Abstract
A smartphone-utilized biosensor was developed for detecting microbial spoilage on ground beef, without using antibodies, microbeads or any other reagents, towards a preliminary screening tool for microbial contamination on meat products, and potentially towards wound infection. Escherichia coli K12 solutions (101–108 CFU/mL) were added to ground beef products to simulate microbial spoilage. An 880 nm near infrared LED was irradiated perpendicular to the surface of ground beef, and the scatter signals at various angles were evaluated utilizing the gyro sensor and the digital camera of a smartphone. The angle that maximized the Mie scatter varied by the E. coli concentration: 15° for 108 CFU/mL, 30° for 104 CFU/mL, and 45° for 10 CFU/mL, etc. SEM and fluorescence microscopy experiments revealed that the antigens and cell fragments from E. coli bonded preferably to the fat particles within meat, and the size and morphologies of such aggregates varied by the E. coli concentration.
Development of rapid, non-destructive, and inexpensive sensing technologies for detection of microbial contamination on meat1,2,3 and potentially on human tissue samples (especially for wound infection)4,5 remain in high demand. The current standard detection methods for this microbial contamination on meat or tissue, described by the U.S. Food and Drug Administration (FDA), are culture-based, which require incubation, inoculation, as well as pre-processing the meat or tissue sample6. Other well-developed (but less standard) methods are immunological or nucleic acid-based methods, such as enzyme-linked immunosorbent assay (ELISA) and methods that utilize polymerase chain reaction (PCR). These current methods are not rapid, destructive, and require time, personnel, and laboratories for the experiments. Delayed identification/diagnosis of such pathogens may lead to empirical, broad-spectrum antibiotic therapy, which may lead to antimicrobial resistance7,8,9.
Several methods that are in development are aiming for non-destructive, rapid, reagentless, and still accurate and sensitive. Electronic noses, Fourier transform infrared (FT-IR) spectroscopy, and Raman scattering are being investigated for microbial contamination on meat10,11,12,13,14. Enzymatic reactions and subsequent spectrophotometric/fluorometric detection have also been investigated for wound infection15,16,17. These developing methods are an improvement in that they are non-destructive, can be real-time, and have the potential to be used as an on-site detection. However, most of these methods still need trained personnel to operate and can be expensive due to the requirement of specific instrumentation.
Previously, our laboratory has investigated the use of microbead immunoagglutination on a lab-on-a-chip platform and subsequent Mie scattering detection towards rapid detection of pathogens. This method has already been proven to have extremely low detection limit (typically at a single-cell level or 10 CFU/mL; CFU = colony forming units), near-real-time detection (less than 5 minutes per assay), as well as portability, towards E. coli detection from lettuce18 and Salmonella detection from poultry package19. However, this method still requires adding the reagent (antibody-conjugated microbeads) to the food/tissue sample via the reagent delivering vehicles – a network of channels and wells, i.e. lab-on-a-chip.
Mie scatter refers to the Mie solution to the scattering problem (Maxwell's equations) on a spherical object, when the object sizes (d) are in a similar order of magnitude to the wavelengths of incident light20. Mie solution describes how much light is scattered and the scatter intensities are changed according to the scattering angles. Immunoagglutination assay is typically conducted by mixing antibody-conjugated microbeads with a target solution; the binding between the antibody and the target antigens causes agglutination of the microbeads. The agglutination is then monitored via Mie scattering measurement at a specific angle where the scattering intensity is maximum among the usable angles of detection. Since the agglutinated microbeads are no longer spherical in shape and its morphology is quite complicated, different scattering centers can constructively and destructively interfere with each other to a greater extent, leading to much stronger scattering intensities at an optimized angle of detection21. The refractive index (n) of microbeads should be substantially higher than that of water and/or food/tissue samples, to successfully monitor this agglutination of microbeads. Previously, polystyrene microbeads (n = 1.59 and d = 920 nm18,19) have successfully been used over water (n = 1.327) and food samples (n ~ 1.40 for typical proteins22).
In fact, E. coli can be identified directly through evaluating Mie scattering, since its refractive index is still higher (n = 1.38823,24) than that of water (n = 1.327). However, the morphologies of the cells, fragments, antigens or their colonies are much simpler than the immunoagglutinated microbeads, and such detection is only possible when the bacterial concentration is very high25. If these cells, fragments, proteins or colonies can interact with some component(s) of meat to form more complicated structure, similar to immunoagglutination, they may be detected through Mie scatter in much lower level of detection. Many foodborne pathogens, including E. coli, are hydrophobic, thus they tend to attach preferentially to fat surfaces. Animal fat cells (typically hydrogenated) generally have an average refractive index of 1.40, which is not very different from other proteins and bacteria, including E. coli. However, n = 1.40 is still higher than that of water (n = 1.327), it may be possible to observe different light scattering characteristics upon association of bacteria to the fats. Pure lipids are slightly better to be distinguished since their refractive index is somewhat higher: n = 1.4626. Based on these facts, we attempted to use Mie scatter detection to measure these pseudo-colonies of E. coli cells, fragments, and antigens (when the concentration is low), or actual colonies (when the concentration is high) around the fats within the meat samples without any microbeads or antibody presence. Since there is no significant difference in the refractive indices between fats (n = 1.40–1.46) and bacteria (n = 1.388), the increase in bacteria concentration may not bring in the overall increase in scatter intensity at a fixed scatter angle. Such overall increases could be observed with polystyrene microbead (n = 1.59) immunoagglutination18,19. However, the angle-dependent Mie scatter characteristics will surely be altered by bacteria concentration, since the optimum angle for detection will change as the sizes and ratios of pseudo-colonies and actual colonies change.
