Abstract
The ability of Ralstonia solanacearum to cause disease on plants depends on its type III secretion system (TTSS) encoded by hrp genes. The expression of hrp genes and known TTSS substrates is coordinately regulated by HrpB, a member of the AraC family of transcriptional regulators. Two HrpB-regulated promoters (hrpY and popABC) were characterized by deletion analysis, and the HrpB-dependent activation of these promoters was found to be conferred by a 25-nucleotide DNA element, the hrpII box (TTCGn16TTCG), which is present in other hrp promoters. The hrpII box element is an imperfect plant inducible promoter box, an element which was originally found in hrp promoters of Xanthomonas campestris (S. Fenselau and U. Bonas, Mol. Plant-Microbe Interact. 8:845-854, 1995) but which was not characterized at the molecular level. Site-directed mutagenesis showed that the hrpII box is essential for hrpY promoter activation in vivo. Functional analysis of the hrpII box element identified critical parameters that are required for HrpB-dependent activity. Further mapping analyses of several other hrpB-dependent promoters also indicated that the position of the hrpII box is conserved, at −70 to −47 bp from the transcriptional start. As a first step toward identifying novel TTSS effectors, we used the hrpII box consensus sequence to search for potential HrpB-regulated promoters in the complete genome sequence of R. solanacearum strain GMI1000. Among the 114 genes identified, a subset of promoters was found to have a structural relationship with hrp promoters, thus providing a pool of candidate genes encoding TTSS effectors.
Type III secretion systems (TTSSs) are used by numerous gram-negative pathogenic bacteria to deliver virulence (effector) proteins directly into the cytoplasm of host cells (24). In bacterial plant pathogens belonging to the genera Erwinia, Pseudomonas, Ralstonia, and Xanthomonas, TTSSs (also referred to as Hrp systems) are encoded by hrc and hrp genes (2, 24). The effector molecules delivered into plant cells through this pathway are believed to act collectively as the primary determinants of pathogenicity and host range and to promote disease by interfering with host cellular signaling pathways (8, 42).
The expression of hrc, hrp, and most type III effector genes is environmentally regulated. These genes are expressed at a low level during growth in complete media, and their expression is induced in plant tissues or in various synthetic minimal media which are thought to mimic conditions found in planta. Despite common regulation patterns, two main groups can be distinguished among plant pathogenic bacteria on the basis of the regulatory components controlling the expression of hrp genes. In fact, this dichotomy illustrates the existence of two lineages of hrp clusters in terms of overall similarity: Erwinia sp., Pantoea stewartii, and Pseudomonas syringae form group I, while Xanthomonas sp. and Ralstonia solanacearum constitute group II (2, 20). In group I organisms, several genes are required for the activation of the hrc, hrp, and TTSS effector genes (see reference 30 and references therein), with the final component of the regulatory cascade being HrpL, a member of the ECF family of alternative sigma factors (48, 49). For bacteria from group II, genetic analyses have shown that transcriptional activation of the TTSS machinery in R. solanacearum relies on an unrelated and complex signaling cascade (see reference 39 for a review), with the final component being an AraC family regulator named HrpB for this bacterium (18) and HrpX for Xanthomonas sp. (47).
R. solanacearum, the causal agent of bacterial wilt, has an extensive host range, including over 450 plant species in tropical and warm temperate zones worldwide (36). In this bacterium, hrpB encodes one of the key regulatory genes controlling pathogenicity functions (18, 39) (Fig. 1). In response to environmental cues (minimal medium or bacterium-plant cell contact), the expression of hrpB is activated (1), and HrpB in turn induces the expression of at least five operons, including the TTSS genes and some of the Hrp-dependent substrates (3, 18). To date, little is known at the molecular level about the interactions that lead from environmental signals to the activation of the genes encoding the TTSS and its secreted substrates. In Xanthomonas campestris, a conserved DNA motif, the so-called plant inducible promoter (PIP) box (14), has been identified in several hrp promoters and proposed to be important for HrpX-dependent regulation, but no functional characterization of this element has been reported. In fact, the role of the PIP box as a control element remains speculative since some HrpX-regulated promoters do not contain PIP boxes (8).
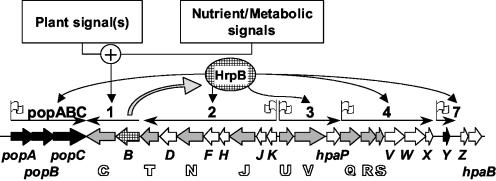
FIG. 1.
General organization of the R. solanacearum hrp gene cluster according to Van Gijsegem et al. (45). Thick arrows symbolize individual genes. Single letters refer to hrp genes, while gray arrows and black outlined letters represent hrc genes. The arrow filled with a grid represents a regulatory gene. Numbers above thin arrows indicate transcriptional units. Flags indicate the presence of a Xanthomonas PIP box-like motif in the respective promoter.
The examination of known hrpB-regulated promoter sequences in R. solanacearum revealed the presence of several PIP box-related motifs at highly variable distances from the translation start codon (up to 330 bp upstream in the case of the popABC operon) (14). Therefore, the aim of this work was to identify the cis-regulatory sequences conferring HrpB-dependent activation and to characterize these elements in order to define a valid consensus for R. solanacearum. We thus concentrated our study on two promoters of known hrpB-regulated genes, namely hrpY, encoding the major constituent of the Hrp pilus (46), an essential component of the TTSS apparatus required for the translocation of effectors into plant cells, and the popABC operon, which encodes three TTSS substrates (3, 21). Our final goal was to perform a bioinformatic screen by using the consensus motif of the putative HrpB operator sequence and the recently completed genome sequence of R. solanacearum strain GMI1000 (37) as a first step for identifying the repertoire of HrpB-dependent pathogenicity effectors in this bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used for this study are described in Table 1. Escherichia coli cells were grown in Luria-Bertani medium (38) at 37°C. R. solanacearum was grown in complete medium B or minimal medium supplemented with 20 mM glutamate (7). Antibiotics were used at the following concentrations: tetracycline, 10 mg/liter; ampicillin, 50 mg/liter; streptomycin, 50 mg/liter; gentamicin, 6 mg/liter. Cocultivation of the bacteria with plant cells was done according to the procedure of Marenda et al. (28).
