Abstract
Tributyltin (TBT) is known to cause developmental defects as endocrine disruptive chemicals (EDCs). At nanomoler concentrations, TBT actions were mediated by genomic pathways via PPAR/RXR. However, non-genomic target of TBT has not been elucidated. To investigate non-genomic TBT targets, we performed comprehensive metabolomic analyses using human embryonic carcinoma NT2/D1 cells. We found that 100 nM TBT reduced the amounts of α-ketoglutarate, succinate and malate. We further found that TBT decreased the activity of NAD-dependent isocitrate dehydrogenase (NAD-IDH), which catalyzes the conversion of isocitrate to α-ketoglutarate in the TCA cycle. In addition, TBT inhibited cell growth and enhanced neuronal differentiation through NAD-IDH inhibition. Furthermore, studies using bacterially expressed human NAD-IDH and in silico simulations suggest that TBT inhibits NAD-IDH due to a possible interaction. These results suggest that NAD-IDH is a novel non-genomic target of TBT at nanomolar levels. Thus, a metabolomic approach may provide new insights into the mechanism of EDC action.
Endocrine disruptive compounds (EDCs) have been studied extensively in environmental biology1. A large number of EDCs are known to cause genomic action via nuclear receptor. For example, xenoestrogens such as bisphenol A, genistein and diethylstilbestrol can bind to the estrogen receptor (ER) in the cell nucleus, followed by the alteration of gene expression2,3. In addition, EDCs induce the activation of non-genomic signaling pathways. For example, xenoestrogens increase intracellular calcium levels, activating eNOS and signaling cascades such as PI3K/AKT and MAPK4,5,6,7. Thus, both genomic and non-genomic pathways are required to understand the mechanism of EDC action.
Organotin compounds, such as tributyltin (TBT) are typical environmental contaminants and well known to cause developmental defects as EDCs. For example, TBT can cause increased fetal mortality, decreased fetal birth weights, and behavioral abnormalities in rat offspring8,9. Although the use of TBT has already been restricted, butyltin compounds, including TBT, can still be found in human blood at concentrations between 50 and 400 nM10. Several studies revealed that TBT activates retinoid X receptor (RXR) and/or peroxisome proliferator-activated receptor γ (PPARγ). These genomic transcriptional activations result in developmental effects, such as the imposex in many marine species11,12,13 and the enhancement of adipocyte differentiation in mammals14,15. These TBT actions involve a higher binding affinity compared to intrinsic ligands at nM concentrations. In addition to the genomic effects, non-genomic action of TBT has been also reported. For example, TBT has been reported to inhibit the steroid biosynthesis pathway, which is responsible for the production of estrogen and androgen16,17,18. Another report has shown that TBT inhibits mitochondrial F1F0 ATP synthase19. These data were obtained at μM concentrations. Thus, the mechanism of nM concentrations of TBT has not been elucidated at a non-genomic level. In a previous study, we reported that treatment with 100 nM TBT resulted in growth arrest by targeting the glycolytic systems of the human embryonic carcinoma cell line NT2/D120. Therefore, we raised the possibility that nM concentrations of TBT may target other non-genomic pathways which are involved in energy metabolism.
In the present study, we investigated the molecular target of TBT at nM levels by comprehensive determination of the intracellular metabolites in NT2/D1 cells after TBT exposure. We found that exposure to 100 nM TBT reduced ATP production via NAD-dependent isocitrate dehydrogenase (NAD-IDH) in the cells. This NAD-IDH inhibition resulted in the reduction of the TCA cycle metabolites. In addition, TBT caused neural differentiation through an NAD-IDH-dependent mechanism. We report here that our metabolomic analysis revealed that NAD-IDH is a novel target of TBT in embryonic carcinoma cells.
Results
Metabolomic analysis of NT2/D1 cells exposed to TBT at nM levels
To investigate the non-genomic effects of a well-known endocrine disruptor TBT in human NT2/D1 embryonic carcinoma cells, we comprehensively determined intracellular metabolites using LC/MS. We found that exposure to 100 nM TBT reduced the amounts of TCA cycle components, such as α-ketoglutarate, succinate and malate (Figure 1a). The amounts of acetyl CoA and isocitrate were not changed. We also found that treatment with 100 nM TBT reduced the ATP content of the cells (Figure 1b). In contrast to TBT, exposure to the less toxic tin acetate (TA) did not affect the amount of each metabolite. These data suggest that TBT exposure decreases the amounts of TCA cycle metabolites, resulting in a reduction of ATP content.
Figure 1. Metabolomic analysis of NT2/D1 cells exposed to TBT.
The cells were exposed to 100 nM TBT or TA for 24 h. (a) The levels of several metabolites, such as acetyl CoA, isocitrate, α-ketoglutarate, succinate and malate, were determined using CE-TOFMS. (b) The intracellular ATP content was determined in the lysed cells. * P < 0.05 compared with the corresponding control group.