Since no antibodies will be utilized in this study, distinguishing similar bacteria species (e.g., E. coli and Salmonella spp.) or assessing pathogenicity will not be possible. The proposed method will serve as a preliminary screening tool for general microbial contamination within meat. In our experimental setup, the digital camera of a smartphone was utilized as an optical detector to quantify the Mie scatter intensities, to replace optical fibers and a spectrometer. Additionally, a smartphone application was programmed in order to eliminate the need of using a benchtop stage or any holders, and to achieve our goal of creating a truly handheld (not just portable), easy-to-use and inexpensive device.
Results and discussion
Smartphone-based detection systems
150 μL of deionized water (negative control) and series of serially diluted Escherichia coli K12 solutions (101–108 CFU/mL) were added to ground beef products to simulate microbial spoilage. An 880 nm near infrared (NIR) LED was irradiated perpendicular to the surface of ground beef, while the digital camera of a smartphone detected the scatter signal angled at 15°, 30°, 40° and 60° from the incident light. Initially, NIR irradiation and scatter detection were made on the positioning stages, shown in Fig. 1. Later, these experiments were duplicated without using the positioning stages, while the distance and angle of a smartphone from the ground beef and the NIR LED were properly fixed through a software application and the built-in gyro sensor of a smartphone (Fig. 2).
Figure 1. The benchtop system consists of an iPhone 4S and its holder, an NIR LED and its holder, and a ground beef sample and its holder.
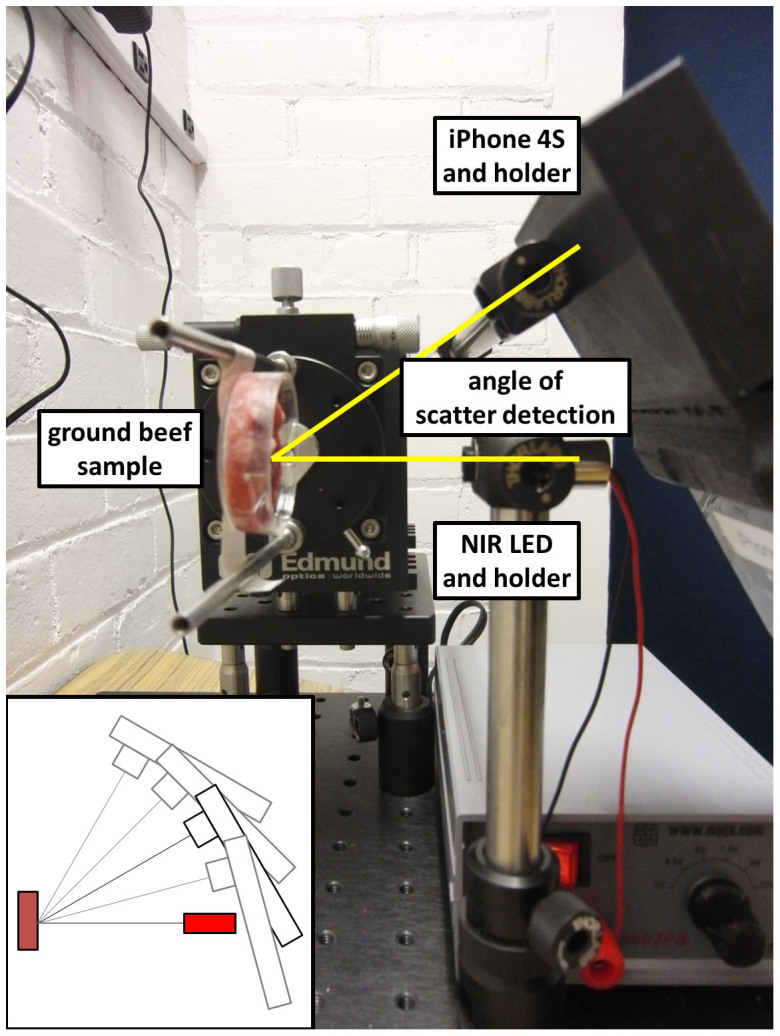
The angle of scatter detection refers to the angle between the iPhone camera and the NIR LED light source.
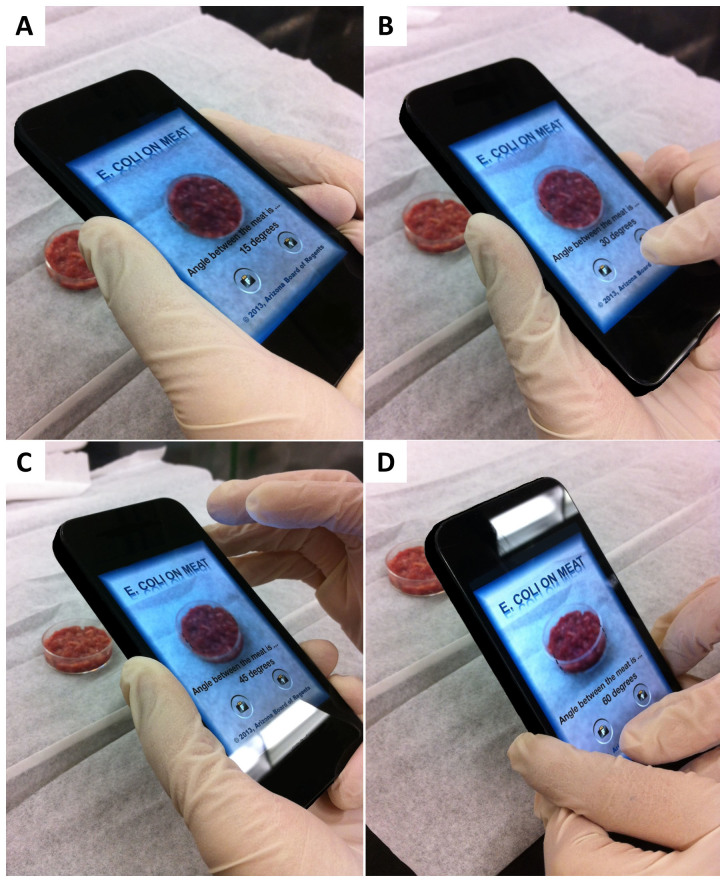
Figure 2. Photographs showing the operation of the smartphone application at the four specific angle of scatter detection: (A) 15°, (B) 30°, (C) 45°, and (D) 60°.
