Abstract
Using partial amino acid sequence data derived from porcine methionyl aminopeptidase (MetAP; methionine aminopeptidase, peptidase M; EC 3.4.11.18), a full-length clone of the homologous human enzyme has been obtained. The cDNA sequence contains 2569 nt with a single open reading frame corresponding to a protein of 478 amino acids. The C-terminal portion representing the catalytic domain shows limited identity with MetAP sequences from various prokaryotes and yeast, while the N terminus is rich in charged amino acids, including extended strings of basic and acidic residues. These highly polar stretches likely result in the spuriously high observed molecular mass (67 kDa). This cDNA sequence is highly similar to a rat protein, termed p67, which was identified as an inhibitor of phosphorylation of initiation factor eIF2 alpha and was previously predicted to be a metallopeptidase based on limited sequence homology. Model building established that human MetAP (p67) could be readily accommodated into the Escherichia coli MetAP structure and that the Co2+ ligands were fully preserved. However, human MetAP was found to be much more similar to a yeast open reading frame that differed markedly from the previously reported yeast MetAP. A similar partial sequence from Methanothermus fervidus suggests that this p67-like sequence is also found in prokaryotes. These findings suggest that there are two cobalt-dependent MetAP families, presently composed of the prokaryote and yeast sequences (and represented by the E. coli structure) (type I), on the one hand, and by human MetAP, the yeast open reading frame, and the partial prokaryotic sequence (type II), on the other.
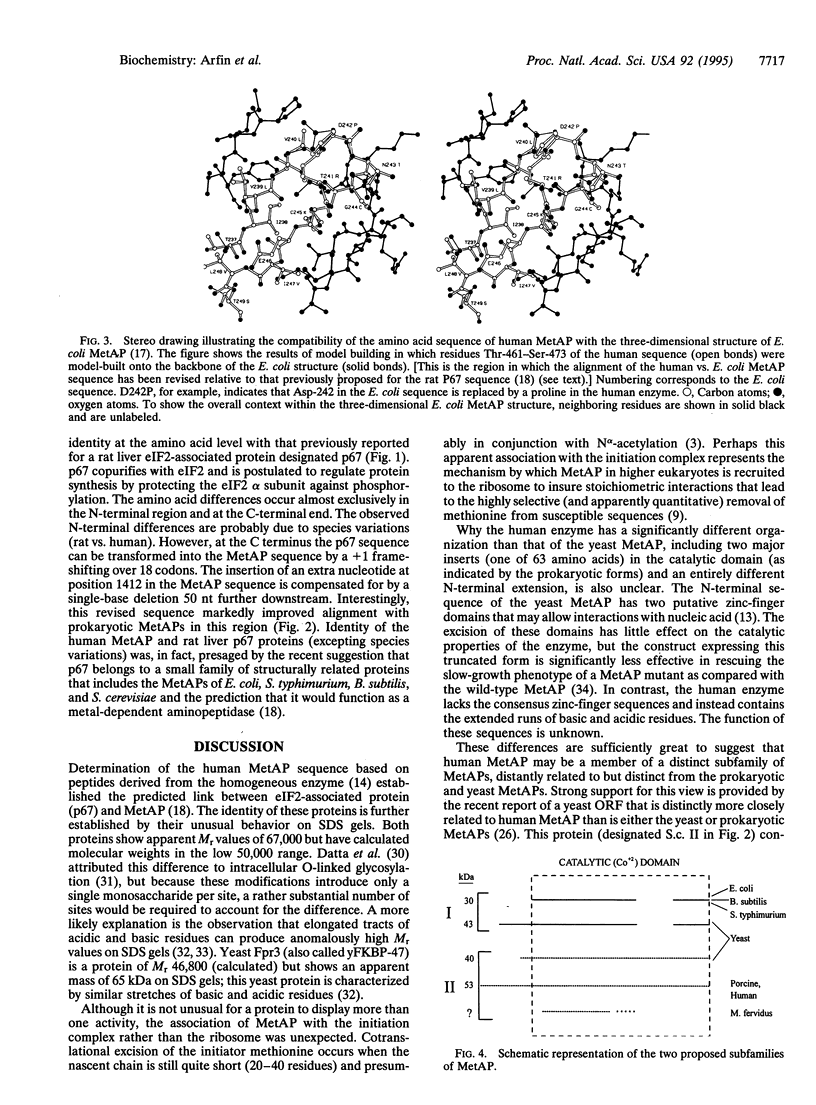
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arfin S. M., Bradshaw R. A. Cotranslational processing and protein turnover in eukaryotic cells. Biochemistry. 1988 Oct 18;27(21):7979–7984. doi: 10.1021/bi00421a001. [DOI] [PubMed] [Google Scholar]
- Bazan J. F., Weaver L. H., Roderick S. L., Huber R., Matthews B. W. Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2473–2477. doi: 10.1073/pnas.91.7.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Bassat A., Bauer K., Chang S. Y., Myambo K., Boosman A., Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J Bacteriol. 1987 Feb;169(2):751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton B. M., Zang J. H., Thorner J. A novel FK506- and rapamycin-binding protein (FPR3 gene product) in the yeast Saccharomyces cerevisiae is a proline rotamase localized to the nucleolus. J Cell Biol. 1994 Nov;127(3):623–639. doi: 10.1083/jcb.127.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissel J. P., Kasper T. J., Bunn H. F. Cotranslational amino-terminal processing of cytosolic proteins. Cell-free expression of site-directed mutants of human hemoglobin. J Biol Chem. 1988 Jun 15;263(17):8443–8449. [PubMed] [Google Scholar]