NAD-IDH enzyme activity of NT2/D1 cells exposed to TBT at nM levels
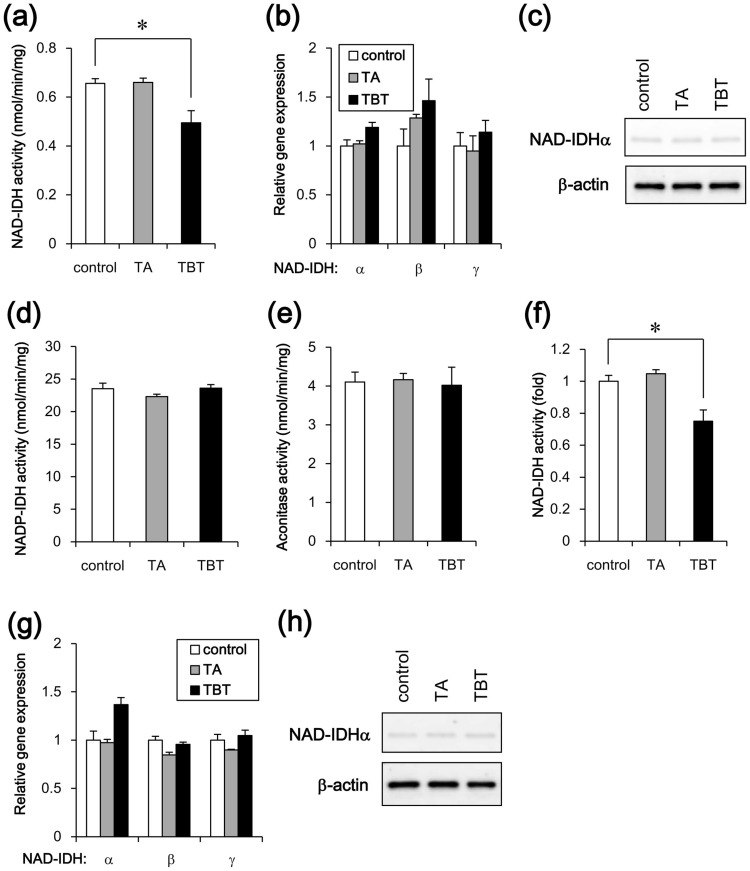
Based on the results of the metabolomic analysis, we focused on isocitrate dehydrogenase, which catalyzes the conversion of isocitrate to α-ketoglutarate in the TCA cycle. Eukaryotes have different types of isocitrate dehydrogenases, such as NAD-dependent form (NAD-IDH; EC 1.1.1.41) and NADP-dependent form (NADP-IDH; EC 1.1.1.42)21. NAD-IDH is first rate-limiting enzyme in the TCA cycle and catalyzes an irreversible reaction, while NADP-IDH is involved in reversible reaction for biosynthesis via production of NADPH. As shown in Figure 2a, NAD-IDH activity was significantly reduced following TBT treatment. Since NAD-IDH is a heterotetramer composed of two α subunits (catalytic subunit), one β subunit and one γ subunit (regulatory subunit), we examined the expression of each subunit gene. Real-time PCR analysis showed that the expression of the NAD-IDHα, β and γ genes was not significantly changed by TBT exposure (Figure 2b). The protein expression of catalytic α subunits was not also changed by TBT exposure (Figure 2c). TA exposure did not affect either the enzyme activity or the NAD-IDH expression. We next examined the effect of TBT on NADP-IDH. The activity of NADP-IDH was not affected by TBT exposure (Figure 2d). We further examined the activity of aconitase (EC 4.2.1.3.), which catalyzes the conversion of citrate to isocitrate in the TCA cycle. Aconitase activity was also not affected by TBT exposure (Figure 2e). Thus, these data suggest that the inhibitory effect of TBT is specific to NAD-IDH in the TCA cycle.
Figure 2. Effect of TBT exposure on NAD-IDH enzyme activity in NT2/D1 cells.
The cells were exposed to 100 nM TBT or TA for 24 h. (a) NAD-IDH activity was determined in the lysed cells. (b) The relative expressions of NAD-IDHα, β, and γ were measured using real-time PCR. The relative changes were normalized to the levels of RPL13. (c) The expression of NAD-IDHα protein was examined by western blot analysis using the anti-NAD-IDHα and anti-β-actin antibodies. Cropped blots were shown and the full-length blots were indicated in Supplementary Fig. 4. (d) NADP-IDH activity was determined in the lysed cells. (e) Aconitase activity was determined in the lysed cells. (f) To induce neuronal differentiation, the NT2/D1 cells were treated with 10 μM retinoic acid (RA) for 14 days. After exposure to 100 nM TBT for 24 h, NAD-IDH activity was determined in the lysed cells. (g) The relative expression levels of NAD-IDHα, β, and γ in the differentiated cells were measured using real-time PCR. The relative changes were normalized to the levels of RPL13. (h) The expression of NAD-IDHα protein was examined by western blot analysis using the anti-NAD-IDHα and anti-β-actin antibodies. Cropped blots were shown and the full-length blots were indicated in Supplementary Fig. 4. * P < 0.05 compared with the corresponding control group.
To investigate whether TBT cytotoxicity was caused by a genomic transcriptional regulation, we tested the effects of the protein synthesis inhibitor cycloheximide in NT2/D1 cells. Treatment with cycloheximide did not alter the inhibitory effects of TBT on NAD-IDH activity (Figure S1a) and intracellular ATP production (Figure S2a). Moreover, the PPARγ agonist rosiglitazone did not reduce NAD-IDH activity (Figure S1b) and ATP content (Figure S2b). These results suggest that transcriptional regulation is not involved in the inhibition of NAD-IDH activity by TBT.
