences. A paired t-test (P ≤ 0.05) was used in the pHi imaging experiments to compare the relative inhibition of ΔpHi/Δt in isolated cells under control conditions and after addition of each pharmacological inhibitor.
RESULTS
Pharmacological inhibition.
Exposure of juvenile rainbow trout to increasing concentrations of DAPI resulted in a concentration-dependent decrease in Na+ uptake, with >90% inhibition at ≥1 μmol/l (Fig. 1A). The DAPI concentration at which there was a 50% inhibition in Na+ uptake (IC50) was calculated to be 0.12 μmol/l. Exposure to diminazene also resulted in a concentration-dependent decrease in Na+ uptake in juvenile trout, with a maximum reduction of Na+ uptake from 303 ± 43 (control) to 51 ± 19 μmol·kg−1·h−1 (79%) after exposure to 3.0 μmol/l diminazene (Fig. 1B). The IC50 for diminazene was calculated to be 0.96 μmol/l.
Fig. 1.
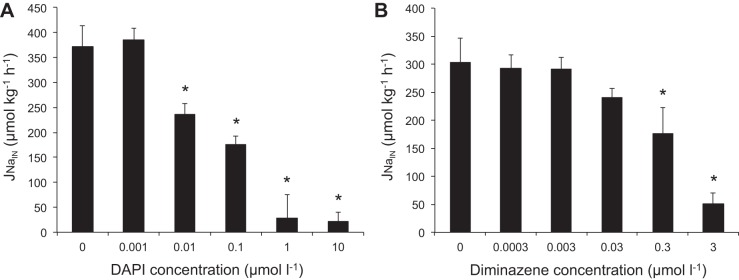
Dose-dependent decreases in Na+ uptake rates (JNa,in) in juvenile trout after exposure to increasing concentrations of 4′,6-diamidino-2-phenylindole (DAPI) and diminazene in low-Na+ (30 μM) and low-pH (pH 6.0) water. Values are means ± SE (n = 6). *P < 0.05 vs. control.
Na+ uptake rates in juvenile rainbow trout were significantly lower in the first 90-min flux time period after exposure to 500 μM amiloride (17 ± 18.5 μmol·kg−1·h−1), 1 μM DAPI (29 ± 18.9 μmol·kg−1·h−1), and 3 μM diminazene (41 ± 33.4 μmol·kg−1·h−1) than in controls (539 ± 170.9 μmol·kg−1·h−1; Fig. 2); while 50 μM phenamil (299 ± 107.9 μmol·kg−1·h−1) did not significantly reduce Na+ uptake. However, for the second consecutive flux period, between 90 and 180 min of exposure to the pharmacological agents, the inhibition was less pronounced but still significant for amiloride (104 ± 33.4 μmol·kg−1·h−1) and DAPI (141 ± 62.7 μmol·kg−1·h−1), while diminazene (297 ± 41.5 μmol·kg−1·h−1) was no longer significantly different from control (393 ± 44.1 μmol·kg−1·h−1; Fig. 2).
Fig. 2.
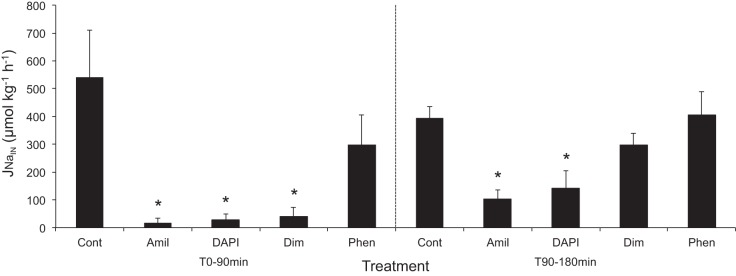
Effects of pharmacological agents [control (Cont, DMSO), amiloride (Amil, 500 μmol/l), DAPI (1 μmol/l), diminazene (Dim, 3 μmol/l), and phenamil (Phen, 50 μmol/l)] during consecutive 90-min measurements of Na+ uptake in juvenile trout in low-Na+ (30 μM) and low-pH (pH 6.0) water. Values are means ± SE (n = 6). *P < 0.05 vs. control.
MRC pHi imaging.
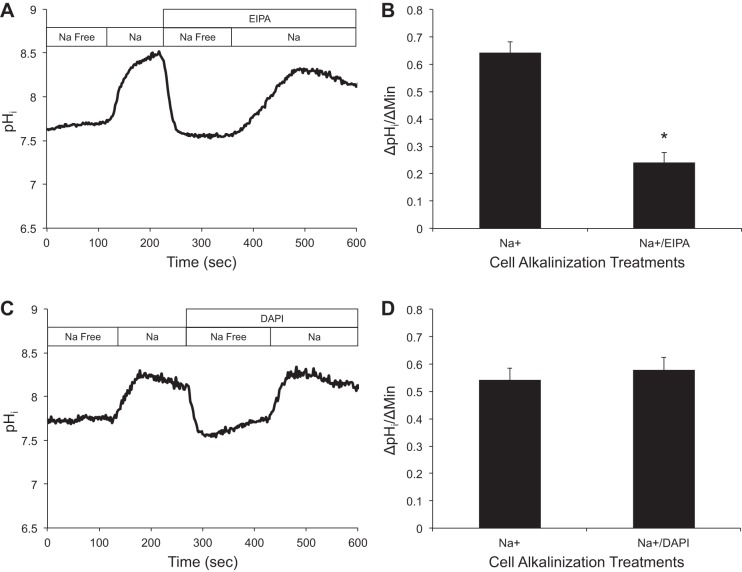
Results presented from the whole-animal flux experiments showed that administration of DAPI at various concentrations significantly inhibited Na+ uptake (Fig. 1A). To demonstrate that the inhibition of Na+ uptake attributed to DAPI was in fact due to the blocking of ASIC-type Na+ channels, rather than affecting NHE activity, we conducted experiments using pHi imaging. NHE activity was measured by examining the rate of pHi alkalization (ΔpHi/Δt) upon exposure of MRCs to a Na+-containing medium (145 mM) in the absence or presence of DAPI (1 μM) or EIPA (100 μM).