Light scatter intensities from ground beef with E. coli
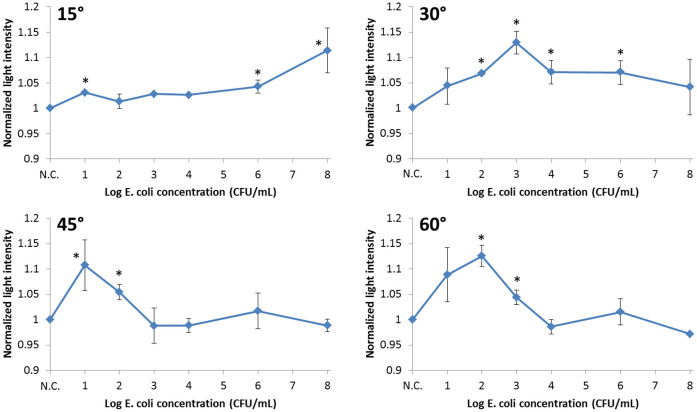
Normalized scatter light intensities from ground beef measured at 15°, 30°, 45°, and 60° were plotted against the log E. coli concentrations (Fig. 3) and they were combined into a surface plot (Fig. 4). To eliminate sample-to-sample variation, all acquired intensities were normalized to the average of the negative control (i.e. ground beef with DI water) readings of the same angle. All data points in Figs. 3 and 4 represent the mean values of three duplicates, each time measured from three different locations (with no overlapping) on a single ground beef sample. A new ground beef sample was used for each E. coli concentration. Two-sample t-tests were performed between each data point and the negative control and the stars (*) indicate significant difference with p < 0.05.
Figure 3. Normalized scatter light intensities plotted against the log concentrations of E. coli on ground beef.
Error bars represent standard errors. The scatter detection angle was 15°, 30°, 45° and 60°. Each data point represents the mean of three replicates (each measured from different locations on a ground beef sample). A new ground beef sample was used for each E. coli concentration. Stars (*) represent the data points that are significantly different from the negative control, with p < 0.05 (two-sample t-tests).
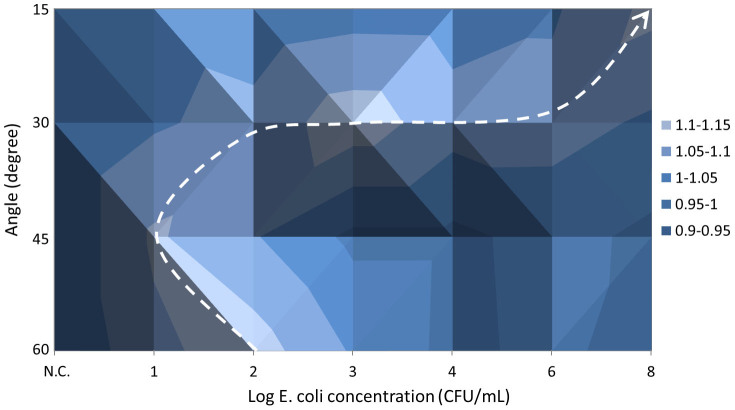
Figure 4. Surface plots that combine the normalized light intensities, the angles of scatter detection, and the log concentrations of E. coli.
The plot measured at 15° does not show significant scatter signals under lower and mid-range concentrations (0–104 CFU/mL) of E. coli. Significant signal can only be seen under very high concentration (108 CFU/mL). The plot measured at 30°, however, shows a linear increase up to 103 CFU/mL, followed by a decrease. The lower limit of detection is 102 CFU/mL with this angle. The plot measured at 45° shows that the peak is shifted from 103 CFU/mL to 10 CFU/mL and the lower limit of detection is 10 CFU/mL, although no linearity can be found beyond this concentration. The plot measured at 60° shows a similar trend, with the peak at 102 CFU/mL and generally higher scatter intensities.
The error bars with 15° detection are generally smaller than the detections at 30°, 45° and 60°, presumably indicating the substantially different character in their signals. Perhaps this 15° detection is primarily reflectance, thus only corresponds to the absorbance of very high E. coli concentration (108 CFU/mL), while the other 30°, 45° and 60° detections are primarily angle-dependent Mie scatter.
Surface plot analysis
Fig. 4 shows a “ridge” moving from 102 CFU/mL at 60° → 101 CFU/mL at 45° → 103 CFU/mL at 30° → 108 CFU/mL at 15°. Fig. 4 clearly shows that the scatter light intensity change over the detection angle has a different trend at each E. coli concentration on the ground beef sample. For example, if the peak in light intensity is found with higher angles (45°–60°), the concentration is likely at the lower range (10–102 CFU/mL); if the peak is found with mid-angles (30°), the concentration is likely at the mid-range (103–106 CFU/mL); if the peak is found with lower angles (15° or less), the concentration is possibly very high (108 CFU/mL).