To examine whether the effect of TBT was selective for embryonic cells, we used NT2/D1 cells that had differentiated in response to retinoic acid. We observed that TBT also inhibited NAD-IDH activity in the differentiated NT2/D1 cells (Figure 2f). Real-time PCR analysis showed that the expression of the NAD-IDHα, β, and γ genes was not significantly affected by TBT exposure (Figure 2g). The protein expression of catalytic α subunits was not also changed by TBT exposure (Figure 2h). TA exposure did not affect either the activity or the NAD-IDH expression. These data suggest that TBT reduces NAD-IDH enzyme activity regardless of the developmental stage of the embryonic carcinoma cells.
Neuronal differentiation of RA-treated NT2/D1 cells exposed to TBT at nM levels
It has been reported that ATP content decreases during the differentiation of human embryonic stem cells into neural stem cells (NSCs)22. Therefore, the reduction of ATP caused by TBT treatment might be involved in neuronal differentiation. Moreover, TBT has been reported to cause cell growth arrest in NT2/D1 cells20. Because cell growth is generally reduced during differentiation, we examined whether TBT affects the neuronal differentiation process in NT2/D1 cells. Real-time PCR analysis revealed that retinoic acid (RA)-treated NT2/D1 cells showed increased expression of the differentiation markers NeuroD and Math1, confirming that neural differentiation had occurred (Figure 3a). Furthermore, we observed that TBT exposure enhanced the expression levels of these neuronal differentiation markers. Treatment with rosiglitazone had little effect on their expression (Figure S3), suggesting that PPARγ is not involved in neuronal differentiation. Taken together, these data suggest that TBT promotes neuronal differentiation.
Figure 3. Effect of TBT exposure or NAD-IDH knockdown on neuronal differentiation in RA-treated NT2/D1 cells.
(a) After treatment with RA for 7 days, the cells were treated with RA in the presence of 100 nM TBT for an additional 7 days. The relative expression levels of the neuronal markers NeuroD and Math1 were measured using real-time PCR. The relative changes were determined following normalization to the levels of RPL13. (b–e) The cells were infected with lentiviruses containing a vector encoding a shRNA directed against NAD-IDHα or a scrambled sequence shRNA (control). The infected cells were subjected to selection with 0.5 μg/ml puromycin for 72 h and were then exposed to TBT at 100 nM for 24 h. (b) The relative expression of NAD-IDHα was measured using real-time PCR. The relative change in expression was normalized to the levels of RPL13. (c) NAD-IDH activity was determined in the lysed cells. (d) The intracellular ATP content was determined in the lysed cells. (e) Infected cells were seeded into 100 mm dishes and cultured for 24 h. Cell count was determined using a hemocytometer. (f) After treatment with RA for 7 days, followed by treatment with RA and the shRNA-containing lentiviruses for an additional 7 days, the relative expression levels of the neuronal markers NeuroD and Math1 were measured using real-time PCR. The relative changes were determined following normalization to the levels of RPL13. * P < 0.05 compared with the corresponding group.
Effect of NAD-IDH knockdown on neuronal differentiation in RA-treated NT2/D1 cells
To further investigate whether the neuronal differentiation triggered by TBT exposure is through an NAD-IDH-dependent mechanism, we performed knockdown (KD) of NAD-IDHα, the catalytic subunit of NAD-IDH, using lentivirus-delivered shRNAs. Real-time PCR analysis showed that KD efficiency was approximately 40% (Figure 3b). Due to the partial KD of the NAD-IDHα gene, NAD-IDH activity decreased to a level (22%) comparable to its level following TBT inhibition (24%) (Figure 3c). Further reduction of NAD-IDH activity was not significantly observed after TBT exposure in the NAD-IDHα KD cells. Similar to the effect of TBT, NAD-IDHα KD also reduced the ATP content of the cells (Figure 3d), and caused cell growth inhibition (Figure 3e). Further inhibition of ATP content and cell growth was not significantly observed after TBT exposure in the NAD-IDHα KD cells, suggesting that the NAD-IDH is a possible target of TBT. In addition, we found that NAD-IDHα KD significantly upregulated the expression of the neuronal differentiation markers NeuroD and Math1 (Figure 3f). These data suggest that NAD-IDH mediates TBT-induced neuronal differentiation in embryonic NT2/D1 cells.
NAD-IDH enzyme activity in the brain of rats orally exposed to TBT at low doses
To examine whether the in vitro inhibitory effect of TBT on NAD-IDH is also observed in vivo, adult rats were orally exposed to TBT at doses of 5 and 50 mg/kg. NAD-IDH activity in the cerebral cortex was significantly reduced following exposure to both doses of TBT (Figure 4a). Real-time PCR analysis showed that the expression of the NAD-IDHα, β, and γ genes was not significantly affected by TBT (Figure 4b). The protein expression of catalytic α subunits was not also affected by TBT (Figure 4c). NADP-IDH and aconitase activities were not affected by exposure to either dose of TBT (Figure 4d and e). These data suggest that TBT inhibits NAD-IDH activity both in vitro and in vivo.