A representative trace (Fig. 3A) shows a Na+-dependent alkalization following Na+ application (1st rise), while addition of EIPA significantly decreased the rate of alkalization (2nd rise). The combined results (n = 19) show a control of 0.642 ± 0.040 ΔpHi/Δmin, while the alkalization rate was reduced by 62% in the presence of EIPA (0. 240 ± 0.036 ΔpHi/Δmin, P < 0.001; Fig. 3B). For DAPI, Na+-dependent alkalization following Na+ application was similar in the presence or absence of DAPI (Fig. 3D). No differences in ΔpHi/Δt were noted between control (0.542 ± 0.042 ΔpHi/Δmin) and DAPI-treated (0.578 ± 0.047 ΔpHi/Δmin) cells (P = 0.149, n = 22; Fig. 3D).
Fig. 3.
Effect of ethylisopropylamiloride (EIPA) and DAPI on Na+-induced alkalinization of peptide nucleic acid-positive (PNA+) cells following Na+-free exposure at resting intracellular pH (pHi). A: representative trace demonstrating inhibition of cell alkalinization by EIPA (100 μM). B: quantitative analysis of cell alkalinization in the absence and presence of EIPA (∼62% inhibition). Values are means ± SE (n = 19 cells). *P < 0.001 (by paired t-test). C: representative trace demonstrating lack of effect on cell alkalinization in the presence of DAPI (1 μM). D: quantitative analysis of cell alkalinization in the absence and presence of DAPI. Values are means ± SE (n = 22). *P > 0.05 (by paired t-test).
Phylogenetic analysis and tissue distribution of ASIC1 and ASIC4.
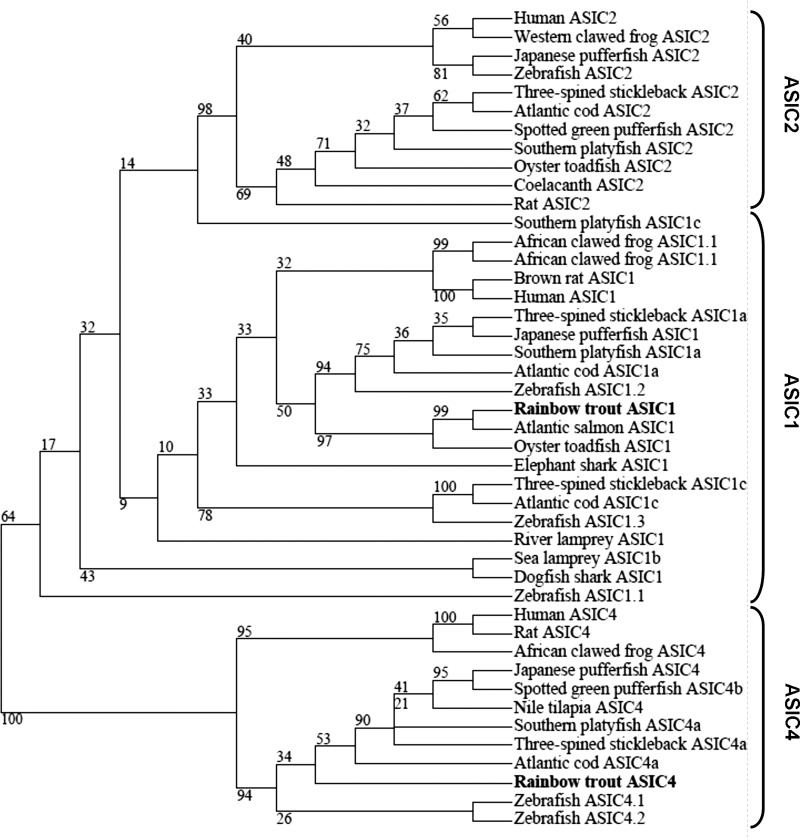
Two distinct DNA fragments, whose derived amino acid sequences exhibit similarity to the ASIC family, were cloned from the gill of rainbow trout. To determine their identity, we conducted a phylogenetic analysis by comparison with known ASIC homologs from a variety of species. The generated phylogenetic tree (Fig. 4) showed that the two cloned rainbow trout ASIC genes (tASICs) were members of the ASIC1 and ASIC4 subunits, respectively (Fig. 4A). The tASIC1 (partial clone) grouped most closely with ASIC1 from salmon (Salmo salar), while tASIC4 grouped with other identified ASIC4 family members from other fish and mammal species.
Fig. 4.