How E. coli interacts with fats towards different light scatter characteristics
It is already known that scattering detection can be used to detect bacteria colonies when the concentration is high (typically 105 CFU/mL and above25). However, when the bacteria concentration is low, such colonies are too small in number to be detected directly. We believe that the cell fragments and proteins from E. coli can interact with and aggregate around the fats within the ground beef sample, to form a pseudo-colony, due to their hydrophobicity. These pseudo-colonies may be substantially smaller than the actual bacteria colonies, perhaps a few microns or less in size. These sizes are comparable to the wavelength of incident light (NIR at 880 nm), where the subsequent Mie scattering is accordingly maximized. In addition, these pseudo-colonies may form more complicated structure, similar to immunoagglutination, which further increase the extent of Mie scatter at a certain specific range of detection angle. The scatter signals that we observed at low bacteria concentrations (at 45–60°) were most likely the light scatter caused by such pseudo-colonies. As the bacteria concentration increases, a larger portion of signals come from actual colonies that were formed prior to their individual attachment to the fat cells. As stated at the end of Introduction, however, this method will not be able to distinguish similar species of bacteria, such as E. coli and Salmonella spp.
Effect of dust particles
Although the experiments were performed to minimize dust deposition (the meat samples were always covered and exposure time to ambient air was minimized), we have nonetheless investigated the potential effect of the dust particles. The difference in absorbance ( = the sum of scattered light in all possible directions of the dust particles plus the true absorbance by the dust particles) was evaluated for the dust particles passively deposited for one hour, on a 645 cm2 desk area in an office space that has about ten student workers. The glass microfiber filter (70 mm diameter; Whatman, GE Healthcare, Waukesha, WI, USA; catalog no. 1823-070) was used to collect the dust particles. The absorbance difference (between the collected sample and a negative control) was maximum at green color (less than 0.02 absorbance) and less than 0.012 at the NIR wavelength (880 nm) that we used in this study. Considering the light scatter signal at a specific angle is only a tiny fraction of the total absorbance, we can conclude that the effect of dust sedimentation is negligible for our angle-specific NIR scatter detection.
SEM imaging
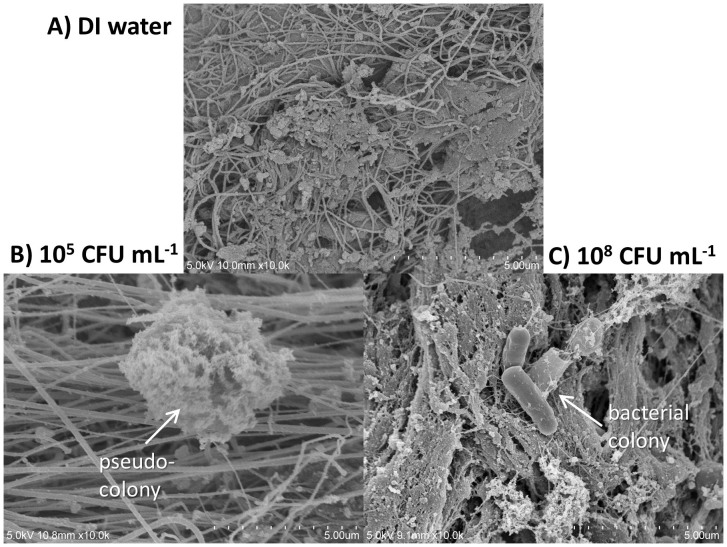
Several scanning electron microscopic (SEM) images of E. coli on ground beef were taken (Fig. 5) to further confirm the above-mentioned E. coli attachment to ground beef. Fig. 5A shows the SEM image of the ground beef with DI water, i.e. negative control. Lots of muscle fibers and smaller particulate matter can be identified, with bigger fat cells occasionally found in between. Smaller particulate matter can be fat particles, protein aggregates, cell fragments, or salt crystals (formed during the sample preparation for SEM). Since the E. coli solutions were serially diluted with DI water, not phosphate buffered saline (PBS), the formation of PBS crystals is not likely to happen during the preparation for SEM imaging. With intermediate E. coli concentration, 105 CFU/mL (Fig. 5B), such “small particulate matter” becomes substantially bigger in size, apparently in an aggregated form. The diameters of such small particulate matter ranged from submicron to 4–5 μm. With much higher E. coli concentration, 108 CFU/mL (Fig. 5C), intact E. coli bacteria (mostly aggregated) can easily be identified, while particulate matter can still be found. These results indirectly indicate the formation of pseudo-colonies between the cell fragments and proteins from E. coli and fat particles within the meat sample.
Figure 5. SEM images of (A) DI water on ground beef, (B) 105 CFU/mL E. coli on ground beef, and (C) 108 CFU/mL E. coli on ground beef.
Each graduation in the lower right corner represents 0.5 μm (10 gradations = 5 μm).
Fluorescence staining for fats and E. coli
Fig. 6 shows fluorescence microscopic images of Hoechst-stained E. coli (stains nucleic acids and appears as blue fluorescence) on Nile Red-stained ground beef (soluble in lipid and appears as yellow fluorescence). Typical dimensions of yellow stains ranged from a few tens of μm (Fig. 6A) to a few hundreds of μm (Fig. 6B). A few tens of μm yellow stains correspond to the smaller lipid particles or potentially the fragments of fat cells, while a few hundreds of μm yellow stains correspond to the intact fact cells. In both cases, blue stains are always found near the yellow stains, but not overlapping them, indicating the E. coli cells are attracted towards the fat regions of the ground beef.