Figure 4. NAD-IDH enzyme activity in the brain of rats orally exposed to TBT at doses of 5 and 50 mg/kg for 6 h.
(a) NAD-IDH activity was determined in the brain lysates. (b) The relative expression levels of NAD-IDHα, β, and γ in rats exposed to 50 mg/kg TBT were measured using real-time PCR. The relative changes were normalized to the levels of RPL13. (c) The expression of NAD-IDHα protein was examined by western blot analysis using the anti-NAD-IDHα and anti-β-actin antibodies. Cropped blots were shown and the full-length blots were indicated in Supplementary Fig. 4. (d) NADP-IDH activity was determined in the brain lysates. (e) Aconitase activity was determined in the brain lysates. * P < 0.05 compared with the corresponding TBT 0 group.
Reduction of recombinant hNAD-IDH enzyme activity in E. coli lysate treated with TBT at nM levels
To investigate the mechanism by which TBT inhibits NAD-IDH activity, we examined whether TBT possibly interacts with NAD-IDH or not. Since NAD-IDHα subunit alone has been reported to show no detectable IDH activity, we used an Escherichia coli co-expression system of recombinant human (h) NAD-IDHα,β,γ subunits23. As shown in Figure 5a, we confirmed the expression of α subunit of hNAD-IDH protein in the extracts of E. coli transformants using western blot analysis. To check the activity of the recombinant hNAD-IDH, we used irreversible and allosteric NAD-IDH regulators. ADP has been reported to activate NAD-IDH allosterically by lowering the Km for the substrate isocitrate24. As expected, ADP increased the activity of hNAD-IDH in our assay system. Conversely, Zn2+ has been reported to inhibit several metabolic enzymes, including NAD-IDH, in hepatocytes25. We confirmed that Zn2+ reduces hNAD-IDH activity. Then, we examined whether TBT directly inhibits hNAD-IDH activity by adding TBT to the E. coli extracts containing hNAD-IDH. Treatment with 100 nM TBT for 1 h significantly reduced the hNAD-IDH activity (Figure 5b). Treatment with TA had little effect. Taken together, these data suggest that TBT inhibits hNAD-IDH activity through its possible interaction, but again we can not be sure that it is through direct binding with the data we have.
Figure 5. Reduction of NAD-IDH enzyme activity by interaction with TBT in an E. coli expression system.
(a) After the expression of pHIDHα, β, γ was induced by culturing E. coli BL21 transformants at 25°C for 20 h, crude extracts were prepared and were subjected to western blot analysis using the anti-NAD-IDHα and anti-GAPDH antibodies. Cropped blots were shown and the full-length blots were indicated in Supplementary Fig. 4. (b) After treatment with 100 nM TBT for 1 h, NAD-IDH activity was determined in the crude extracts. ADP (100 μM) and Zn2+ (2 mM) were used as positive and negative controls, respectively. * P < 0.05 compared with the corresponding control group.
In silico docking simulation analysis
To further consider this possible interaction between TBT and hNAD-IDH, we estimate TBT accessibility into hNAD-IDH (EC 1.1.1.41) α and hNADP-IDH (EC 1.1.1.42) homodimers by homology modeling and docking studies. We show the overlaid structure of the calculated hNAD-IDHα and hNADP-IDH (Figure 6a). The ligand binding pocket of hNAD-IDHα was larger than that of hNADP-IDH (Figure 6b). In our docking simulation, TBT was able to access the hNAD-IDHα ligand-binding pocket, whereas the hNADP-IDH pocket was not spacious enough to accommodate TBT (Figure 6c and d). Thus, these studies suggest that the selective inhibition of NAD-IDH by TBT may be due to differences in the pocket volumes between hNAD-IDHα and hNADP-IDH.
Figure 6. In silico docking simulation analysis.
(a) Overlaid structures of the calculated hNAD-IDHα (orange) and hNADP-IDH (cyan) homodimers bound to isocitric acid (Space-filling model). (b) The ligand binding pockets of the hNAD-IDHα (solid) and hNADP-IDH (wireframe) proteins. (c, d) Docking structure of hNAD-IDHα containing TBT (Space-filling model).
Discussion
In the present study, we demonstrate that NAD-IDH is a novel non-genomic target of TBT at nM levels both in vitro and in vivo. We showed that exposure to nM concentrations of TBT reduced the activity of NAD-IDH due to its possible interaction. We also found that TBT exposure caused both inhibition of cell growth and enhancement of neuronal differentiation through its inhibitory effect of NAD-IDH.
Our data suggest that NAD-IDH is a novel target molecule of TBT action. NAD-IDH is a NAD-dependent form of IDH found in NT2/D1 cells and the rat brain (Figure 2–4). Because NAD-IDH is ubiquitously expressed, the toxicity of TBT might be observed in various cell types. Several TCA cycle enzymes have been reported to contribute to cell proliferation. For example, NADP-IDH plays a role for cell growth under hypoxic conditions in human glioblastoma cells26. Another study has shown that aconitase mediated cell proliferation via ATP production in human prostate carcinoma cells27. NAD-IDH has been shown to regulate the metabolic fluxes and the generation of ATP in the TCA cycle28. Therefore, it is likely that NAD-IDH is a target of TBT cytotoxicity and regulates cell growth in embryonic carcinoma cells.