Phylogenetic analysis of amino acid sequences of cloned rainbow trout acid-sensing ion channels ASIC1 and ASIC4 and ASICs from various fish species. Phylogenetic tree was constructed using the maximum-likelihood method, and numbers indicate bootstrap values for 700 resamplings. GenBank or Ensembl accession numbers are as follows: human ASIC2, NP_001085.2; Western clawed frog ASIC2, XP_002933086.2; Japanese pufferfish ASIC2, XP_003964871.1; zebrafish ASIC2, NP_999953.1; three-spined stickleback ASIC2, ENSGACG00000009558; Atlantic cod ASIC2, ENSGMOT00000019145; spotted green pufferfish ASIC2, ENSTNIG00000014943; Southern platyfish ASIC2, ENSXMAG00000008283, oyster toadfish ASIC2, AAP45633.1; coelacanth ASIC2, ENSLACG00000007191; rat ASIC2, NP_037024.2; Southern platyfish ASIC1c, ENSXMAG00000018841; African clawed frog ASIC1.1, AEC48665.1; African clawed frog ASIC1.2, AEC48667.1; brown rat ASIC1, NP_077068.1; human ASIC1, NP_064423.2; three-spined stickleback ASIC1a, ENSGACG00000003075; Japanese pufferfish ASIC1, XP_003963520.1; Southern platyfish ASIC1a, XP_005801801.1; Atlantic cod ASIC1a, ENSGMOG00000009659; zebrafish ASIC1.2, NP_999955.1; rainbow trout ASIC1, KF964646; Atlantic salmon ASIC1, NP_001133456.1; oyster toadfish ASIC1, Q7T1N4.1; elephant shark ASIC1, AEC48668.1; three-spined stickleback AIC1c, ENSGACG00000017974; Atlantic cod ASIC1c, ENSGMOG00000003105; zebrafish ASIC1.3, NP_999954.1; river lamprey ASIC1, AAY28983.1; sea lamprey ASIC1b, ENSPMAG00000003813; dogfish shark ASIC1, AAY28985.1; zebrafish ASIC1.1, NP_999956.1; human ASIC4, NP_878267.2; rat ASIC4, NP_071570.2; African clawed frog ASIC4, AEC48666.1; Japanese pufferfish ASIC4, XP_003962127.1; spotted green pufferfish ASIC4b, ENSTNIG00000010994; Nile tilapia ASIC4, XP_003445446.1; Southern platyfish ASIC4a, ENSXMAG00000003624; three-spined stickleback ASIC4a, ENSGACG00000007052; Atlantic cod ASIC4a, ENSGMOG00000012740; rainbow trout ASIC4, KF964645; zebrafish ASIC4.1, NP_999952.1; zebrafish ASIC4.2, NP_999951.1.
Furthermore, we conducted a tissue distribution analysis of tASIC1 and tASIC4 by RT-PCR. We observed mRNA expression of tASIC1 in all tested tissues: gill, MRCs, head and trunk kidney, brain, and blood. tASIC4 had a slightly different distribution: it was present in the perfused gill, MRCs, brain, and trunk kidney, but it was absent in head kidney and blood tissues.
Immunoprecipitation and immunostaining with anti-zASIC4.2.
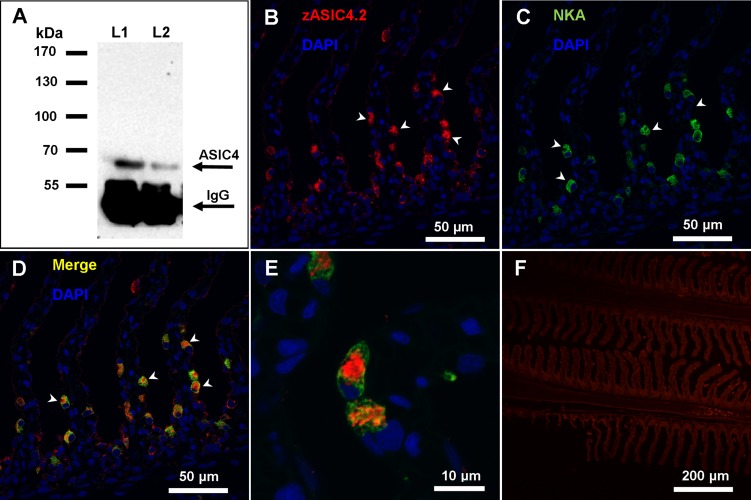
To determine if ASIC4 was present in the fish gill and to validate our antibody for later immunocytochemistry, we performed an immunoprecipitation using an antibody raised against zASIC4.2 on the basis of sequence similarly of ∼60% between zASIC4.2 (aa 146–160: PKSRKGHRPSELQYPP) and the cloned tASIC4 (aa 142–156: PKNREGHKPTDLDYPA). Using Western blotting, we analyzed the precipitates for the presence of ASIC4 (see Fig. 6A) and found a single band of ∼65 kDa, which approximates the predicted size for tASIC4 (60.4 kDa). The strong band at ∼55 kDa corresponds to the IgG heavy chain.
Fig. 6.
Immunoreactivity of anti-zebrafish ASIC4.2 (zASIC4.2) antibody in the gill of rainbow trout. A: immunoprecipitation Western blot from whole gill homogenates with zASIC4.2 showing a distinct band of ∼65 kDa. Lane 1 (L1) contains 300 mg of gill tissue, and lane 2 (L2) contains 200 mg of gill tissue. B–E: confocal images of gill sections labeled for ASIC4 and Na+-K+-ATPase (NKA) proteins. B and C: double-labeling with DAPI and anti-zASIC4.2 (B) and DAPI and anti-NKA (C). D: merged image of B and C. E: higher-magnification image of a MRC showing that ASIC4 and NKA are present in distinct regions within the MRC. F: control micrograph with DAPI and no primary antibody. Arrowheads (B–D) indicate positive staining.