- Boissel J. P., Kasper T. J., Shah S. C., Malone J. I., Bunn H. F. Amino-terminal processing of proteins: hemoglobin South Florida, a variant with retention of initiator methionine and N alpha-acetylation. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8448–8452. doi: 10.1073/pnas.82.24.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. L., Roberts W. K. Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J Biol Chem. 1976 Feb 25;251(4):1009–1014. [PubMed] [Google Scholar]
- Butler M. J., Bergeron A., Soostmeyer G., Zimny T., Malek L. T. Cloning and characterisation of an aminopeptidase P-encoding gene from Streptomyces lividans. Gene. 1993 Jan 15;123(1):115–119. doi: 10.1016/0378-1119(93)90549-i. [DOI] [PubMed] [Google Scholar]
- Chakraborty A., Saha D., Bose A., Chatterjee M., Gupta N. K. Regulation of eIF-2 alpha-subunit phosphorylation in reticulocyte lysate. Biochemistry. 1994 May 31;33(21):6700–6706. doi: 10.1021/bi00187a041. [DOI] [PubMed] [Google Scholar]
- Chang Y. H., Teichert U., Smith J. A. Molecular cloning, sequencing, deletion, and overexpression of a methionine aminopeptidase gene from Saccharomyces cerevisiae. J Biol Chem. 1992 Apr 25;267(12):8007–8011. [PubMed] [Google Scholar]
- Chang Y. H., Teichert U., Smith J. A. Purification and characterization of a methionine aminopeptidase from Saccharomyces cerevisiae. J Biol Chem. 1990 Nov 15;265(32):19892–19897. [PubMed] [Google Scholar]
- Datta B., Ray M. K., Chakrabarti D., Wylie D. E., Gupta N. K. Glycosylation of eukaryotic peptide chain initiation factor 2 (eIF-2)-associated 67-kDa polypeptide (p67) and its possible role in the inhibition of eIF-2 kinase-catalyzed phosphorylation of the eIF-2 alpha-subunit. J Biol Chem. 1989 Dec 5;264(34):20620–20624. [PubMed] [Google Scholar]
- Driessen H. P., de Jong W. W., Tesser G. I., Bloemendal H. The mechanism of N-terminal acetylation of proteins. CRC Crit Rev Biochem. 1985;18(4):281–325. doi: 10.3109/10409238509086784. [DOI] [PubMed] [Google Scholar]
- Endo F., Tanoue A., Nakai H., Hata A., Indo Y., Titani K., Matsuda I. Primary structure and gene localization of human prolidase. J Biol Chem. 1989 Mar 15;264(8):4476–4481. [PubMed] [Google Scholar]
- Feldmann H., Aigle M., Aljinovic G., André B., Baclet M. C., Barthe C., Baur A., Bécam A. M., Biteau N., Boles E. Complete DNA sequence of yeast chromosome II. EMBO J. 1994 Dec 15;13(24):5795–5809. doi: 10.1002/j.1460-2075.1994.tb06923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller R. S., Brake A., Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1434–1438. doi: 10.1073/pnas.86.5.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart G. W. Glycosylation. Curr Opin Cell Biol. 1992 Dec;4(6):1017–1023. doi: 10.1016/0955-0674(92)90134-x. [DOI] [PubMed] [Google Scholar]
- Henrich B., Monnerjahn U., Plapp R. Peptidase D gene (pepD) of Escherichia coli K-12: nucleotide sequence, transcript mapping, and comparison with other peptidase genes. J Bacteriol. 1990 Aug;172(8):4641–4651. doi: 10.1128/jb.172.8.4641-4651.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffken H. W., Knof S. H., Bartlett P. A., Huber R., Moellering H., Schumacher G. Crystal structure determination, refinement and molecular model of creatine amidinohydrolase from Pseudomonas putida. J Mol Biol. 1988 Nov 20;204(2):417–433. doi: 10.1016/0022-2836(88)90586-4. [DOI] [PubMed] [Google Scholar]