Figure 6. Fluorescence microscopic images of Hoechst-stained E. coli (blue) on Nile Red-stained ground beef sample (yellow) with the E. coli (102 and 105 CFU/mL).
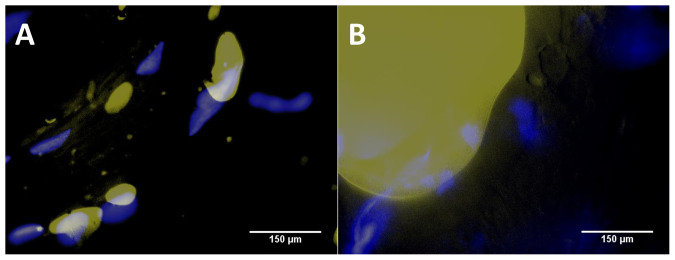
The SEM images together with the fluorescence microscopic images strongly support our theory about the formation of pseudo-colonies at lower concentrations which make the Mie scattering possible at a concentration that is well below the previously shown detection limit.
Mie scatter simulations
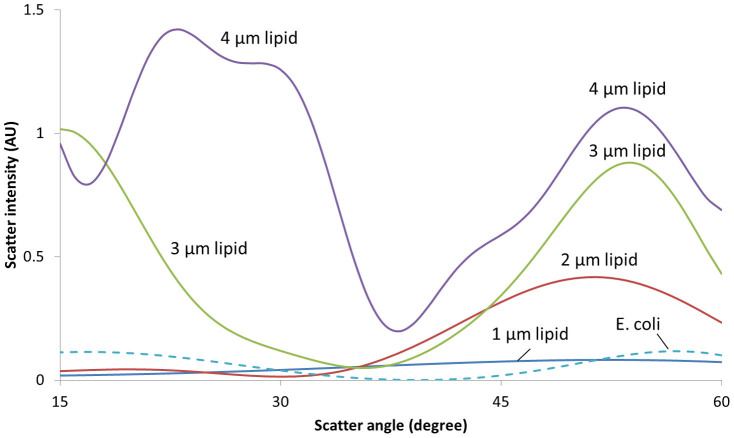
To better interpret the results shown in Figs. 3 and 4, a series of Mie scatter simulations were performed using the publicly available software (MiePlot v4.2.11), and the results are shown in Fig. 7. Refractive indices were set to 1.327 for water, 1.388 for E. coli24, and 1.46 for lipids26. Four different diameters of lipid particles were used: 1, 2, 3 and 4 μm, following the results obtained from the SEM images (Fig. 5). Fig. 5A shows that lipid particles are mostly 1 μm in size whereas in Fig. 5B, as the concentration increases to 105 CFU/mL, the pseudo-colony size goes up to 4 μm. The dimension of E. coli was set to 2.83 μm24. Simulations were performed for the scatter angles from 15° to 60°.
Figure 7. Mie scatter simulations for lipid particles and E. coli.
As shown in Fig. 7, scatter is insignificant and essentially angle-independent for the 1 μm lipid particles (i.e. negative control). For the 2 μm lipid particles, however, a noticeable rise in scatter can be observed for 45–60°. This result corresponds very well to the high scatter intensities for 10–102 CFU/mL E. coli detection at 45° and 60°: lower E. coli concentration corresponds to the smaller pseudo-colony (lipid particle) formation. As the lipid particle size increases, the scatters at 45–60° continue to increase and another bigger peak emerges at around 30°, especially with 4 μm lipid particles. This secondary peak at 30° corresponds to the 105 CFU/mL SEM image from Fig. 5, as well as the high scatter intensities for 103–106 CFU/mL E. coli detection at 30° from Fig. 3: mid-range E. coli concentration corresponds to the bigger pseudo-colony (lipid particle) formation. Finally, for E. coli, scatter is insignificant but highly angle-dependent, showing two small peaks at 15° and 60°. If E. coli concentration is very high, e.g., 108 CFU/mL, it may scatter significantly at both 15° and 60°, but not at 30° and 45°. The results shown in Figs. 3 and 4 show the significantly scatter only at 15°, there must be some other factor contributing to the augmented scatter at that angle. Since 15° is very close to back scatter (0°), which can be related to the absorbance, the 15° peak can be explained by the direct NIR absorbance by highly concentrated E. coli.
Handheld device with smartphone application
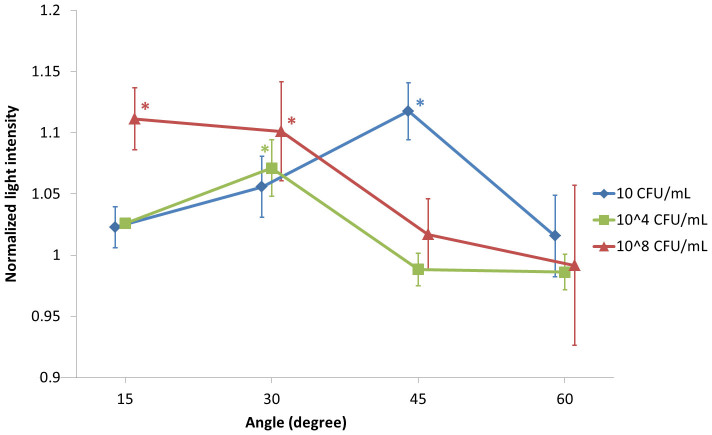
Using the handheld device with smartphone application, normalized scatter light intensities were evaluated and plotted against the angles, with varying E. coli concentration (Fig. 8). Actual images taken by a smartphone are shown in Fig. 9. Most digital cameras are capable of recognizing NIR, as demonstrated in this work. Results obtained here are similar to what we received with a benchtop system: With lower E. coli concentration the peak in light intensity is found with higher detection angles. With mid-range E. coli concentration, the peak is found with mid-range detection angles. With higher E. coli concentration, the peak is found with lower detection angles. The characteristic peaks can be found at 45° with 10 CFU/mL, 30° with 104 CFU/mL, and 15° with 108 CFU/mL, which are all significantly different from the negative control (normalized light intensity of 1). The error bars of the 108 CFU/mL series are larger than the other two series, which can be explained by the uneven distribution of bacterial colonies (found in very high E. coli concentration; refer to the SEM imaging section and Fig. 5). Overall, this result suggests that we can use a smartphone application to replace the benchtop system and make the device truly handheld and inexpensive, while maintaining the low detection limit.