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant genotype or characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−recA lacZΔM15 | BRL |
| R. solanacearum | ||
| GMI1000 | Wild-type strain | 7 |
| GMI1525 | hrpB::Ω mutant | 18 |
| Plasmids | ||
| pGEM-T | Cloning vector, Ampr | Promega |
| pBluescript II SK(−) | Cloning vector, Ampr | Stratagene |
| pLAFR6 | pLAFR1 with trp terminators, Tcr | 25 |
| pCZ227 | popABCp::lacZ fusion, −643 to +272 PCR fragment cloned on a pUC19 backbone, Ampr | This work |
| pSC131 | Promoterless lacZ reporter gene vector, pVS1ori, Genr | This work |
| pCZ388 | Promoterless lacZ reporter gene cloned in pLAFR6 | This work |
| pCZ209 | popABCp::lacZ fusion, −643 to +272 PCR fragment, cloned in pLAFR6 | This work |
| pCZ368 | popABCp::lacZ fusion, −215 to + 272 ExoIII fragment, cloned in pLAFR6 | This work |
| pSC151 | popABCp::lacZ fusion, −70 to +272 PCR fragment, cloned in pLAFR6 | This work |
| pCZ353 | popABCp::lacZ fusion, −57 to +272 ExoIII fragment, cloned in pLAFR6 | This work |
| pCZ369 | popABCp::lacZ fusion, +154 to +272 ExoIII fragment, cloned in pLAFR6 | This work |
| pSC146 | hrpYp::lacZ fusion, −121 to +122 PCR fragment cloned in pSC131 | This work |
| pSC152 | hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pSC157 | hrpYp::lacZ fusion, −44 to +122 PCR fragment cloned in pSC131 | This work |
| pSC156 | hrpYp::lacZ fusion, +4 to +122 PCR fragment cloned in pSC131 | This work |
| pSC143 | Mutated hrpYp::lacZ fusion, −121 to +122 PCR fragment cloned in pSC131 | This work |
| pSC144 | Mutated hrpYp::lacZ fusion, −121 to +122 PCR fragment cloned in pSC131 | This work |
| pSC145 | Mutated hrpYp::lacZ fusion, −121 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ475 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ472 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ473 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ478 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ474 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ444 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ460 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ445 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ446 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ476 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
| pCZ494 | Mutated hrpYp::lacZ fusion, −70 to +122 PCR fragment cloned in pSC131 | This work |
Numbering is relative to the transcription start point of the corresponding gene. Ampr, ampicillin resistance; Genr, gentamicin resistance; Tcr, tetracycline resistance.
DNA manipulations.
Standard recombinant DNA techniques were performed as described previously (38). Restriction enzymes, DNA ligase, the Klenow fragment, and other DNA enzymes were used according to the manufacturer's recommendations. PCR amplifications were done in 50-μl volumes with the Expand Long Template PCR system (Roche Molecular Systems), using genomic DNA of R. solanacearum strain GMI1000, unless otherwise stated. Sequences of oligonucleotide primers are available upon request.
Construction of reporter plasmids. (i) popABCp reporter constructs.
The popABC promoter region (915 bp) corresponding to positions −643 to +272 (relative to the transcription start point) was PCR amplified with primers DPOP1 and DPOPRev and cloned into the pGEM-T vector (Promega). This upstream region was subsequently cloned as a HindIII-XbaI fragment into a pBluescript II SK(−)-based vector containing the lacZ coding sequence from the Tn5-B20 transposon (40) as an XbaI-XhoI fragment, thus generating a popABCp::lacZ transcriptional fusion in the resulting plasmid, pCZ227. For the creation of progressive 5′ deletions of the popABC promoter, an exonuclease III-based Erase-a-Base kit (Promega) was used on pCZ227 digested with SphI and SalI according to the manufacturer's instructions. After sequencing, HindIII-KpnI fragments encompassing deleted promoter sequences fused to lacZ from pCZ227 and three selected derivatives of this plasmid were cloned into pLAFR6 to give pCZ209, pCZ368, pCZ353, and pCZ369. Plasmid pCZ388 corresponds to a control gene vector that was created by cloning lacZ alone in pLAFR6. For the construction of a popABC promoter fragment that started at the first nucleotide of the hrpII box, pSC151 was obtained by the same procedure as that used for pCZ209, except that the upstream primer was DPOP2.
(ii) hrpYp reporter constructs.
Plasmid pSC131 was created by cloning the lacZ reporter cassette from pCZ205 (27) as a HindIII-KpnI fragment in a pPROBE-GT backbone (31). 5′ deletions of the hrpY promoter were constructed by using PCR to amplify fragments with a shared 3′ end and different lengths of 5′ promoter sequence. Forward PCR primers DY1, DY3, DY5, and DY8 associated with DYRev as the reverse primer amplified hrpYp fragments with 5′ ends corresponding, respectively, to positions −121, −70, −44, and +4 relative to the transcription start point of hrpY (as described below) and with identical 3′ ends corresponding to the second nucleotide of the initiation codon (+122 relative to the transcription start), followed immediately by a BamHI site.
PCR fragments were first cloned into the pGEM-T plasmid, and their sequences were verified. Inserts were subcloned into the polylinker of pSC131, giving rise to pSC146, pSC152, pSC157, and pSC156, in which the first codon of hrpY was fused with the lacZ open reading frame.
(iii) hrpYp site-directed mutagenesis.
Point mutations in the −121 to +122 fragment of the hrpY promoter were generated by a recombinant PCR approach (23) using mutagenic primers. In the first round of PCR, pSC146 served as the DNA template. Vector-specific primer pPROBE-F with DY16 (wild-type sequence) or DY14 (mutated sequence) amplified the upstream promoter region. Similarly, LacZ-R with DY13 or DY15 amplified the downstream promoter region. Partially overlapping upstream and downstream promoter regions were mixed in a second PCR that used LacZ-R and pPROBE-F to amplify recombinant full-length −121 to +122 fragments of the hrpY promoter with the desired mutations. Engineered DNA fragments were gel purified, digested with HindIII and BamHI, and cloned into pSC131 to generate pSC143, pSC144, and pSC145 (see Tables 1 and 2 for details on individual plasmids).
TABLE 2.