In addition to cell growth inhibition, we also showed that neural differentiation is enhanced by TBT exposure or NAD-IDH inhibition (Figure 3). Consistent with our data, overexpression of NAD-IDHα has been shown to reduce neuronal differentiation and neurite outgrowth through the inactivation of MAPK phosphorylation in PC12 cells29. Because TBT exposure has been reported to induce neurotoxicity via ERK and p38MAPK phosphorylation in cultured rat cortical neurons30, TBT exposure might cause cytotoxicity through the MAPK pathways. Thus, a non-genomic pathway plays a role in TBT toxicity. Indeed, the genomic target PPARγ and treatment with cycloheximide did not alter the effects of TBT (Figure S1–3). It is unlikely that transcriptional regulation is involved in NAD-IDH activity and the enhancement of neuronal differentiation. The downstream pathway of TBT-NAD-IDH should be determined in embryonic carcinoma cells.
Our data suggest that TBT regulates NAD-IDH activity through possible interaction. However, we can not conclude that it is through direct binding. Previous reports have suggested that TBT can bind to multiple target proteins, such as PPARγ, RXR, F1F0 ATP synthase and 11β-hydroxysteroid dehydrogenase (11β-HSD) type 2, with broad specificity19,31. For example, TBT binds the RXRα ligand-binding domain through a covalent bond between the tin atom and the Cys residue32. TBT also binds 11β-HSD type 2 by interacting with several Cys residues in the active site31. Because the ligand-binding pocket of hNAD-IDHα contains several Cys residues33 and has enough space to accommodate TBT, TBT might bind to hNAD-IDHα via Cys residues. Future conformational analysis, including X-ray crystallography or computer simulation, and mutagenesis studies should be performed to determine whether TBT binds to hNAD-IDHα or not.
Our metabolomic analysis showed that TBT inhibited cell growth and enhanced neuronal differentiation through possible direct inhibition of NAD-IDH activity in human embryonic carcinoma cells. Thus, comprehensive approach of non-genomic metabolic pathway might be a powerful tool to elucidate the mechanism of EDC action.
Methods
Chemicals and reagents
TBT was obtained from Tokyo Chemical Industry (Tokyo, Japan). Tin acetate (TA) and rosiglitazone were obtained from Sigma-Aldrich (St. Louis, MO, USA). All other reagents were of analytical grade and were obtained from commercial sources.
Cell culture
NT2/D1 cells were obtained from the American Type Culture Collection. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Ashrat, Israel) and 0.05 mg/ml penicillin-streptomycin mixture (Life Technologies, Carlsbad, CA, USA) at 37°C and 5% CO2. For neural differentiation, all-trans retinoic acid (RA; Sigma-Aldrich) was added to the medium twice a week at a final concentration of 10 μM.
Determination of TCA cycle metabolites
Intracellular metabolites were extracted and used for subsequent capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) analysis, as previously described. The amounts of the metabolites were determined using an Agilent CE capillary electrophoresis system (Agilent Technologies, Waldbronn, Germany) equipped with an Agilent G3250AA LC/MSD TOF system (Agilent Technologies, Palo Alto, CA), an Agilent 1100 series isocratic HPLC pump, a G1603A Agilent CE-MS adapter kit, and a G1607A Agilent CE-electrospray ionization 53-MS sprayer kit. For system control and data acquisition, the G2201AA Agilent ChemStation software was used for CE, and the Agilent TOF (Analyst QS) software was used for the TOFMS.
Measurement of intracellular ATP levels
The intracellular ATP content was measured using the ATP Determination Kit (Life Technologies), according to the manufacturer's protocol. Briefly, the cells were washed and lysed with 0.1% Triton X-100/PBS. The resulting cell lysates were added to a reaction mixture containing 0.5 mM D-luciferin, 1 mM DTT, and 1.25 μg/ml luciferase and incubated for 30 min at room temperature. Luminescence was measured using a Wallac1420ARVO fluoroscan (Perkin-Elmer, Waltham, MA, USA). The luminescence intensities were normalized to the total protein content.
Isocitrate dehydrogenase (IDH) activity assay
IDH activity was determined using the commercial Isocitrate Dehydrogenase Activity Colorimetric Assay Kit (Biovision, Mountain View, CA, USA), according to the manufacturer's instructions. Briefly, NT2/D1 cells were lysed in an assay buffer provided in the kit. The lysate was centrifuged at 14,000 g for 15 min, and the cleared supernatant was used for the assay. NADP or NAD was used as the substrate for the NADP-IDH or NAD-IDH assay, respectively.