Figure 8. Normalized light intensities plotted against scatter detection angles with different E. coli concentrations, using the handheld device.
Each plot was shifted by 1° to better indicate the error bars (standard errors). The data points with 10 CFU/mL and 108 CFU/mL E. coli concentrations are the means of seven replicates, while those with 104 CFU/mL are the means of three replicates, each measured from different ground beef sample. Stars (*) represent the data points that are significantly different from the negative control, with p < 0.05 (two-sample t-tests).
Figure 9. Typical images obtained with the iPhone camera.
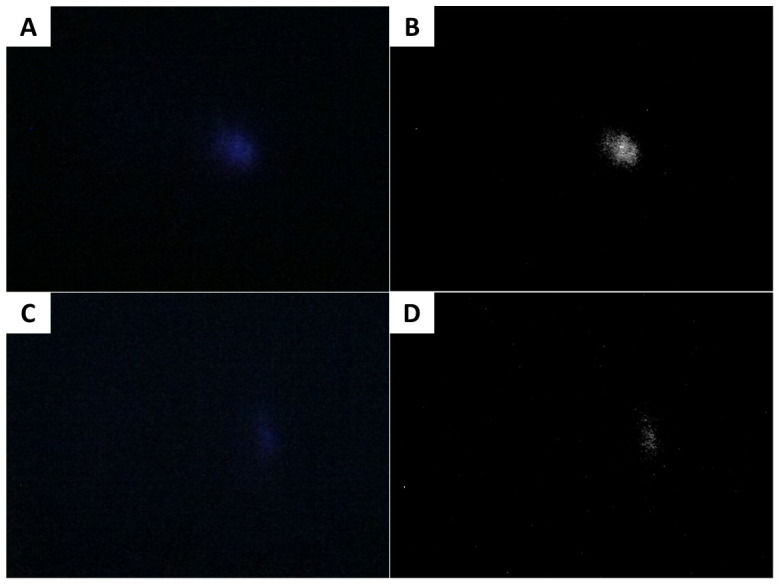
(A) Raw image showing higher light intensity. (B) Processed image of (A). (C) Raw image showing lower light intensity. (D) Processed image of (C).
Conclusion
A smartphone-based biosensor was developed to detect and quantify microbial contamination on ground beef, consisting of an 880 nm NIR LED and a smartphone (utilizing its digital camera, software application, and an internal gyro sensor). Mie scatter measurements were made at four different angles (15°, 30°, 45° and 60°), and the concentrations of E. coli (from 101 CFU/mL to 108 CFU/mL) could be determined by the “pattern” of such scatter intensities over the angles, i.e., at which angle the peak/valley intensity was observed or whether the scatter intensities monotonically decreased/increased over the angles, etc. The lower limits of detection were 101 CFU/mL at 45° and 102 CFU/mL at 30° and 60°, which are comparable to the antibody-based microparticle Mie scatter assay18,19 This superior detection limit is substantially lower than the infectious dose of typical E. coli (106–108) or Salmonella spp. (105) and comparable to the highly pathogenic bacteria such as E. coli O157:H7 (10)27,28. The proposed smartphone-based biosensor does not require any antibodies, microbeads or any other reagents, and is handheld, easy-to-use, rapid, and inexpensive. The proposed device can be used as a preliminary screening tool to monitor microbial contamination on meat products, and potentially towards wound infection.
Methods
E. coli K12 solutions
Escherichia coli K12 lyophilized cell powder (Sigma-Aldrich, St. Louis, MO, USA; catalog no. EC1) was cultured in lysogeny broth (LB-Miller; Growcells, Irvine, CA, USA; catalog no. MBPE-1050) at 37°C for 18 hours. The cultured solutions were serially diluted with autoclaved, deionized (DI) water. The colony-forming units (CFU) of each serial dilution were quantified by spread plate method, in which a 0.1 mL aliquot was uniformly spread on top of an LB agar plate (VWR International, Radnor, PA, USA; catalog no. 29447-000). The plate was incubated at 37°C for 24 hours before the colonies were counted.
Ground beef samples
Packages of 80% lean ground beef were purchased from local grocery stores one week before the experiments. The ground beef were made into 5 gram aliquots and each aliquot was flattened and placed on a cell culture dish (Corning Inc., Corning, NY, USA; catalog no. 3294). The meat dishes were then covered and sealed with a paraffin film (Parafilm “M”; Pechiney Plastic Packaging, Menash, WI, USA) and put in a freezer (−20°C). 12 hours before the experiments, dishes were moved to a refrigerator (4°C) to thaw. During the experiments, the 150 μL of each diluted E. coli solution was applied to the flattened 80% lean meat at the center. The dishes were covered and sealed again, and put on a nutator mixer (VWR International; catalog no. 15172-203) for 30 minutes.