Mutational analysis of the hrpII box direct repeats of hrpYpa
| Plasmid | hrpII box and flanking region sequenceb | β-Galactosidase activity (Miller units [SD])c
|
Induction ratiod | |
|---|---|---|---|---|
| Wild type | hrpB | |||
| pSC146 | TTCGTACGCTTGCACAAGGTTTCGG | 740 (31) | 43 (4) | 17.2 |
| pSC143 | AG....................... | 100 (24) | 59 (4) | 1.7 |
| pSC144 | ....................AG... | 81 (15) | 46 (6) | 1.8 |
| pSC145 | AG..................AG... | 55 (9) | 56 (12) | 1.0 |
Replacement of the thymine doublets with AG in either one or both hrpII box repeats of hrpYp was introduced into pSC146-based lacZ reporter plasmids, and the plasmids were assayed for β-galactosidase activity in a wild-type or hrpB mutant strain of R. solanacearum after overnight culturing in inducing minimal medium.
Nucleotides in bold indicate conserved residues of hrpII box tandem repeats. Dots indicate conserved residues at the given positions in pSC146-derived plasmids.
β-Galactosidase activities assayed in the wild-type strain (GMI1000) and a hrpB mutant (GMI1525) are means of at least three experiments.
Ratio of activity in the wild-type strain to activity in the hrpB mutant strain.
With pSC152 as a template, mutated −70 to +122 fragments of the hrpY promoter were PCR amplified with a forward mutagenic primer and the LacZ-R reverse primer to hybridize between 60 and 80 bp downstream of the lacZ start codon. Derivatives of pSC152, namely pCZ475, pCZ472, pCZ473, pCZ478, pCZ474, pCZ444, pCZ460, pCZ445, pCZ446, pCZ476, and pCZ494, were obtained by cloning of these PCR fragments after they were digested by PstI and BamHI into pSC131 (see Tables 1 and 3 for details on individual plasmids). Each promoter had its insert sequenced on both strands to ensure the proper nucleotide sequence.
TABLE 3.
Mutational analysis of the hrpYp hrpII box flanking regionsa
| Location of mutation(s) | Plasmid | hrpII box and flanking region sequenceb | β-Galactosidase activity (Miller units [SD])c
|
Induction ratiod | |
|---|---|---|---|---|---|
| Wild type | hrpB | ||||
| 5 10 15 20 25 30 | |||||
| ....|....|....|....|....|....|.... | |||||
| Flanking sequences | pSC152 | TTCGTACGCTTGCACAAGGTTTCGGGGCAGCGG | 728 (49) | 18 (1) | 40.4 |
| pCZ475 | ........................A........ | 155 (5) | 20 (1) | 7.8 | |
| pCZ472 | ....A............................ | 91 (8) | 18 (2) | 5.1 | |
| pCZ473 | ..............A.................. | 372 (16) | 21 (4) | 17.7 | |
| pCZ478 | ...................G............. | 583 (42) | 32 (10) | 18.2 | |
| pCZ474 | .......CT.GTGC..GC............... | 371 (25) | 17 (1) | 21.8 | |
| Repeat spacing | pCZ444 | TTCGTA--CTTGCACAAGGTTTCGGGGCAGCGG | 15 (2) | 16 (1) | 0.9 |
| pCZ460 | TTCGTAC-CTTGCACAAGGTTTCGGGGCAGCGG | 54 (8) | 19 (5) | 2.8 | |
| pCZ445 | TTCGTACGGCTTGCACAAGGTTTCGGGGCAGCGG | 141 (17) | 18 (1) | 7.8 | |
| pCZ446 | TTCGTACGCGCTTGCACAAGGTTTCGGGGCAGCGG | 19 (3) | 20 (2) | 1.0 | |
| Distance between hrpII and −10 boxes | pCZ476 | TTCGTACGCTTGCACAAGGTTTCGGATATGGCAGCGG | 21 (2) | 19 (3) | 1.1 |
| pCZ494 | TTCGTACGCTTGCACAAGGTTTCGGGG---CGG | 28 (3) | 18 (3) | 1.6 | |
Derivatives of the pSC152 reporter plasmid with mutations in DNA regions downstream of the hrpII box repeats of minimal hrpYp were obtained by PCR and tested for transcriptional activity in R. solanacearum.
Nucleotides in bold indicate conserved residues of hrpII box tandem repeats. Dots symbolize the conserved residues in the pSC152-derived plasmids. Dashes indicate that the corresponding nucleotide in the wild-type sequence has been deleted from the synthetic promoter construct. Underlined nucleotides have been added in the regions flanking hrpII box repeats.
β-Galactosidase activities assayed for the wild-type strain (GMI1000) and a hrpB mutant (GMI1525) are the means of at least three experiments.
Ratio of activity in the wild-type strain to activity in the hrpB mutant strain.
Bacterial transformation.
pLAFR6- and pSC131-based plasmid constructs were introduced into R. solanacearum strains by electroporation (2.5 kV, 200 Ω, 25 μF, 0.2-cm cuvette gap).
β-Galactosidase assays.
β-Galactosidase assays were performed in R. solanacearum cultures grown under activating (minimal medium supplemented with 20 mM glutamate) or nonactivating (complete medium) conditions as described previously (3).
Extraction of total RNA from R. solanacearum.
The total RNA was extracted from R. solanacearum strain GMI1000 grown in complete medium or minimal medium or cocultured with an Arabidopsis thaliana Col-0 cell suspension. Bacterial pellets from 20 ml of culture at an optical density at 600 nm of 0.6 were homogenized in 1 ml of the commercial buffer Extract All (Eurobio Laboratories) and were treated as recommended by the manufacturer. Five microliters of DNAse I from Boehringer Mannheim and 12 μl of buffer provided with the enzyme were added to the total RNA, which was resuspended in 100 μl of water. After 30 min of incubation at 28°C, an extra 2 μl of enzyme was added and the reaction proceeded for 30 min. After phenol-chloroform purification and ethanol precipitation, the RNA pellet was resuspended in 100 μl of distilled water.
Primer extension analysis.