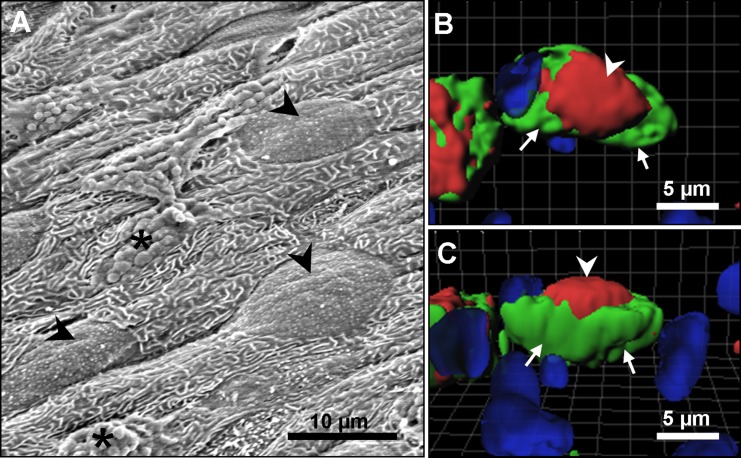
To determine the location of ASIC4 protein within the rainbow trout gill tissue, fixed sections were double-labeled with anti-zASIC4.2 antibody (see Fig. 6B) and anti-NKA and then counterstained with the nuclear stain DAPI. Immunoreactivity was observed in cells located in the lamellae and interlamellar region (see Fig. 6B, arrowhead). Moreover, the majority of cells positive for zASIC4.2 also colocalized with NKA protein (Fig. 6, C and D, arrowhead) a known marker of MRCs, suggesting that, in gill epithelium, ASIC4 protein is mainly expressed in MRCs. SEM analysis of the gill filament demonstrated that MRCs are clearly distinguished on the surface of the epithelium and have a broad apical convex opening to the environment (see Fig. 7A, arrowheads). The generated three-dimensional reconstructions of the staining immunoreactivity pattern for ASIC4 and NKA proteins revealed the presence of ASIC4 and NKA in the same cells, but in distinct regions of the cells. NKA was present on the basolateral membrane (arrows, green staining), while ASIC4 was localized to the broad apical surface of the cell (arrowheads, red staining). Figure 7B shows a generated image with a top view, while Fig. 7C is the same cell viewed from the side.
Fig. 7.
Three-dimensional views of freshwater (FW) rainbow trout MRCs. A: scanning electron micrograph of FW rainbow trout gill epithelium showing broad apical openings of MRCs (arrowheads) and mucous cells (*). B and C: 3-dimensional reconstruction of z-stack images showing apical localization of ASIC4 protein (red; arrowhead) and basolateral localization of NKA (green; arrow). B: top view. C: side view.
DISCUSSION
The existence of an apical Na+ channel electrochemically coupled to a VHA mediating Na+ uptake from dilute FW has been postulated on theoretical thermodynamic grounds for a number of decades (1, 36, 34). However, an inability to determine the molecular identity of the Na+ channel has been a major drawback in the general acceptance of the model. Using a combination of pharmacological blockade, isotopic fluxes, immunocytochemistry, and molecular biology, we propose that an ASIC family member is potentially the long-sought-after apical Na+ channel responsible for Na+ uptake by rainbow trout acclimatized to water with low Na+ and low pH. Our study provides the first evidence for a nonneuronal role for ASICs in any vertebrate, whereby in gills from FW fishes ASICs are associated with the MRCs, and tASIC4 localizes to the apical membrane of NKA positive cells.
ASICs are a broad family of Na+-specific ion channels that have been extensively studied in mammalian models (for review see Ref. 23), with only a few studies performed in any fish species (35, 42, 46). ASICs have primarily been associated with neuronal tissues, with studies focusing on a neuronal function for ASICs (23). In this study, ASIC1 and ASIC4 were cloned in rainbow trout using PCR, and they were expressed in a variety of tissues, both neuronal and nonneuronal in origin (Fig. 5). Importantly, tASIC1 gene was present in all five investigated tissues, including whole blood. It appears that the tASIC4 gene has a more defined tissue distribution than tASIC1, with expression in the ionoregulatory tissues (trunk kidney and perfused gill) and the brain. Unlike tASIC1, there was no apparent expression of tASIC4 in the head kidney or the blood. Therefore, tASIC4 expression in MRCs and perfused gill could not be attributed to blood contamination.
Fig. 5.
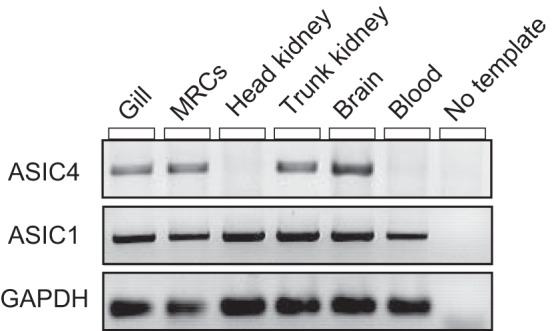
Tissue distribution analysis of ASIC4 and ASIC1 genes in adult rainbow trout as determined by PCR. GAPDH was used as a reference gene. MRCs, mitochondrion-rich cells.
Both tASIC1 and tASIC4 genes were expressed in the gill epithelium and, more specifically, were present in MRCs of rainbow trout gills. It is believed that, similar to ENaCs, functional ASICs are trimers (20), although their configuration has not been resolved. It has been demonstrated that, with a few exceptions, ASICs function as homomers (forming channels of identical subunits) or heteromers (forming channels with other ASIC subunits) (4, 23). ASICs have been characterized by using oocyte expression in zebrafish (4, 35) and dogfish shark (ASIC1b only) (42). It has been found that coexpression of zASIC4.1 with zASIC1.3 in Xenopus oocytes increased the current amplitude and abundance (∼15-fold) of the channel at the cell surface, indicating that these two ASIC subunits form a functional channel (4). Moreover, this ASIC4.1/1.3 heteromeric channel was more efficiently trafficked to the plasma membrane and had increased affinity for H+. Additionally, zASIC4.1, when heterologously expressed in Xenopus oocytes, has been shown to be also gated open by decreases in extracellular Ca2+. The signal responsible for gating of zASIC4.2 was unable to be determined in that study (4). It is therefore possible that tASIC1 and tASIC4 form a heteromeric channel in MRCs, but this cannot be determined at this time and would require confirmation by immunohistochemistry with an anti-ASIC1 antibody that is immunoreactive for rainbow trout. It is also possible that other ASIC subunits and/or regulatory elements may be involved in the function of ASICs in the FW trout gill. A full characterization of tASIC4 by oocyte expression and electrophysiology is required to understand the mechanisms of gating in low-Na+, low-Ca2+ waters.