Benchtop system for optimizing Mie scatter detection
Positioning stages (Edmund Optics, NJ, USA) with two holders were developed to expose the samples at varying angles of incident light and detector device (the digital camera of a smartphone) (Fig. 1). An 880 nm 10 mW near infrared light emitting diode (NIR LED; AixiZ LLC, Houston, TX, USA; catalog no. AIX-880-10) was placed in the incident light holder, and an iPhone 4S (Apple, CA, USA) was placed in the detector holder. The NIR LED light source enables us to obtain the scatter light signals without being affected by the natural meat color differences (in the visible light range), and provides a sufficiently strong signal to the iPhone digital camera which makes the final device portable. A support for precisely attaching the iPhone to the benchtop system was designed using AutoCAD 2000i (Autodesk, CA, USA) and was then stereolithographically printed using a uPrint 3D printer (Stratasys, MN, USA).
Assay procedure
The E. coli-applied samples, together with a negative control (150 μL of DI water was applied to the meat instead of E. coli solutions), were then placed perpendicular to the incident light at a distance of 6.35 cm (2.5 inch), and the attached iPhone 4S took pictures in a dark environment (i.e. minimum ambient light) at four different angles from the incident light (i.e. 15°, 30°, 45°, and 60°) (Fig. 1). The light scatter intensities were identified by analyzing the pictures taken by the iPhone using ImageJ version 1.44p (National Institutes of Health, Bethesda, MD, USA). All images were circular-cropped to eliminate the noise around the edge of the picture, and were converted to 8-bit (gray scale) pictures. The processed pictures were then analyzed to get the median intensity value (between 0–255) among all pixels.
SEM imaging
SEM images of E. coli on 80% lean ground beef were taken using an ultra-high resolution field emission scanning electron microscope (Hitachi, Schaumburg, IL, USA; model S-4800 Type II). 100 μL of water (negative control) or E. coli solution (105 or 108 CFU/mL) was applied to 1.5 g of thawed ground beef in a petri dish and mixed on a nutator mixer for 30 minutes. After mixing, the fixative was added to fix the sample overnight. The fixative consisted of 4% v/v paraformaldehyde and 1% v/v glutaraldehyde with phosphate buffer. The fixed samples were then dried in a Polaron Critical Point Dryer (Quorum Technologies, East Sussex, UK; model E3000) and platinum-coated using a Hammer 6.2 Sputter Coater (Anatech, Union City, CA, USA) before SEM imaging.
Fluorescence staining for fats and bacteria
Less than 1 g of 80% lean ground beef sample was placed on a glass slide. 10 μL solution of Nile red (Sigma-Aldrich; catalog no. 72485) dissolved in acetone (1 mg/mL) was added to the ground beef and the samples were lightly pressed down with coverslips. The samples were stained overnight in a refrigerator (4°C) before adding 20 μL of Hoechst-stained E. coli solutions. Nile red is very soluble in lipid and has an emission peak between 620–660 nm when excited in the range 525–575 nm. Each serial-diluted E. coli solution was stained with Hoechst dye that stains nucleic acids (NucBlue Live ReadyProbes Reagent, Life Technologies Corporation, Carlsbad, CA, USA; catalog no. R37605) for 30 minutes before it was applied to the Nile red-stained ground beef sample. Hoechst dye has a blue emission (around 460 nm) when excited in the ultraviolet (UV) range (around 350 nm). Once the Hoechst-stained E. coli solutions were applied to the Nile red-stained ground beef samples, they were firmly pressed down with the coverslips and were observed using a fluorescent microscope (Nikon, Tokyo, Japan). Images of each sample were taken under UV light and green light separately, and were overlayed using ImageJ (National Institutes of Health).
Smartphone application for handheld detection
A smartphone application was programmed using Xcode (Apple, CA, USA), which allows users to take pictures at the four angles at a fixed distance (Fig. 2). The application shows the angle by using the built-in gyro sensor. Two dotted lines appear on the screen for the users to match the outline width of a meat dish; therefore the distances between the camera and the sample are consistent. The application further analyzes and compares the pictures through implementing an image processing algorithm, and displays the bacteria concentration on the smartphone screen.
Author Contributions
P.S.L. and J.Y.Y. conceived the concept and designed the experiment. T.S.P. designed, fabricated and assembled the positioning stages, and also coded the smartphone application. P.S.L. conducted all microbial detection experiments from ground beef. J.Y.Y. conducted and analyzed the Mie scatter simulations. P.S.L. and J.Y.Y. analyzed the data and wrote the manuscript.
Acknowledgments
The authors thank Steven Hernandez at the University Spectroscopy and Imaging Facilities at the University of Arizona for his help in SEM sample processing and subsequent imaging.
References
- Shillam P. et al. Multistate outbreak of Escherichia coli O157:H7 infections associated with eating ground beef - United States, June--July 2002. Morb. Mortal. Wkly. Rep. 51, 637–639 (2002). [PubMed] [Google Scholar]
- Scallan E. et al. Foodborne illness acquired in the United States - major pathogens. Emerg. Infect. Dis. 17, 7–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J. A. et al. Estimating the burden of acute gastroenteritis, foodborne disease, and pathogens commonly transmitted by food: an international review. Clin. Infect. Dis. 41, 698–704 (2005). [DOI] [PubMed] [Google Scholar]
- Bowler P. G., Duerden B. I. & Armstrong D. G. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 14, 244–269 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D. G. & Lavery L. A. Negative pressure wound therapy after partial diabetic food amputation: a multicenter, randomised controlled trial. Lancet 366, 1704–1710 (2005). [DOI] [PubMed] [Google Scholar]
- Feng P., Weagant S. D., Grant M. A. & Burkhardt W. Bacteriological Analytical Manual: Enumeration of Escherichia coli and the Coliform Bacteria. http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm064948.htm (2013) (Date of access: 01/02/2014).