Oligonucleotide hrpY + 1 (400 ng) was 5′ end labeled with 5 U of T4 polynucleotide kinase (Invitrogen) and 555 × 1010 Bq of [γ-32P]ATP. An equivalent of 5 × 105 cpm of MicroSpin (Sephadex G50)-purified oligonucleotide was hybridized to 10 μg of total RNA in 5 μl of a solution containing 50 mM Tris-HCl (pH 8.2), 10 mM dithiothreitol (DTT), and 60 mM NaCl by heating at 90°C for 1 min on a thermoblock and then cooling on the bench. When the temperature reached 40°C, 1 μl of MgCl2 (36 mM) was added to the mix. For reverse transcription (RT) reactions, 2.5 μl of hybrid solution was extended for 30 min at 45°C with 200 U of SuperScript II (Invitrogen), 1 μl of deoxynucleoside triphosphates (25 mM), 0.5 μl DTT (100 mM), and 1 μl of the buffer provided with the enzyme. Extension products were analyzed in a 6% polyacrylamide sequencing gel together with dideoxy chain termination reactions made with a T7 sequencing kit (USB Corp.), oligonucleotide primer hrpY + 1, and a preparation of plasmid DNA template pGMI1989 (45).
5′-RACE.
To determine the 5′ ends of mRNA transcripts, we also performed rapid amplification of cDNA 5′ ends-PCR (5′-RACE-PCR) with a modified protocol from Tillett et al. (43). Briefly, RT reactions were performed in 50-μl volumes containing a 0.2 mM concentration of each deoxynucleoside triphosphate, 10 mM DTT, 126 pmol of random hexamer primers (except for popABC, for which 20 pmol of the gene-specific oligonucleotide pop + 1 was used), and 2 μg of total RNA from R. solanacearum GMI1000 grown in minimal medium. Initial experiments with hrpY and hrp transcription unit 2 used a thermally cycled RT procedure (43) in which 200 U of SuperScript II (Invitrogen) was added to the reaction after stepwise annealing of the primers, followed by incubation at 42°C for 30 min and five cycles at 50°C for 1 min, 53°C for 1 min, and 56°C for 1 min. More stringent temperature parameters (50°C for 30 min and five cycles at 53°C for 1 min, 56°C for 1 min, and 59°C for 1 min) and the addition of 300 U of reverse transcriptase were used in the case of popABC and hrp transcription unit 3. All experiments included an RT negative control in which no enzyme was added to the reaction mix to ensure that RACE-PCR products were indeed derived from mRNA.
RNAs were removed by RNase H digestion at 37°C for 30 min. cDNAs were purified through MicroSpin S-400 HR (Amersham Pharmacia) columns. Ligation of the anchor oligonucleotide DT88 (43) was done overnight at 18°C with 10 μl of cDNA sample and 3 U of T4 RNA ligase (Promega).
Ligation-anchored PCRs were performed with 10 μl of DNA template ligation products, using anchor-specific oligonucleotide DT89 (10 pmol) (43) and gene-specific primer hrpY + 1, pop + 1c, U2-R1, or U3-R1 (10 pmol), corresponding to hrpY, popABC, hrp unit 2, and hrp unit 3, respectively. For the latter transcriptional unit, a second heminested PCR using primer pair DT89 and U3-R2 and 5 μl of a 500-fold dilution of the first PCR was required to obtain a specific amplification product. Thermal cycler program parameters were as follows: a 94°C initial denaturation for 2 min before the addition of Taq DNA polymerase; 15 cycles at 94°C for 10 s, 70°C − 1°C/cycle for 30 s, and 68°C for 30 s; 25 cycles at 94°C for 10 s, 55°C for 30 s, and 68°C for 30 s; and a final extension at 68°C for 7 min. PCR amplification products were separated in a 2.5% agarose gel. The DNA bands that were absent from negative control reactions were gel purified and cloned into the pGEM-T vector. Transcription starts were derived from insert sequences of at least seven independent clones that were randomly selected after plating of E. coli DH5α cells transformed with ligation products.
Computer analysis.
An inventory of the hrpII box motif (TTCGN16TTCG) in the genome sequence of R. solanacearum strain GMI1000 was performed with PatScan software (http://www-unix.mcs.anl.gov/compbio/PatScan/HTML/patscan.html). DNA sequence manipulation, alignment, editing, and formatting for publication were done with Bioedit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
RESULTS
5′ deletions of the popABC and hrpY promoters identify regions essential for HrpB-dependent activation.
For identification of the cis-acting regions responsible for the regulation of the transcription of hrpB-regulated genes, the promoter regions of the popABC operon (popABCp) and the hrpY gene (hrpYp) were cloned upstream of a promoterless lacZ gene in a reporter plasmid. The β-galactosidase activity was assayed for each construct in the wild-type strain GMI1000 or in the hrpB mutant strain GMI1525 after growth in complete medium (noninductive) or minimal medium (inductive) (Fig. 2). We first checked that the reporter plasmids pCZ209 and pSC146, which contain popABCp (915 bp) and hrpYp (241 bp), respectively, were both activated in minimal medium in a strictly hrpB-dependent manner. The promoter sequence in pSC146 corresponds to the DNA fragment that was used for trans-complementation experiments with the hrpY mutant strain (45). 5′ nested deletions of popABCp were generated by exonuclease III digestion of pCZ209 DNA, except for one construct (pSC151) carrying a promoter fragment starting at position −70, which was synthesized by PCR. 5′ deletions of hrpYp were also generated by PCR amplification.
FIG. 2.
Transcriptional activities of the popABC (A) and hrpY (B) promoters and deletion fragments. 5′ nested deletions of popABCp and hrpYp were cloned into a promoter-probe plasmid, and their abilities to promote transcription of the lacZ reporter gene were monitored by β-galactosidase assays. For each deletion, the coordinate of its 5′ end relative to the transcription start point of the relevant gene and the corresponding reporter plasmid name are shown to the left. Light gray bars delineate promoter regions, black bars indicate hrpII box locations, and dark gray bars represent lacZ reporter sequences. β-Galactosidase activity values for each construct in R. solanacearum wild-type strain GMI1000 and a hrpB mutant (GMI1525) after overnight growth in minimal or complete medium are shown to the right and are the means of at least three independent experiments, with error bars representing standard deviations.