In our study, ASIC4 protein was colocalized with NKA and was present on the apical surface of cells, suggesting that it is found primarily in MRCs (Fig. 6, D and E). Previous studies on ASICs in fishes have suggested that they are primarily associated with neuronal tissue and sensory cells (27, 35, 42, 46). In adult and developing zebrafish, morphological studies using confocal immunofluorescence techniques demonstrated that gill MRCs come into direct contact with nerve fibers in the basolateral region (21, 22). However, in our study, ASIC4 protein was clearly localized to the apical region of the MRCs, as demonstrated by three-dimensional reconstruction of confocal images. ASIC4 immunoreactivity did not overlap with the standard basolateral immunoreactive pattern associated with NKA (Fig. 7, B and C). Therefore, we conclude that neuronally derived tissues cannot account for the expression of ASIC4 protein in MRCs. We also corroborate the ASIC4 staining pattern with the MRC external morphology, which shows that, in FW rainbow trout raised in Edmonton tap water (∼500 μM Na+), the MRCs have wide apical openings (Fig. 7A). This apical localization of ASIC4 protein in the cells of a transporting epithelium is the first evidence for a nonneuronal function for any ASIC member and suggests that ASIC4, as a Na+ channel, may be responsible for apical uptake of Na+ in FW fish gill.
Pharmacological profiling has long been used to differentiate the mechanisms of ion transport across epithelial tissues. For Na+ transport, classical analysis has usually involved amiloride and its substituted derivatives (e.g., EIPA, mercaptoisobutyric acid, and phenamil) as a means to differentiate between the involvement of NHE isoforms and ENaC in Na+ transport (25). We investigated two newly described ASIC-specific inhibitors, diminazene and DAPI (5), for their potency in inhibiting unidirectional Na+ uptake in fish. Diminazene and DAPI exhibited a dose-dependent inhibitory relationship, and both were very effective at low concentrations (Fig. 1). Na+ uptake in rainbow trout acclimatized to low-ionic-strength, acidic water was clearly inhibited by these diarylamidines at concentrations similar to those reported in a previous study of cultured rat hippocampal neurons (5). However, in our study, DAPI proved to be more effective than diminazene, in contrast to observations in rat neurons. The inhibition of 22Na+ uptake by DAPI and diminazene was also compared with other common Na+ transport inhibitors: amiloride and phenamil. Amiloride was observed to be an effective inhibitor of Na+ uptake in rainbow trout, but only at relatively high concentrations (500 μM), which is consistent with previous studies (2, 37). Recently, amiloride was also demonstrated to completely block ASIC-mediated currents (5, 35, 42); however, it is also known to block many other Na+ transporters, including NHE and ENaC (25). Amiloride, therefore, cannot be used as a distinguishing pharmacological agent.
While DAPI, diminazene, and amiloride almost completely blocked Na+ transport in the present study, phenamil had no significant effect on Na+ flux in rainbow trout. This suggests that phenamil is a poor inhibitor of Na+ uptake in trout exposed to low-ionic-strength water (30 μmol/l Na+). Past studies of the effects of phenamil on Na+ uptake in a variety of FW fishes have reported inconsistent results. For example, in neon tetras acclimatized to low Na+ (50 μmol/l), phenamil had no effect on Na+ transport (38), whereas it caused >70% reduction in Na+ influx in Cyprinidon variegatus hubbsi (100 μmol/l Na+) and 60% reduction in goldfish (300 μmol/l Na+) (2, 38). These conflicting results from various phenamil studies suggest that sensitivity to phenamil might be species- and/or environment-specific. Collectively, the pharmacological analysis suggests that the Na+ uptake in FW rainbow trout is most likely not mediated by ENaC, given the lack of inhibition by phenamil, but may be attributed to ASIC, since Na+ uptake was almost completely blocked by DAPI and diminazene, both of which inhibit ASICs, but not ENaC.
In our Na+ flux study with various pharmacological agents, we made an interesting observation that inhibition of 22Na+ uptake was less pronounced for all tested inhibitors during the second consecutive measurement (90–180 min) (Fig. 2). To our knowledge, this phenomenon, where potency of the tested drug diminishes over time, has not been reported in any previous study on fish. This observation may be explained by a decreased stability/efficacy of the tested pharmacological agents in aqueous media, metabolism of the drug by the animal, or activation of compensatory Na+ uptake mechanisms not sensitive to amiloride, DAPI, or diminazene. We believe that the decrease of the potency of amiloride, DAPI, and diminazene over time demonstrates the importance of proper experimental design and choice of period of study when ion fluxes are examined using pharmacological inhibitors.
While the two novel ASIC inhibitors have been tested for their potency in inhibiting ASICs and ENaC in neurons (5), they have not been tested for their potential effects on other Na+ transporters implicated in Na+ uptake in fishes, most notably, the NHEs. To verify that the Na+ uptake inhibition caused by DAPI and diminazene in rainbow trout was due to inhibition of ASICs, rather than NHEs, we tested the effect of DAPI on MRCs isolated from the rainbow trout gill epithelium. Our laboratory previously showed that rainbow trout possess two populations of MRCs: peanut lectin agglutinin (PNA) positive (PNA+) and PNA negative (PNA−) MRCs (10). It has been demonstrated that NHE3 and NHE2 are expressed in PNA+ cells isolated from branchial epithelium of FW rainbow trout (19). Therefore, we investigated the effect of DAPI on PNA+ cells and compared it with the effect of EIPA, a potent and highly selective inhibitor of NHE (25). Earlier studies in our laboratory using cell imaging determined that PNA+ cells undergo NHE-dependent alkalinization (32, 33). Here we demonstrate that while EIPA was effective at inhibiting NHE function in PNA+ cells (Fig. 3A), there was no effect of high concentrations of DAPI on Na+-dependent pHi elevation, demonstrating that NHE was not inhibited by DAPI (Fig. 3B). Therefore, the measured inhibition of Na+ uptake by DAPI that we see in whole animals (Fig. 1) cannot be attributed to inhibition of NHE.