- Huang S., Elliott R. C., Liu P. S., Koduri R. K., Weickmann J. L., Lee J. H., Blair L. C., Ghosh-Dastidar P., Bradshaw R. A., Bryan K. M. Specificity of cotranslational amino-terminal processing of proteins in yeast. Biochemistry. 1987 Dec 15;26(25):8242–8246. doi: 10.1021/bi00399a033. [DOI] [PubMed] [Google Scholar]
- Kendall R. L., Bradshaw R. A. Isolation and characterization of the methionine aminopeptidase from porcine liver responsible for the co-translational processing of proteins. J Biol Chem. 1992 Oct 15;267(29):20667–20673. [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991 Oct 25;266(30):19867–19870. [PubMed] [Google Scholar]
- Miller C. G., Strauch K. L., Kukral A. M., Miller J. L., Wingfield P. T., Mazzei G. J., Werlen R. C., Graber P., Movva N. R. N-terminal methionine-specific peptidase in Salmonella typhimurium. Proc Natl Acad Sci U S A. 1987 May;84(9):2718–2722. doi: 10.1073/pnas.84.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerschell R. P., Hosokawa Y., Tsunasawa S., Sherman F. The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. Processing of altered iso-1-cytochromes c created by oligonucleotide transformation. J Biol Chem. 1990 Nov 15;265(32):19638–19643. [PubMed] [Google Scholar]
- Nakamura K., Nakamura A., Takamatsu H., Yoshikawa H., Yamane K. Cloning and characterization of a Bacillus subtilis gene homologous to E. coli secY. J Biochem. 1990 Apr;107(4):603–607. doi: 10.1093/oxfordjournals.jbchem.a123093. [DOI] [PubMed] [Google Scholar]
- Roderick S. L., Matthews B. W. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli: a new type of proteolytic enzyme. Biochemistry. 1993 Apr 20;32(15):3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Tsunasawa S. Methionine or not methionine at the beginning of a protein. Bioessays. 1985 Jul;3(1):27–31. doi: 10.1002/bies.950030108. [DOI] [PubMed] [Google Scholar]
- Tsunasawa S., Stewart J. W., Sherman F. Amino-terminal processing of mutant forms of yeast iso-1-cytochrome c. The specificities of methionine aminopeptidase and acetyltransferase. J Biol Chem. 1985 May 10;260(9):5382–5391. [PubMed] [Google Scholar]
- Wu S., Gupta S., Chatterjee N., Hileman R. E., Kinzy T. G., Denslow N. D., Merrick W. C., Chakrabarti D., Osterman J. C., Gupta N. K. Cloning and characterization of complementary DNA encoding the eukaryotic initiation factor 2-associated 67-kDa protein (p67). J Biol Chem. 1993 May 25;268(15):10796–10801. [PubMed] [Google Scholar]
- Yoshimoto T., Tone H., Honda T., Osatomi K., Kobayashi R., Tsuru D. Sequencing and high expression of aminopeptidase P gene from Escherichia coli HB101. J Biochem. 1989 Mar;105(3):412–416. doi: 10.1093/oxfordjournals.jbchem.a122678. [DOI] [PubMed] [Google Scholar]
- Zuo S., Guo Q., Ling C., Chang Y. H. Evidence that two zinc fingers in the methionine aminopeptidase from Saccharomyces cerevisiae are important for normal growth. Mol Gen Genet. 1995 Jan 20;246(2):247–253. doi: 10.1007/BF00294688. [DOI] [PubMed] [Google Scholar]