The identification of deletions that abolished lacZ expression revealed that the element conferring HrpB-dependent activation encompasses a 13-bp fragment in popABCp and a 26-bp region in hrpYp (Fig. 2). A DNA sequence comparison of these two regions led to the identification of a conserved motif consisting of two perfect four-nucleotide direct repeats (TTCGn16TTCG), which in fact corresponds to an imperfect PIP box motif (TTCGCn15TTCGC) as originally described for Xanthomonas (14). In popABCp, the loss of HrpB-dependent activity was associated with the pCZ353 construct starting at the ninth nucleotide after the first TTCG repeats (Fig. 2A). The deletion of this 26-bp motif from the pSC157 construct clearly abolished the HrpB-dependent activity of hrpYp observed in pSC146 (Fig. 2B). This demonstrated that this 26-nucleotide element, henceforth referred to as a hrpII box (for reasons detailed below), is required for HrpB-activated transcription.
Mutational analysis reveals the importance of hrpII box direct repeats for hrpY promoter function.
For an investigation of the potential role of the hrpII box in the regulation of hrpYp, mutations leading to the replacement of each TT repeat with an AG doublet were generated. The β-galactosidase activities expressed by recombinant plasmids (pSC143, pSC144, and pSC145) were compared to that for plasmid pSC146, which carries a similar promoter fragment with an intact hrpII box (Table 2). These assays indicated that each conserved TT repeat of the hrpII box is essential for the activation of hrpYp since each TT→AG substitution totally abolished the β-galactosidase activity of the reporter fusion. These mutations prevented the ability of hrpYp to respond to HrpB.
In order to more accurately define the important positions in the hrpII box motif, we generated site-directed mutations that affected the nucleotides in the vicinity of the conserved TTCG repeats (T-to-A change at position 5, T-to-G change at position 20, and G-to-A change at position 25). In addition, we created two other constructs to evaluate the importance of nucleotide positions within the 16-bp spacer region in between the TTCG repeats: one carried a single C-to-A substitution at position 15 and the second had eight internal nucleotides changed (positions 8, 9, 11, 12, 13, 14, 17, and 18). The activities of the hrpYp-lacZ constructs with the various mutated hrpII boxes described above are shown in Table 3. A single substitution in pCZ472 (T to A, at position 5) or pCZ475 (G to A, at position 25) affected the hrpB-dependent regulation of hrpYp but did not abolish its potential to activate transcription since an induction ratio of five- to eightfold was still observed. In contrast, constructs pCZ473, pCZ474, and pCZ478 did not reach the level of activity of the wild-type promoter but retained the ability to confer hrpB dependency (induction ratio ranging from 18- to 22-fold). We concluded, therefore, that the 16-nucleotide spacer region does not contain crucial residues mediating HrpB activation, except maybe positions 5 and 25 immediately downstream of each conserved TTCG repeat.
Spacing of TTCG direct repeats is critical for hrpY promoter function.
We then addressed the question of whether the 16-bp spacing between the TTCG repeats was an important parameter for the HrpB-dependent activation of hrpYp. For this purpose, derivatives of pSC152 carrying 14 (deletion of positions 7 and 8)-, 15 (deletion of position 8)-, 17-, and 18-bp spacer regions were tested (Table 3).
The β-galactosidase activities monitored in these constructs clearly showed that alterations of the 16-bp spacing dramatically influenced the ability of hrpYp to respond to HrpB (Table 3). HrpB-dependent activation was lost in constructs harboring a hrpII box with 14-, 15-, or 18-bp spacing. However, the 17-bp-spacing construct (pCZ445) retained a residual activation potential (induction ratio of eightfold), although it was significantly decreased compared to that with wild-type 16-bp spacing.
Four HrpB-regulated promoters have related structures.
For determination of the transcription start point of hrpYp, RNA was prepared from the wild-type strain grown under inductive (minimal medium, cocultivation with plant cell suspension) or noninductive (complete medium) conditions and was used for primer extension analysis. We used a primer complementary to the region flanking the hrpY translation start site for an RT reaction. Two primer extension products of equal intensities and differing by 37 nucleotides in length were detected when cells were grown under inductive conditions, but no product was found under repressive conditions (Fig. 3A). The shorter cDNA may have originated from an mRNA transcribed from a second promoter. However, in the absence of an obvious sigma factor binding element within this region, we hypothesized that this cDNA came from the premature termination of RT due to the presence of some secondary structure in the RNA transcript 5′-untranslated region or from RT of a hrpY mRNA major degradation product. We therefore believe that the longer product most likely represents the legitimate transcription start site.
FIG. 3.
Mapping of the transcription start sites of the hrpY, popABC, and hrp transcription unit 2 and 3 promoters. (A) Primer extension products of the hrpY promoter. The sequence indicated is the antisense (bottom) strand. Asterisks indicate the transcription start sites of the two extension products. RT was done with RNA from the wild-type strain grown in minimal medium (lane 1), in the presence of Arabidopsis plant cells (lane 2), or in complete medium (lane 3). An ethidium bromide-stained denaturing agarose gel loaded with the samples of the total RNA extracts used for RT is inserted below the appropriate gel tracks. (B) Sequence alignment of the hrpY, popABC, and hrp transcription unit 2 and 3 promoters. The last adenine in each sequence (underlined) is the transcription start as determined by 5′-RACE-PCR (for hrpY, the transcription start corresponds to the longest primer extension product) and was used as the origin for numbering of the sequence ruler. Numbers to the right of sequences represent distances from the initiation codons. The gray shaded box shows the position of the −10 hexamer. Open boxes indicate the conserved direct repeats of the hrpII box.
5′-RACE allowed the transcription initiation site for most HrpB-regulated operons, namely hrpY, popABC, and hrp transcription units 2 and 3, to be mapped (Fig. 3B). Regarding the 5′ end of the hrpY mRNA, this technique gave results identical to those obtained by primer extension analysis: the nested PCR resulted in two bands and the subsequent cloning and sequencing of these fragments revealed the same transcription start sites as those found above. The popABC mRNA was found to have a long 5′-untranslated region (272 bp), while for hrp transcription units 2 and 3, this region spans 63 and 55 bp, respectively. The alignment shown in Fig. 3B shows that for all mapped promoters, the distance between the transcription start and the hrpII box is well conserved, with a spacing of 47 ± 1 bp. A second region of homology was also apparent which resembled the −10 binding element of the RNA polymerase σ70 factor. These observations support the view that HrpB-regulated promoters share a common architecture. Remarkably, no −35 consensus motif could be predicted from the sequence alignment.
Functionality of the hrpYp hrpII box depends on its position relative to the −10 element.