On the basis of the mRNA expression of tASIC1 and tASIC4 genes in the gill epithelium and isolated MRCs, localization of ASIC4 protein to the apical region of the MRCs, and pharmacological blockade of Na+ influx by ASIC inhibitors in adult rainbow trout, we propose a revised model for transepithelial Na+ uptake in FW rainbow trout gills (Fig. 8) whereby one apical mode of Na+ entry is via an ASIC4-mediated mechanism.
Fig. 8.
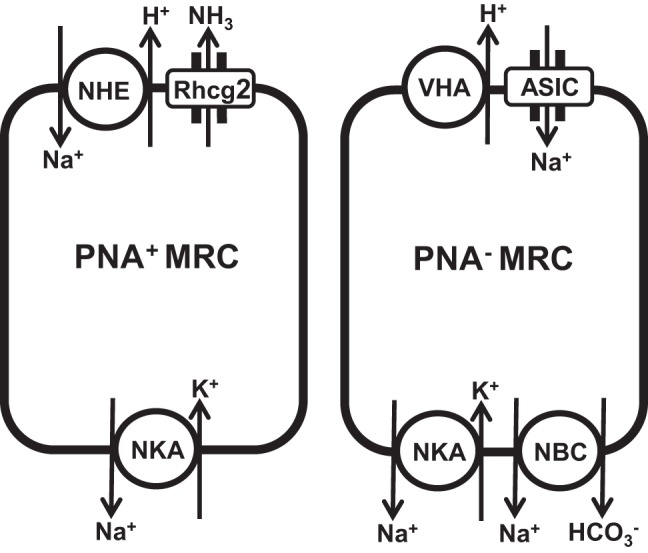
Model for Na+ uptake in PNA+ and PNA− MRCs in gill epithelium of rainbow trout showing placement of proposed ASIC-mediated Na+ uptake mechanisms. Rhcg2, rhesus glycoprotein; NHE, Na+/H+ exchanger; VHA, vacuolar H+-ATPase; NBC, Na+-HCO3− cotransporter.
A number of recent studies on Na+ uptake in FW fishes have demonstrated the presence of the NHE/Rh metabolon in the gills of various species (17, 30, 31). As mentioned previously, we specifically targeted the rainbow trout as a model for this study, as it is native to oligotrophic, Na+-poor mountain streams with low buffering capacity. This type of environment presents key thermodynamic challenges to the NHE/Rh metabolon, as Na+ restrictions cannot be alleviated by the presence of an Rh protein (6). Moreover, searches of rainbow trout genomes and transcriptomes have failed to identify any ENaC subunits (α, β, or γ), despite the fact that they are known to actively take up Na+ from water with <50 μM Na+ (49). Furthermore, other research studies have suggested and demonstrated an apical localization of the VHA in the gills of rainbow trout (44, 48), and bafilomycin, a VHA-specific inhibitor, has been demonstrated to reduce Na+ uptake in zebrafish (2), tilapia, and carp (9). Previous studies have proposed VHA to be a part of the NHE/Rh metabolon (41, 50, 51); however, the placement of an apically oriented VHA is counterintuitive to the function of an NHE, as it would exacerbate the noted thermodynamic constraints. Placement of a VHA in concert with an apical ASIC is more plausible, since it would not only facilitate Na+ uptake from very low ionic strength water but could also provide a mechanism for gating of ASIC-mediated Na+ conductance. It remains to be determined if ASIC and VHA proteins colocalize to the same type of MRCs, and this warrants further investigation.
Perspectives.
In summary, we propose that ASCI4 is involved in mediating Na+ uptake from FW in rainbow trout acclimatized to low ionic strength, low pH media. This is the first demonstration of a nonneuronal function for any ASIC family member and simultaneously provides an attractive solution to the debate surrounding the thermodynamic limitations of the other proposed Na+ transport mechanisms and resolves the conundrum of the role of an apical VHA in Na+ transport. The possibility that other fish models may use similar mechanisms under different environmental conditions where models such as the NHE/Rh metabolon cannot be expected to function remains to be explored.
GRANTS
This study was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant to G. G. Goss and an Alberta Ingenuity Student Scholarship to A. K. Dymowska.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.K.D., A.G.S., and G.G.G. are responsible for conception and design of the research; A.K.D., A.G.S., S.D.B., and D.C. performed the experiments; A.K.D., A.G.S., S.D.B., D.C., and G.G.G. analyzed the data; A.K.D., A.G.S., S.D.B., D.C., and G.G.G. interpreted the results of the experiments; A.K.D., A.G.S., S.D.B., and G.G.G. prepared the figures; A.K.D., A.G.S., S.D.B., D.C., and G.G.G. drafted the manuscript; A.K.D., A.G.S., S.D.B., D.C., and G.G.G. edited and revised the manuscript; A.K.D., A.G.S., S.D.B., and G.G.G. approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank the staff in the Biosciences Aquatic Facilities, the Molecular Biology Service Unit, and the Cross Cancer Institute Cell Imaging Facility at the University of Alberta for outstanding help throughout the study. We also thank Alex Clifford for advice and help with phylogenetic analysis and James Ede for help with confocal microscopy.