- Lipsky B. A. & Berendt A. R. Principles and practice of antibiotic therapy of diabetic foot infections. Diabetes Metab. Res. Rev. 16, S42–S46 (2000). [DOI] [PubMed] [Google Scholar]
- Baumgart A. M. K., Molinari M. A. & Silveira A. C. D. O. Prevalence of carbapenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii in high complexity hospital. Braz. J. Infect. Dis. 14, 433–436 (2010). [DOI] [PubMed] [Google Scholar]
- Lucignano B. et al. Multiplex PCR allows rapid and accurate diagnosis of bloodstream infections in newborns and children with suspected sepsis. J. Clin. Microbiol. 49, 2252–2258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D. I., Broadhurst D., Kell D. B., Rowland J. J. & Goodacre R. Rapid and quantitative detection of the microbial spoilage of meat by Fourier transform infrared spectroscopy and machine learning. Appl. Environ. Microbiol. 68, 2822–2828 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D. I. & Goodacre R. Rapid and quantitative detection of the microbial spoilage of muscle foods: current status and future trends. Trends Food Sci. Technol. 12, 414–424 (2001). [Google Scholar]
- Kalasinsky K. S. et al. Raman chemical imaging spectroscopy reagentless detection and identification of pathogens: signature development and evaluation. Anal. Chem. 79, 2658–2673 (2007). [DOI] [PubMed] [Google Scholar]
- Sowoidnich K., Schmidt H., Kronfeldt H. D. & Schwägele F. A portable 671 nm Raman sensor system for rapid meat spoilage identification. Vib. Spectrosc. 62, 70–76 (2012). [Google Scholar]
- Sundaram J., Park B., Kwon Y. & Lawrence K. C. Surface enhanced Raman scattering (SERS) with biopolymer encapsulated silver nanosubstrates for rapid detection of foodborne pathogens. Int. J. Food Microbiol. 167, 67–73 (2013). [DOI] [PubMed] [Google Scholar]
- Hasmann A. et al. Novel peptidoglycan-based diagnostic devices for detection of wound infection. Diagn. Microbiol. Infect. Dis. 71, 12–23 (2011). [DOI] [PubMed] [Google Scholar]
- Heinzle A. et al. Novel protease-based diagnostic devices for detection of wound infection. Wound Rep. Reg. 21, 482–489 (2013). [DOI] [PubMed] [Google Scholar]
- Tang E. N., Nair A., Baker D. W., Hu W. & Zhou J. In vivo imaging of infection using a bacteria-targeting optical nanoprobe. J. Biomed. Nanotechnol. 10, 856–863 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- You D. J., Geshell K. J. & Yoon J. Y. Direct and sensitive detection of foodborne pathogens within fresh produce samples using a field-deployable handheld device. Biosens. Bioelectron. 28, 399–406 (2011). [DOI] [PubMed] [Google Scholar]
- Fronczek C. F., You D. J. & Yoon J. Y. Single-pipetting microfluidic assay device for rapid detection of Salmonella from poultry package. Biosens. Bioelectron. 40, 342–439 (2013). [DOI] [PubMed] [Google Scholar]
- van de Hulst H. C. [Chapter 9: Rigorous scattering theory for spheres of arbitrary size (Mie theory)]. Light scattering by small particle [114–130] (John Wiley and Sons, Mineola, NY, 1983). [Google Scholar]
- Heinze B. C. & Yoon J. Y. Nanoparticle immunoagglutination Rayleigh scatter assay to complement microparticle immunoagglutination Mie scatter assay in a microfluidic device. Colloids Surf. B 85, 168–173 (2011). [DOI] [PubMed] [Google Scholar]
- Vörös J. The density and refractive index of adsorbing protein layers. Biophys. J. 87, 553–561 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaev A. E., Dvoretski K. N. & Doubrovski V. A. Refractive index of Escherichia coli cells. Proc. SPIE 4707, 253–260 (2002). [Google Scholar]
- Liu Y. et al. A single living bacterium's refractive index measurement by using optofluidic immersion refractometry. 17th Int. Conf. Miniaturized Syst. Chem. Life Sci., Freiburg, Germany. pp.263–265 (2013). [Google Scholar]
- Hirleman E. D. Jr, Guo S., Bhunia A. K. & Bae E. System and method for rapid detection and characterization of bacterial colonies using forward light scattering. United States Patent, No. 7,465,560 (2008). [Google Scholar]
- Board N. Modern Technology of Oils, Fats and Its Derivatives (Asia Pacific Business Press, Delhi, India, 2002). [Google Scholar]
- Tutte J. et al. Lessons from a large outbreak of Escherichia coli O157:H7 infections: insights into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 122, 185–192 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Ray C. G., Ahmad N., Drew W. L. & Plorde J. J. [Chapter 33: Enterobacteriaceae] Sherris Medical Microbiology: An Introduction to Infectious Diseases, 5th Edn. [579–608] (McGraw-Hill, New York, NY, 2010). [Google Scholar]