To test whether the position of the hrpII box in the hrpYp promoter (−47 bp) is important for gene expression, we generated two constructions in which the position of the hrpII box relative to the +1 transcription start point was modified. pCZ476, a derivative of pSC152, carries a 4-bp insertion just downstream of the hrpII box (thus positioned at −51 bp), and pCZ494 carries a 3-bp deletion leading to the positioning of the hrpII box at −44 bp from the transcription start site. As shown in Table 3, such alterations of the position of the hrpII box in hrpYp dramatically reduced the HrpB-dependent activation of the promoter. The hrpYp hrpII box activating element must thus be exactly positioned relative to the transcription initiation site and hence to the presumed core promoter regions.
Computer-assisted identification of putative HrpB-regulated genes in the R. solanacearum genome sequence.
R. solanacearum depends on the Hrp TTSS for the successful invasion of its hosts. Effector proteins translocated into host plant cells through this system may act collectively to promote disease or may be required for specific host infections. HrpB is known to control several Hrp secretion system substrates at the transcriptional level (3, 21). In order to enlarge our knowledge of hrpB regulon members and thus to enrich the candidate effector repertoire for this bacterium (37), we used the hrpII box motif to perform a systematic search for this element in the genome sequence of R. solanacearum strain GMI1000. We selected the hits matching the hrpII box consensus sequence which were located in a region extending from 50 to 450 bp upstream of the start codon of each predicted coding sequence. In addition to the five known HrpB-dependent hrp or popABC promoters, the search produced a total of 95 hits. Hits were more frequent on the megaplasmid (2.1 Mb; 57 hits) than on the chromosome (3.7 Mb; 39 hits). These 95 transcriptional units were estimated to comprise 114 genes (the complete list is given in Table S1 in the supplemental material). Four genes corresponded to apparently nonfunctional pseudogenes, as they included coding sequence frameshifts or transposase insertions; interestingly, one of them was predicted to encode a member of the AvrRxv/YopJ family of TTSS effectors (42; M. Lavie, B. Sennes, P. Prior, and C. Boucher, submitted for publication).
An analysis of the homologies displayed by the 110 genes (excluding pseudogenes) which were found to be driven by the putative promoter regions containing a hrpII box indicated that 19 are most likely to be involved in pathogenicity-related functions. They code for proteins homologous to several type III-associated proteins that were already described for other bacterial pathogens, such as the avr- and hop-encoded proteins of P. syringae (11). Two genes encoding homologs of HrpF, a probable type III translocon protein from X. campestris pv. vesicatoria (9, 10), also contain a hrpII box in their upstream DNA regions. Interestingly, 14 gene products encoding R. solanacearum candidate type III effectors, identified previously on the basis of the presence of domains which could be predictive of eukaryotic functions (37), were also identified in our in silico screen. For example, five genes encoding predicted leucine-rich repeat domains, protein-protein interaction domains commonly found in various pathogenicity determinants in the prokaryotic kingdom (4, 21, 41), contained a hrpII box in their promoter.
DISCUSSION
We have provided experimental evidence that the 25-bp bipartite nucleotide element TTCGn16TTCG, which corresponds to an imperfect PIP box originally described for a Xanthomonas sp. (14), is required for the hrpB-dependent activation of both hrpY and popABC promoters. This nucleotide element was named the hrpII box (for hrp box in group II hrp clusters, i.e., R. solanacearum and Xanthomonas sp.), referring to the term “hrp box” which is used for the consensus motif found in group I hrp promoters controlled by the alternative sigma factor HrpL (15, 49). The hrpII box is found in all of the other HrpB-dependent promoters known so far. Sequence alignments of these HrpB-regulated promoters clearly illustrated the strict conservation of the direct TTCG repeats, which suggests that these four nucleotide repeats are crucial for functioning. The functional analysis of the hrpYp hrpII box confirmed this hypothesis: substitutions of the conserved TT nucleotides abolished hrpYp activation, while other substitutions of nonconserved nucleotides only had a minor effect on the promoter activity. Mutations at the first nucleotide 3′ of either of the two hrpII box repeats, however, measurably altered the promoter responsiveness. These residues correspond to the last conserved cytosine of the PIP box repeats of X. campestris, which suggests that although there is little conservation in R. solanacearum, this position is likely to play a modulating role either on DNA topology or on the transcription activator affinity for its target sequence, thereby altering hrpB-dependent transcriptional stimulation.
We hypothesize that the tandem direct repeats of the hrpII box comprise a HrpB binding site. This hypothesis is supported by the fact that several AraC family regulators are known to activate transcription by binding to target sequences featuring direct repeats and located adjacent to or overlapping the −35 regions of regulated class I promoters (16), such as MixE and HilD, which are AraC-like TTSS substrate-related regulators in Shigella flexneri (29) and Salmonella enterica serovar Typhimurium (6), respectively. However, since we have not yet been able to demonstrate the direct binding of HrpB to the hrpY promoter sequence (S. Cunnac, unpublished data), we cannot exclude a more complex mechanism in which another partner participates to produce an active binding complex. A similar observation was made for the orthologous Xanthomonas regulator HrpX, for which no binding to PIP box-containing promoters could be detected (8). Both HrpB and HrpX are AraC family regulators (13, 44), and the defining characteristic of this family of proteins is a C-terminal amino acid region of homology comprising the DNA binding domain which exhibits two helix-turn-helix (HTH) motifs (16). In the case of AraC and XylS, the two best characterized regulators of the family, it has been shown that each HTH motif binds to specific submotifs which are separated by 6 or 7 bp in the target sequence (19, 32). On the basis of these results, although the spacing between the two direct TTCG repeats is more important, it is tempting to speculate that each HTH motif in HrpB (located at positions 390 to 413 and 443 to 467) recognizes one-half of the motif of the hrpII box.
We investigated the hrpII box function by using site-directed mutagenesis in order to establish a robust consensus and by using this nucleotide motif to search for candidate HrpB-regulated genes in the GMI1000 genome. The strictly conserved nature of the hrpII box consensus in R. solanacearum hrp and pop promoters enabled us to scan the complete genome of strain GMI1000 for candidate HrpB-regulated genes by using this sequence. Remarkably, 14 of 20 candidate type III-dependent effectors originally identified on the basis of protein sequence homology (37) harbor a canonical hrpII box in their promoter regions. This observation is in agreement with our results showing that the 16-bp spacing of the TTCG repeats is important for the full activation of hrpYp. In contrast, in Xanthomonas sp., more variable spacing between the PIP box direct repeats can be observed for some HrpX-dependent promoters (26, 33). However, we showed that a 17-bp spacing strongly reduces but does not abolish hrpYp expression, and it is therefore possible that we overlooked other potential HrpB-regulated genes. Such variations in the structure of the hrpII box may conceivably represent a way of modulating the expression of certain type III effector genes.