REFERENCES
- 1.Avella M, Bornancin M. A new analysis of ammonia and sodium transport through the gills of the freshwater rainbow trout (Salmo gairdneri). J Exp Biol 142: 155–175, 1989 [Google Scholar]
- 2.Boisen AM, Amstrup J, Novak I, Grosell M. Sodium and chloride transport in soft water and hard water acclimatized zebrafish (Danio rerio). Biochim Biophys Acta 1618: 207–218, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Brix KV, Grosell M. Comparative characterization of Na+ transport in Cyprinodon variegatus variegatus and Cyprinodon variegatus hubbsi: a model species complex for studying teleost invasion of freshwater. J Exp Biol 215: 1199–1209, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Polleichtner G, Kadurin I, Grunder S. Zebrafish acid-sensing ion channel (ASIC). 4. Characterization of homo- and heteromeric channels and identification of regions important for activation by H+. J Biol Chem 282: 30406–30413, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Qiu L, Li M, Durrnagel S, Orser BA, Xiong ZG, MacDonald JF. Diarylamidines: high potency inhibitors of acid-sensing ion channels. Neuropharmacology 58: 1045–1053, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dymowska AK, Hwang PP, Goss GG. Structure and function of ionocytes in the freshwater fish gill. Respir Physiol Neurobiol 184: 282–292, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Edwards SL, Tse CM, Toop T. Immunolocalization of NHE3-like immunoreactivity in the gills of the rainbow trout (Oncorhynchus mykiss) and the blue-throated wrasse (Pseudolabrus tetrious). J Anat 195: 465–469, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans DH, Piermarini PM, Choe KP. The multifunctional gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85: 97–177, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Fenwick JC, Bonga SE, Flik G. In vivo bafilomycin-sensitive Na+ uptake in young freshwater fish. J Exp Biol 202: 3659–3666, 1999 [DOI] [PubMed] [Google Scholar]
- 10.Galvez F, Reid SD, Hawkings G, Goss GG. Isolation and characterization of mitochondria-rich cell types from the gill of freshwater rainbow trout. Am J Physiol Regul Integr Comp Physiol 282: R658–R668, 2002 [DOI] [PubMed] [Google Scholar]
- 11.Goss G, Orlowski J, Grinstein S. Coimmunoprecipitation of a 24-kDa protein with NHE1, the ubiquitous isoform of the Na+/H+ exchanger. Am J Physiol Cell Physiol 270: C1493–C1502, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Goss GG, Wood CJ. Na+ and Cl− uptake kinetics, diffusive effluxes and acidic equivalent fluxes across the gills of rainbow trout. 1. Responses to environmental hyperoxia. J Exp Biol 152: 521–547, 1990 [Google Scholar]
- 13.Grunder S, Geissler HS, Bassler EL, Ruppersberg JP. A new member of acid-sensing ion channels from pituitary gland. Neuroreport 11: 1607–1611, 2000 [DOI] [PubMed] [Google Scholar]
- 14.Harvey BJ. Energization of sodium absorption by the H+-ATPase pump in mitochondria-rich cells of frog skin. J Exp Biol 172: 289–309, 1992 [DOI] [PubMed] [Google Scholar]
- 15.Hirata T, Kaneko T, Ono T, Nakazato T, Furukawa N, Hasegawa S, Wakabayashi S, Shigekawa M, Chang MH, Romero MF, Hirose S. Mechanism of acid adaptation of a fish living in low pH 3.5 lake. Am J Physiol Regul Integr Comp Physiol 284: R1199–R1212, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Holzer P. Acid-sensitive ion channels and receptors. Hand Exp Pharmacol 194: 283–332, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hung CY, Tsui KN, Wilson JM, Nawata CM, Wood CM, Wright PA. Rhesus glycoprotein gene expression in the mangrove killifish Kryptolebias marmoratus exposed to elevated environmental ammonia levels and air. J Exp Biol 210: 2419–2429, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Hwang PP, Lee TH, Lin LY. Ion regulations in fish gills: recent progresses in the cellular and molecular mechanisms. Am J Physiol Regul Integr Comp Physiol 301: R28–R47, 2011 [DOI] [PubMed] [Google Scholar]
- 19.Ivanis G, Esbaugh AJ, Perry SF. Branchial expression and localization of SLC9A2 and SLC9A3 sodium/hydrogen exchangers and their possible role in acid-base regulation in freshwater rainbow trout (Oncorhynchus mykiss). J Exp Biol 211: 2467–2477, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9A resolution and low pH. Nature 449: 316–323, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Jonz MG, Nurse CA. Epithelial mitochondria-rich cells and associated innervation in adult and developing zebrafish. J Comp Neurol 497: 817–832, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Jonz MG, Nurse CA. New developments on gill innervation: insights from a model vertebrate. J Exp Biol 211: 2371–2378, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Kellenberger S. Epithelial sodium and acid-sensing ion channels. In: Sensing With Ion Channels, edited by Martinac B. New York: Springer, 2008, chapt. 11, p. 225–246 [Google Scholar]
- 24.Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev 82: 735–767, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Kleyman TR, Cragoe EJ. Amiloride and its analogs as tools in the study of ion-transport. J Membr Biol 105: 1–21, 1988 [DOI] [PubMed] [Google Scholar]
- 26.Krogh A. The active absorption of ions in some freshwater animals. Z Vergl Physiol 25: 335–350, 1938 [Google Scholar]
- 27.Levanti MB, Guerrera MC, Calavia MG, Ciriaco E, Montalbano G, Cobo J, Germana A, Vega JA. Acid-sensing ion channel 2 (ASIC2) in the intestine of adult zebrafish. Neurosci Lett 494: 24–28, 2011 [DOI] [PubMed] [Google Scholar]