Real-time PCR
Total RNA was isolated from NT2/D1 cells using the TRIzol reagent (Life Technologies), and quantitative real-time reverse transcription (RT)-PCR with the QuantiTect SYBR Green RT-PCR Kit (QIAGEN, Valencia, CA, USA) was performed using an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA, USA), as previously reported. The relative changes in the transcript amounts of each sample were normalized to the mRNA levels of ribosomal protein L13 (RPL13). The following primer sequences were used for real-time PCR analysis: human NAD-IDHα: forward, 5′-ATCGGAGGTCTCGGTGTG-3′, reverse, 5′-AGGAGGGCTGTGGGATTC-3′; human NAD-IDHβ: forward, 5′-GCCTCAGCCGCATATCATAG-3′, reverse, 5′-GAGCAGGTGCTGAGTTCCAT-3′; human NAD-IDHγ: forward, 5′-TTAGCGGACGGAGGAATTGT-3′, reverse, 5′-CAGCCCTTCTCTGCCGT-3′; human NeuroD: forward, 5′-GGAAACGAACCCACTGTGCT-3′, reverse, 5′-GCCACACCAAATTCGTGGTG-3′; human Math1: forward, 5′-GTCCGAGCTGCTACAAACG-3′, reverse, 5′-GTGGTGGTGGTCGCTTTT-3′; human RPL13: forward, 5′-CATCGTGGCTAAACAGGTACTG-3′, reverse, 5′-GCACGACCTTGAGGGCAGCC-3′; rat NAD-IDHα: forward, 5′-TGGGTGTCCAAGGTCTCTC-3′, reverse, 5′-CTCCCACTGAATAGGTGCTTTG-3′; rat NAD-IDHβ: forward, 5′-AGGCACAAGATGTGAGGGTG-3′, reverse, 5′-CAGCAGCCTTGAACACTTCC-3′; rat NAD-IDHγ: forward, 5′-TGGGCGGCATACAGTGACTA-3′, reverse, 5′-TTGGAGCTTACATGCACCTCT-3′; rat RPL13: forward, 5′-GGCTGAAGCCTACCAGAAAG-3′, reverse, 5′-CTTTGCCTTTTCCTTCCGTT-3′.
Aconitase activity assay
Aconitase activity was determined using the commercial Aconitase Activity Colorimetric Assay Kit (Biovision), according to the manufacturer's instructions. Briefly, NT2 cells were lysed in an assay buffer provided in the kit. The lysate was centrifuged at 14,000 g for 15 min, and the cleared supernatant was used for the aconitase assay.
NAD-IDHα knockdown experiment
Transient gene knockdown was performed using NAD-IDHα shRNA lentiviruses from Sigma-Aldrich (MISSION® shRNA) according to manufacturer's protocol. A scrambled hairpin sequence was used as a negative control. Briefly, the cells were infected with the viruses at a multiplicity of infection of 10, in the presence of 8 μg/ml hexadimethrine bromide (Sigma-Aldrich), for 24 h, and were then subjected to selection with 0.5 μg/ml puromycin for 72 h prior to functional analyses.
Tissue preparation
The present study was approved by the animal ethics committee and was conducted in accordance with the regulations on the use of living modified organisms of Hiroshima University. Male Slc:Wistar/ST rats (8 weeks old) were purchased from Japan SLC, Inc. (Shizuoka, Japan). They were housed under controlled temperature, 12 h light/dark cycle, and humidity (75 ± 5%) for at least 1 week prior to experiments. Standard pellet food and water were provided ad libitum. TBT solution (5 and 50 mg/kg body weight) was orally administered to rats. TBT was dissolved in polyethylene glycol. The whole brain was exposed by the use of fine scissors and forceps, and the frontal part of cerebral cortex was excised from the brain.
Transformation and Expression of Recombinant Human NAD-IDH Proteins in E. coli
pHIDHαβ1γ plasmid DNA (a kind gift from Dr. T. L. Huh) was used to transform E. coli BL21 (DE3) ultracompetent cells (BioDynamics, Tokyo, Japan). The colonies with positive inserts were subcultured and grown overnight at 37°C in LB medium (2 ml) supplemented with ampicillin (0.1 mg/ml). To express the enzyme, 15 ml tubes containing 2 ml of LB medium with 0.1 mg/ml ampicillin were inoculated with freshly grown E. coli cells (1% v/v), and these cultures were grown at 37°C while being shaken at 220 rpm for 4 h. The flasks were then temporarily placed in chilled water to lower the culture temperature to 25°C. Protein expression was induced in the cells by shaking at a lower speed of 140 rpm with minimal aeration at 25°C for 20 h. Then, 2 ml of the cell culture was centrifuged at 6,000 g for 15 min to separate the cells from the media, and the pellet was suspended in a total volume of 300 μl of cold assay buffer, a component of Isocitrate Dehydrogenase Activity Assay Kit. The suspended cells were subjected to 3 cycles of freeze-thaw before sonication (5 cycles of 15 sec sonication and 45 sec rest) and lysed. The cell lysate was centrifuged at 14,000 g for 15 min, and the cleared supernatant (crude extract) was separated. Recombinant human NAD-IDH activity was determined by subtracting basal activity in the crude extract from control BL21 cells.
Western blot analysis
Western blot analysis was performed as previously reported34. Briefly, the cells were lysed with Cell Lysis Buffer (Cell Signaling Technology, Danvers, MA, USA), and the proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and electrophoretically transferred to Immobilon-P (Millipore, Billerica, MA, USA). The membranes were probed with an anti-NAD-IDH (IDH3) α polyclonal antibody (1:1,000; Abcam, Cambridge, UK), an anti-β-actin monoclonal antibody (1:5,000; Sigma-Aldrich), and an anti-GAPDH monoclonal antibody (1:2,000; Abcam). The membranes were then incubated with secondary antibodies against rabbit or mouse IgG conjugated to horseradish peroxidase (Cell Signaling Technology). The bands were visualized using the ECL Western Blotting Analysis System (GE Healthcare, Buckinghamshire, UK), and the images were acquired using a LAS-3000 Imager (FUJIFILM UK Ltd., Systems, Bedford, UK). The density of each band was quantified using the ImageJ software (NIH, Bethesda, MD, USA).