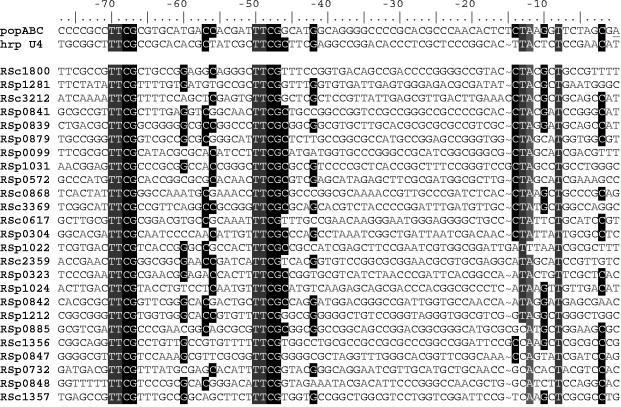
The 95 candidate HrpB-regulated promoters identified in this study were aligned by using the hrpII motif as an anchor. Seventy-three sequences contained the last conserved thymine of the −10 box at a distance of 37 ± 1 nucleotides from the fourth residue of the hrpII box. For this subset of promoters, 68% had a pyrimidine nucleotide at the first position of the −10 box hexamer and 71% had an adenine at the second position. Figure 4 shows the sequence alignment of two known hrp promoters (with popABCp and hrp transcription unit 4 anchoring the alignment) with 25 candidate HrpB-regulated promoters identified in this study, a pool composed of 7 genes encoding products homologous to known type III effectors and 18 genes encoding conserved hypothetical or unknown proteins. In addition to the hrpII box, this alignment highlights another conserved region, namely an appropriately positioned putative −10 region (as determined for promoters for which the transcription start point was available). This conserved region is homologous to the −10 sequence found for the R. solanacearum AW1 epsA promoter (TACACT), in which both conserved thymine residues are essential for its function (17). This finding is in agreement with the results from our hrpYp analysis showing that the position of the hrpII box relative to the −10 element is a critical parameter for promoter activation. The alignment in Fig. 4 also supports the observation that there is no clear nucleotide preference in positions outside of the conserved TTCG repeats, except maybe for position 5 (never adenine) and position 25 (never adenine and mainly cytosine).
FIG. 4.
Alignment of upstream regions of several hrpB-regulated candidate genes. Nucleotides that are highlighted at a given position in the alignment are conserved in at least 50% of the sequences.
The global search for regulatory targets thus appears to be an interesting tool for the identification of novel virulence-related genes. The functional analysis of most of these novel candidate genes has been undertaken and we now have experimental evidence that several of them indeed belong to the HrpB regulon (11a). This and other genome-based studies (5, 15, 22, 34, 35, 50) mark the beginning of the next step in research, which is aimed at making a complete inventory of pathogenicity factors in bacterial plant pathogens. Because various virulence functions are known to be subjected to complex regulatory cascades in R. solanacearum (12, 39), similar genome-wide searches for regulatory gene targets will certainly allow our understanding of this network to be developed. Altogether, this information will contribute to the identification of novel pathogenicity factors and will permit a far more developed global picture of the multiple determinants contributing to bacterial wilt disease.
Supplementary Material
Acknowledgments
We thank Steve Lindow for the gift of the pPROBE-GT vector, Anne-Marie Garnerone for help with primer extension experiments, and Pierre Boistard, Nemo Peeters, and Nigel Grimsley for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aldon, D., B. Brito, C. Boucher, and S. Genin. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19:2304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlat, M., F. Van Gijsegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bierne, H., and P. Cossart. 2002. InlB, a surface protein of Listeria monocytogenes that behaves as an invasin and a growth factor. J. Cell Sci. 115:3357-3367. [DOI] [PubMed] [Google Scholar]
- 5.Boch, J., V. Joardar, L. Gao, T. L. Robertson, M. Lim, and B. N. Kunkel. 2002. Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44:73-88. [DOI] [PubMed] [Google Scholar]
- 6.Boddicker, J. D., B. M. Knosp, and B. D. Jones. 2003. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J. Bacteriol. 185:525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher, C. A., P. A. Barberis, A. P. Trigalet, and D. A. Démery. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449-2457. [Google Scholar]
- 8.Buttner, D., and U. Bonas. 2002. Getting across—bacterial type III effector proteins on their way to the plant cell. EMBO J. 21:5313-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buttner, D., D. Nennstiel, B. Klusener, and U. Bonas. 2002. Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casper-Lindley, C., D. Dahlbeck, E. T. Clark, and B. J. Staskawicz. 2002. Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA 99:8336-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collmer, A., M. Lindeberg, T. Petnicki-Ocwieja, D. J. Schneider, and J. R. Alfano. 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10:462-469. [DOI] [PubMed] [Google Scholar]
- 11a.Cunnac, S., A. Occhialini, P. Barberis, C. Boucher, and S. Genin. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol., in press. [DOI] [PubMed]
- 12.Denny, T. P. 1999. Autoregulator-dependent control of extracellular polysaccharide production in phytopathogenic bacteria. Eur. J. Plant Pathol. 105:417-430. [Google Scholar]
- 13.Egan, S. M. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenselau, S., and U. Bonas. 1995. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol. Plant-Microbe Interact. 8:845-854. [DOI] [PubMed] [Google Scholar]
- 15.Fouts, D. E., R. B. Abramovitch, J. R. Alfano, A. M. Baldo, C. R. Buell, S. Cartinhour, A. K. Chatterjee, M. D'Ascenzo, M. L. Gwinn, S. G. Lazarowitz, N. C. Lin, G. B. Martin, A. H. Rehm, D. J. Schneider, K. van Dijk, X. Tang, and A. Collmer. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99:2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg, R. P., J. Huang, W. Yindeeyoungyeon, T. P. Denny, and M. A. Schell. 2000. Multicomponent transcriptional regulation at the complex promoter of the exopolysaccharide I biosynthetic operon of Ralstonia solanacearum. J. Bacteriol. 182:6659-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genin, S., C. L. Gough, C. Zischek, and C. A. Boucher. 1992. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol. Microbiol. 6:3065-3076. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Perez, M. M., J. L. Ramos, M. T. Gallegos, and S. Marques. 1999. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 274:2286-2290. [DOI] [PubMed] [Google Scholar]
- 20.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151-163. [DOI] [PubMed] [Google Scholar]
- 21.Guéneron, M., A. C. Timmers, C. Boucher, and M. Arlat. 2000. Two novel proteins, PopB, which has functional nuclear localization signals, and PopC, which has a large leucine-rich repeat domain, are secreted through the hrp-secretion apparatus of Ralstonia solanacearum. Mol. Microbiol. 36:261-277. [DOI] [PubMed] [Google Scholar]
- 22.Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler, and J. T. Greenberg. 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295:1722-1726. [DOI] [PubMed] [Google Scholar]
- 23.Higuchi, R. 1989. Using PCR to engineer DNA, p. 61-70. In H. A. Erlich (ed.), PCR technology: principles and applications for DNA amplification. M. Stock Press, Inc., New York, N.Y.