- 28.Lin H, Randall DJ. Evidence for the presence of an electrogenic proton pump on the trout gill epithelium. J Exp Biol 161: 119–134, 1991 [Google Scholar]
- 29.Maetz J, Garcia Romeu F. The mechanism of sodium and chloride uptake by the gills of a fresh-water fish, Carassius auratus. II. Evidence for NH4+/Na+ and HCO3−/Cl− exchanges. J Gen Physiol 47: 1209–1227, 1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakada T, Hoshijima K, Esaki M, Nagayoshi S, Kawakami K, Hirose S. Localization of ammonia transporter Rhcg1 in mitochondrion rich cells of yolk sac, gill and kidney of zebrafish and its ionic strength-dependent expression. Am J Physiol Regul Integr Comp Physiol 293: R1743–R1753, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Nawata CM, Hung CC, Tsui TK, Wilson JM, Wright PA, Wood CM. Ammonia excretion in rainbow trout (Oncorhynchus mykiss): evidence for Rh glycoprotein and H+-ATPase involvement. Physiol Genomics 31: 463–474, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Parks SK, Tresguerres M, Galvez F, Goss GG. Intracellular pH regulation in isolated trout gill mitochondrion-rich (MR) cell subtypes: evidence for Na+/H+ activity. Comp Biochem Physiol A 155: 139–145, 2010 [DOI] [PubMed] [Google Scholar]
- 33.Parks SK, Tresguerres M, Goss GG. Interactions between Na+ channels and Na+-HCO3− cotransporters in the freshwater fish gill MRC: a model for transepithelial Na+ uptake. Am J Physiol Cell Physiol 292: C935–C944, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Parks SK, Tresguerres M, Goss GG. Theoretical considerations underlying Na+ uptake mechanisms in freshwater fishes. Comp Biochem Physiol C 148: 411–418, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Paukert M, Sidi S, Russell C, Siba M, Wilson SW. A family of acid-sensing ion channels from zebrafish: widespread expression in the central nervous system suggests a conserved role in neuronal communication. J Biol Chem 279: 18783–18791, 2004 [DOI] [PubMed] [Google Scholar]
- 36.Perry SF. The chloride cell: structure and function in the gill of freshwater fish. Annu Rev Physiol 59: 325–347, 1997 [DOI] [PubMed] [Google Scholar]
- 37.Perry SF, Randall DJ. Effects of amiloride and SITS on branchial ion fluxes in rainbow trout, Salmo gairdneri. J Exp Zool 215: 225–228, 1981 [DOI] [PubMed] [Google Scholar]
- 38.Preest MR, Gonzales RJ, Wilson RW. Pharmacological examination of Na+ and Cl− transport in two species of freshwater fish. Physiol Biochem Zool 78: 259–272, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Reid SD, Hawkings GS, Galvez F, Goss GG. Localization and characterization of phenamil-sensitive Na+ influx in isolated rainbow trout gill epithelial cells. J Exp Biol 206: 551–559, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Sakai H, Lingueglia E, Champigny G, Mattei MG, Lazdunski M. Cloning and functional expression of a novel degenerin-like Na+ channel gene in mammals. J Physiol 519: 323–333, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shih TH, Horng JL, Liu ST, Hwang PP, Lin LY. Rhcg1 and NHE3b are involved in ammonium-dependent sodium uptake by zebrafish larvae acclimated to low-sodium water. Am J Physiol Regul Integr Comp Physiol 302: R84–R93, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Springauf A, Grunder S. An acid-sensing ion channel from shark (Squalus acanthias) mediates transient and sustained responses to protons. J Physiol 588: 809–820, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spry DJ, Wood CM. Ion flux rates, acid-base status, and blood gases in rainbow trout, Salmo gairdneri, exposed to toxic zinc in natural soft water. Can J Fish Aquat Sci 42: 1332–1337, 1985 [Google Scholar]
- 44.Sullivan GV, Fryer JN, Perry SF. Immunolocalization of proton pumps (H+-ATPase) in pavement cells of rainbow trout gill. J Exp Biol 198: 2619–2629, 1996 [DOI] [PubMed] [Google Scholar]
- 45.Tresguerres M, Parks SK, Katoh F, Goss GG. Microtubule-dependent relocation of branchial V-H+-ATPase to the basolateral membrane in the Pacific spiny dogfish (Squalus acanthias): a role in base secretion. J Exp Biol 209: 599–609, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Vina E, Parisi V, Cabo R, Laura R, Lopez-Velasco S, Lopez-Muniz A, Garcia-Suarez O, Germana A, Vega JA. Acid-sensing ion channels (ASICs) in the taste buds of adult zebrafish. Neurosci Lett 536: 35–40, 2013 [DOI] [PubMed] [Google Scholar]
- 47.Waldmann R, Lazdunski M. H+-gated cation channels: neuronal acid sensors in the NaC/DEG family of ion channels. Curr Opin Neurobiol 8: 418–424, 1998 [DOI] [PubMed] [Google Scholar]
- 48.Wilson JM, Laurent P, Tufts BL, Benos DJ, Donowitz M, Vogl AW, Randall DJ. NaCl uptake by the branchial epithelium in freshwater teleost fish: an immunological approach to ion-transport protein localization. J Exp Biol 203: 2279–2296, 2000 [DOI] [PubMed] [Google Scholar]
- 49.Wood CM, Randall DJ. Sodium balance in rainbow-trout (Salmo-gairdneri) during extended exercise. J Comp Physiol 82: 235–256, 1973 [Google Scholar]
- 50.Wright PA, Wood CM. A new paradigm for ammonia excretion in aquatic animals: role of rhesus (Rh) glycoproteins. J Exp Biol 212: 2303–2312, 2009 [DOI] [PubMed] [Google Scholar]
- 51.Yan JJ, Chou MY, Kaneko T, Hwang PP. Gene expression of Na+/H+ exchanger in zebrafish H+-ATPase-rich cells during acclimation to low-Na+ and acidic environments. Am J Physiol Cell Physiol 293: C1814–C1823, 2007 [DOI] [PubMed] [Google Scholar]