In silico docking simulation studies
Homology modeling and docking studies of the human NAD-IDHα (hNAD-IDHα) and NADP-IDH (hNADP-IDH) homodimers were performed using Molecular Operating Environment (MOE) 2012.10. The models of hNAD-IDHα and hNADP-IDH were constructed based on the crystallographic structure of the porcine NADP-IDH homodimer (PDB: 1LWD)35 using the standard protocols in MOE 2012.10. The docking simulations of the TBT-bound hNAD-IDHα and hNADP-IDH were carried out using ASEDock36. The TBT ligand was assigned in ASEDock, and the conformations were calculated using MMFF94S force field37.
Statistical analysis
Results are shown as mean ± S.D. Statistical analysis was performed using one-way ANOVA followed by Dunnett's test. Differences at P < 0.05 were considered to be significant.
Author Contributions
S.Y. performed most of the experiments. Y.Ka. planned the project. S.Y., Y.S. and Y.Ka. wrote the manuscript. Y.Ko. performed the animal experiments. Y.D. and M.K. performed In silico docking simulation analysis. All authors reviewed the manuscript.
Supplementary Material
Supplementary figures
Acknowledgments
We thank Ms. Mami Kohno, Mr. Kyoichi Masuda, and Ms. Saki Tanaka (Graduate School of Biomedical and Health Sciences, Hiroshima University) for technical assistance in the animal experiment. This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (#23590322 to Y.K.), a Health and Labour Sciences Research Grant from the Ministry of Health, Labour and Welfare, Japan (Y.K.), and a grant from the Smoking Research Foundation (Y.K.).
References
- Toppari J. et al. Male reproductive health and environmental xenooestrogens. Environ. Health Perspect. 104, 741–803 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. M. & Korach K. S. Analysis of the molecular mechanisms of human estrogen receptors alpha and beta reveals differential specificity in target promoter regulation by xenoestrogens. J. Biol. Chem. 277, 44455–44461 (2002). [DOI] [PubMed] [Google Scholar]
- Newbold R. R., Padilla-Banks E. & Jefferson W. N. Adverse Effects of the Model Environmental Estrogen Diethylstilbestrol Are Transmitted to Subsequent Generations. Endocrinology 147, s11–17 (2006). [DOI] [PubMed] [Google Scholar]
- Bulayeva N. N., Gametchu B. & Watson C. S. Quantitative measurement of estrogen-induced ERK 1 and 2 activation via multiple membrane-initiated signaling pathways. Steroids 69, 181–192 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Homan L. L. & Dillon J. S. Genistein acutely stimulates nitric oxide synthesis in vascular endothelial cells by a cyclic adenosine 5′-mono-phosphate-dependent mechanism. Endocrinology 145, 5532–5539 (2004). [DOI] [PubMed] [Google Scholar]
- Watson C. S., Alyea R. A., Jeng Y. J. & Kochukov M. Y. Nongenomic actions of low concentration estrogens and xenoestrogens on multiple tissues. Mol. Cell Endocrinol. 274, 1–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredfeldt T. G. et al. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Mol. Endocrinol. 24, 993–1006 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardlund A. T. et al. Effects of prenatal exposure to tributyltin and trihexyltin on behavior in rats. Neurotoxicol. Teratol. 13, 99–105 (1991). [DOI] [PubMed] [Google Scholar]
- Noda T. et al. Teratogenicity study of tri-n-butyltin acetate in rats by oral administration. Toxicol. Lett. 55, 109–115 (1991). [DOI] [PubMed] [Google Scholar]
- Whalen M. M., Loganathan B. G. & Kannan K. Immunotoxicity of environmentally relevant concentrations of butyltins on human natural killer cells in vitro. Environ Res. 81, 108–116 (1999). [DOI] [PubMed] [Google Scholar]
- Matthiessen P. & Gibbs P. E. Critical appraisal of the evidence for tributyltin mediated endocrine disruption in mollusks. Environ. Toxicol. Chem. 17, 37–43 (1998). [Google Scholar]
- McAllister B. G. & Kime D. E. Early life exposure to environmental levels of the aromatase inhibitor tributyltin causes masculinisation and irreversible sperm damage in zebrafish (Danio rerio). Aquat. Toxicol. 65, 309–316 (2003). [DOI] [PubMed] [Google Scholar]
- Nishikawa J. et al. Involvement of the retinoid X receptor in the development of imposex caused by organotins in gastropods. Environ. Sci. Technol. 38, 6271–6276 (2004). [DOI] [PubMed] [Google Scholar]
- Kanayama T., Kobayashi N., Mamiya S., Nakanishi T. & Nishikawa J. Organotin compounds promote adipocyte differentiation as agonists of the peroxisome proliferator-activated receptor gamma/retinoid X receptor pathway. Mol. Pharmacol. 67, 766–774 (2005). [DOI] [PubMed] [Google Scholar]