- 24.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynh, T. V., D. Dahlbeck, and B. J. Staskawicz. 1989. Bacterial blight of sojabean: regulation of a pathogen gene determining host cultivar specificity. Science 245:1374-1377. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J. G., B. K. Park, C. H. Yoo, E. Jeon, J. Oh, and I. Hwang. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavie, M., E. Shillington, C. Eguiluz, N. Grimsley, and C. Boucher. 2002. PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant-Microbe Interact. 15:1058-1068. [DOI] [PubMed] [Google Scholar]
- 28.Marenda, M., B. Brito, D. Callard, S. Genin, P. Barberis, C. Boucher, and M. Arlat. 1998. PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol. Microbiol. 27:437-453. [DOI] [PubMed] [Google Scholar]
- 29.Mavris, M., P. J. Sansonetti, and C. Parsot. 2002. Identification of the cis-acting site involved in activation of promoters regulated by activity of the type III secretion apparatus in Shigella flexneri. J. Bacteriol. 184:6751-6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merighi, M., D. R. Majerczak, E. H. Stover, and D. L. Coplin. 2003. The HrpX/HrpY two-component system activates hrpS expression, the first step in the regulatory cascade controlling the Hrp regulon in Pantoea stewartii subsp. stewartii. Mol. Plant-Microbe Interact. 16:238-248. [DOI] [PubMed] [Google Scholar]
- 31.Miller, W. G., J. H. Leveau, and S. E. Lindow. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant-Microbe Interact. 13:1243-1250. [DOI] [PubMed] [Google Scholar]
- 32.Niland, P., R. Huhne, and B. Muller-Hill. 1996. How AraC interacts specifically with its target DNAs. J. Mol. Biol. 264:667-674. [DOI] [PubMed] [Google Scholar]
- 33.Noel, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okinaka, Y., C. H. Yang, N. T. Perna, and N. T. Keen. 2002. Microarray profiling of Erwinia chrysanthemi 3937 genes that are regulated during plant infection. Mol. Plant-Microbe Interact. 15:619-629. [DOI] [PubMed] [Google Scholar]
- 35.Petnicki-Ocwieja, T., D. J. Schneider, V. C. Tam, S. T. Chancey, I. Shan, Y. Jamir, L. M. Schechter, M. D. Janes, C. R. Buell, X. Tang, A. Collmer, and J. R. Alfano. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99:7652-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prior, P., C. Allen, and J. Elphinstone (ed.). 1997. Bacterial wilt disease. Springer-Verlag, Berlin, Germany.
- 37.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Schell, M. A. 2000. Regulation of virulence and pathogenicity genes in Ralstonia solanacearum by a complex network. Annu. Rev. Phytopathol. 38:263-292. [DOI] [PubMed] [Google Scholar]
- 40.Simon, R., J. Quandt, and W. Klipp. 1989. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene 80:161-169. [DOI] [PubMed] [Google Scholar]
- 41.Skrzypek, E., T. Myers-Morales, S. W. Whiteheart, and S. C. Straley. 2003. Application of a Saccharomyces cerevisiae model to study requirements for trafficking of Yersinia pestis YopM in eucaryotic cells. Infect. Immun. 71:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staskawicz, B. J., M. B. Mudgett, J. L. Dangl, and J. E. Galan. 2001. Common and contrasting themes of plant and animal diseases. Science 292:2285-2289. [DOI] [PubMed] [Google Scholar]
- 43.Tillett, D., B. P. Burns, and B. A. Neilan. 2000. Optimized rapid amplification of cDNA ends (RACE) for mapping bacterial mRNA transcripts. BioTechniques 28:448-456. [DOI] [PubMed] [Google Scholar]
- 44.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Gijsegem, F., C. Gough, C. Zischek, E. Niqueux, M. Arlat, S. Genin, P. Barberis, S. German, P. Castello, and C. Boucher. 1995. The hrp locus of Pseudomonas solanacearum, which controls production of a type III secretion system, encodes eight proteins related to components of bacterial flagellar biogenesis complex. Mol. Microbiol. 15:1095-1114. [DOI] [PubMed] [Google Scholar]
- 46.Van Gijsegem, F., J. Vasse, J. C. Camus, M. Marenda, and C. Boucher. 2000. Ralstonia solanacearum produces hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol. 36:249-260. [DOI] [PubMed] [Google Scholar]
- 47.Wengelnik, K., and U. Bonas. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao, Y., S. Heu, J. Yi, Y. Lu, and S. W. Hutcheson. 1994. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J. Bacteriol. 176:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao, Y., and S. W. Hutcheson. 1994. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J. Bacteriol. 176:3089-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zwiesler-Vollick, J., A. E. Plovanich-Jones, K. Nomura, S. Bandyopadhyay, V. Joardar, B. N. Kunkel, and S. Y. He. 2002. Identification of novel hrp-regulated genes through functional genomic analysis of the Pseudomonas syringae pv. tomato DC3000 genome. Mol. Microbiol. 45:1207-1218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.