- Grün F. et al. Endocrine-disrupting organotin compounds are potent inducers of adipogenesis in vertebrates. Mol. Endocrinol. 20, 2141–2155 (2006). [DOI] [PubMed] [Google Scholar]
- Cooke G. M. Effect of organotins on human aromatase activity in vitro. Toxicol. Lett. 126, 121–130 (2002). [DOI] [PubMed] [Google Scholar]
- Doering D. D., Steckelbroeck S., Doering T. & Klingmüller D. Effects of butyltins on human 5alpha-reductase type 1 and type 2 activity. Steroids 67, 859–867 (2002). [DOI] [PubMed] [Google Scholar]
- McVey M. J. & Cooke G. M. Inhibition of rat testis microsomal 3beta-hydroxysteroid dehydrogenase activity by tributyltin. J. Steroid Biochem. Mol. Biol. 86, 99–105 (2003). [DOI] [PubMed] [Google Scholar]
- von Ballmoos C., Brunner J. & Dimroth P. The ion channel of F-ATP synthase is the target of toxic organotin compounds. Proc. Natl. Acad. Sci. U S A 101, 11239–11244 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Kotake Y., Sekino Y. & Kanda Y. AMP-activated protein kinase-mediated glucose transport as a novel target of tributyltin in human embryonic carcinoma cells. Metallomics 5, 484–491 (2013). [DOI] [PubMed] [Google Scholar]
- Vinekar R., Verma C. & Ghosh I. Functional relevance of dynamic properties of Dimeric NADP-dependent Isocitrate Dehydrogenases. BMC Bioinformatics 13, S2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birket M. J. et al. A reduction in ATP demand and mitochondrial activity with neural differentiation of human embryonic stem cells. J. Cell Sci. 124, 348–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. O. et al. Identification and functional characterization of a novel, tissue-specific NAD(+)-dependent isocitrate dehydrogenase β subunit isoform. J. Biol. Chem. 274, 36866–36875 (1999). [DOI] [PubMed] [Google Scholar]
- Cohen P. F. & Colman R. F. Diphosphopyridine nucleotide dependent isocitratedehydrogenase from pig heart. Charactgerization of the active substrate and modes of regulation. Biochemistry 11, 1501–1508 (1972). [DOI] [PubMed] [Google Scholar]
- Lemire J., Mailloux R. & Appanna V. D. Zinc toxicity alters mitochondrial metabolism and leads to decreased ATP production in hepatocytes. J. Appl. Toxicol. 28, 175–182 (2008). [DOI] [PubMed] [Google Scholar]
- Wise D. R. et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl. Acad. Sci. U S A 108, 19611–19616 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juang H. H. Modulation of mitochondrial aconitase on the bioenergy of human prostate carcinoma cells. Mol. Genet. Metab. 81, 244–252 (2004). [DOI] [PubMed] [Google Scholar]
- Gabriel J. L., Zervos P. R. & Plaut G. W. Activity of purified NAD-specific isocitrate dehydrogenase at modulator and substrate concentrations approximating conditions in mitochondria. Metabolism 35, 661–667 (1986). [DOI] [PubMed] [Google Scholar]
- Cho S. A. et al. Up-regulation of Idh3alpha causes reduction of neuronal differentiation in PC12 cells. BMB Rep. 43, 369–374 (2010). [DOI] [PubMed] [Google Scholar]
- Nakatsu Y. et al. Glutamate excitotoxicity is involved in cell death caused by tributyltin in cultured rat cortical neurons. Toxicol. Sci. 89, 235–242 (2006). [DOI] [PubMed] [Google Scholar]
- Atanasov A. G., Nashev L. G., Tam S., Baker M. E. & Odermatt A. Organotins disrupt the 11beta-hydroxysteroid dehydrogenase type 2-dependent local inactivation of glucocorticoids. Environ. Health Perspect. 113, 1600–1606 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire A. et al. Activation of RXR-PPAR heterodimers by organotin environmental endocrine disruptors. EMBO Rep. 10, 367–373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. O. et al. Characterization of a cDNA clone for human NAD(+)-specific isocitrate dehydrogenase alpha-subunit and structural comparison with its isoenzymes from different species. Biochem. J. 308, 63–68 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda Y. & Watanabe Y. Adrenaline increases glucose transport via a Rap1-p38MAPK pathway in rat vascular smooth muscle cells. Br. J. Pharmacol. 151, 476–482 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli C., Grodsky N. B., Ariyaratne N., Colman R. F. & Bahnson B. J. Crystal structure of porcine mitochondrial NADP+-dependent isocitrate dehydrogenase complexed with Mn2+ and isocitrate. Insights into the enzyme mechanism. J. Biol. Chem. 277, 43454–43462 (2002). [DOI] [PubMed] [Google Scholar]
- Goto J., Kataoka R., Muta H. & Hirayama N. ASEDock-docking based on alpha spheres and excluded volumes. J. Chem. Inf. Model. 48, 583–590 (2008). [DOI] [PubMed] [Google Scholar]
- Halgren T. A. MMFF VI. MMFF94s option for energy minimization studies. J. Comput. Chem. 20, 720–729 (1999). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures