Abstract
Gene expression profiling of psoriasis has driven research advances and may soon provide the basis for clinical applications. For expression profiling studies, RNA-seq is now a competitive technology, but RNA-seq results may differ from those obtained by microarray. We therefore compared findings obtained by RNA-seq with those from eight microarray studies of psoriasis. RNA-seq and microarray datasets identified similar numbers of differentially expressed genes (DEGs), with certain genes uniquely identified by each technology. Correspondence between platforms and the balance of increased to decreased DEGs was influenced by mRNA abundance, GC content, and gene length. Weakly expressed genes, genes with low GC content, and long genes were all biased toward decreased expression in psoriasis lesions. The strength of these trends differed among array datasets, most likely due to variations in RNA quality. Gene length bias was by far the strongest trend and was evident in all datasets regardless of the expression profiling technology. The effect was due to differences between lesional and uninvolved skin with respect to the genome-wide correlation between gene length and gene expression, which was consistently more negative in psoriasis lesions. These findings demonstrate the complementary nature of RNA-seq and microarray technology and show that integrative analysis of both data types can provide a richer view of the transcriptome than strict reliance on a single method alone. Our results also highlight factors affecting correspondence between technologies, and we have established that gene length is a major determinant of differential expression in psoriasis lesions.
Keywords: degradation, gene expression, ribonuclease, RNase 7, RNase L
psoriasis is characterized by the development of skin lesions that arise from interactions of keratinocytes (KCs) with resident and infiltrating immune cells. During the last decade, great progress has been made toward understanding molecular-level features that differentiate psoriasis lesions from normal skin. This work has been facilitated by an accumulation of microarray datasets, which have allowed investigators to identify protein-coding genes with robustly altered expression between psoriasis lesions and uninvolved skin (9, 30, 43, 58, 62, 75). More recently, RNA-seq has been used to analyze the psoriasis transcriptome, providing an alternative strategy for characterizing expression shifts in psoriasis (and other skin diseases) (28, 31, 37, 73). Compared with microarrays, RNA-seq offers improved dynamic range and can quantify expression for the full complement of coding and noncoding mRNAs. For many diseases, therefore, the number of patient samples generated by RNA-seq should soon rival or exceed that available for microarrays. Nevertheless, statistical analysis methods for RNA-seq are still evolving (14, 16, 35, 36), and each expression profiling strategy (microarray and RNA-seq) entails a distinct set of biases, strengths, and weaknesses. To fully leverage these technologies, therefore, investigators will need to integrate findings from new RNA-seq datasets with existing microarray data (15), while identifying factors influencing consistency between the two expression profiling platforms.
Studies have compared RNA-seq and microarrays as alternative expression profiling strategies, but there remains uncertainty as to whether RNA-seq should be viewed as a general replacement of microarray technology (10). Direct comparison studies have often indicated good correspondence with respect to protein-coding genes (12, 44, 59, 74), leading some investigators to advance a balanced perspective in which RNA-seq and microarrays are viewed as complementary expression profiling strategies (10, 34). Others, however, have concluded that RNA-seq offers definite advantages in terms of accuracy and sensitivity, suggesting or implying that RNA-seq can effectively replace microarrays for most purposes (63, 77). A consensus point is that RNA-seq offers improved dynamic range and is better suited for evaluating transcripts with weak expression, in addition to high-abundance transcripts that would otherwise saturate signal intensities on microarray platforms (28, 37, 59). Still, in some contexts, microarray analysis may prove advantageous. First, for microarrays, preprocessing and low-level analyses are simplified, and since bioinformatic analysis effort is finite in practice, this means that bioinformatics work can be more targeted toward specific scientific questions (e.g., pathway analysis, comparisons with other data sources, etc.). Second, a decade's worth of microarray data are now available in public databases, and new microarray datasets can still be combined with such existing data without the introduction of platform-specific heterogeneity (15). It has also been suggested that, in the absence of cross-hybridization, expression estimates obtained for different genes by microarray are more independent of one another (51). This is due to the fact that, for microarrays, expression estimates for different genes are derived from separate probe sets and the influence of high-expressed genes is attenuated by signal saturation. For RNA-seq, this same saturation does not occur, and expression of any one gene may depend more heavily on other genes (particularly those with high expression). This would impact the validity of statistical methods that may assume expression is independently measured for each gene (e.g., multiple test correction, gene set enrichment analysis and coexpression analysis) (72).
Psoriasis provides a uniquely informative context for exploring the performance of RNA-seq compared with microarray analysis (28, 37). Several microarray datasets have been independently generated over the last decade, which include lesional skin samples from psoriasis patients (PP), uninvolved skin from psoriasis patients (PN), and/or normal skin from control subjects (NN) (9, 30, 43, 58, 62, 75). Several of these have utilized the same full-genome Affymetrix platform, and prior work has shown that correspondence among these datasets is strong, even though studies have differed with respect to patient recruitment criteria and hybridization protocols (9). Two comparison studies have been performed to assess correspondence between RNA-seq and microarray data in psoriasis (28, 37). In both cases, it was concluded that RNA-seq provides a more sensitive platform for detection of differentially expressed genes (DEGs), with RNA-seq identifying more DEGs, in part due to increased sensitivity for quantifying weakly expressed genes (28, 37). Neither study, however, investigated whether RNA-seq/array correspondence or differential expression trends are influenced by GC content and/or gene length (7, 46, 65). RNA-seq may provide inaccurate quantification of transcripts with extreme GC content, due to PCR amplification bias in favor of GC-neutral transcripts, which can result in sequencing libraries with poor representation of GC- and AT-rich transcripts (7, 17, 65). Additionally, RNA-seq data are expected to exhibit gene length bias, since more reads will map to longer transcripts, leading to increased read counts and improved statistical power for differential expression analyses (46).
We here provide a comprehensive evaluation of gene expression in the lesional and uninvolved skin of psoriasis patients based upon integrative analysis of RNA-seq and microarray datasets (9, 28, 30, 43, 58, 62, 75). This setting allowed us to compare RNA-seq findings with those from eight microarray datasets, and we show that RNA quality appears to be an important factor distinguishing array datasets and their correspondence to RNA-seq results. We further analyze each dataset to characterize differences with regard to the number of DEGs identified and to determine which gene characteristics influence correspondence between technologies. Unexpectedly, our findings reveal that psoriasis DEGs are subject to previously undescribed biases, in which the balance of increased to decreased DEGs is influenced by mRNA abundance, GC content, and gene length. These results provide new insights into factors that determine differential expression in psoriasis and have implications regarding mechanisms that control gene transcript levels in lesional skin.
MATERIALS AND METHODS
Ethics statement.
Lesional and uninvolved skin biopsies were obtained from volunteer patients in accordance with Declaration of Helsinki principles. Each participant provided informed written consent under protocols approved by an ethics committee or institutional review board (IRB) (Alfred Hospital, Melbourne, Australia; Chesapeake Research Review, Columbia, MD; Research Review Board, Richmond Hill, Ontario, Canada; Rockefeller University, New York, NY, IRB no. AMA-0674; Royal Adelaide Hospital, Adelaide, Australia; University of Michigan, Ann Arbor, MI, IRB no. HUM00037994).
Microarray analysis of lesional and uninvolved skin samples.
Our analysis includes eight microarray datasets from prior studies that had utilized the same commercial microarray platform (Affymetrix Human Genome U133 Plus 2.0 array, Supplementary Table S1) (9, 30, 43, 58, 62, 75). CEL files were downloaded from Gene Expression Omnibus (GEO) (accession IDs: GSE13355, GSE14905, GSE30999, GSE34248, GSE41662, GSE41663, GSE47751, and GSE50790). The initial dataset included samples from 229 patients (i.e., 458 paired PP and PN samples). We evaluated Affymetrix quality control (QC) metrics, such as average background, scale intensity factor, posthybridization RNA degradation score, percentage of probe sets called present, and four metrics derived from probe level models (NUSE median, NUSE IQR, RLE median, RLE IQR) (11). Posthybridization RNA degradation scores were calculated based upon genome-wide differences in signal intensities of probe sets targeting the 5′-end of genes compared with 3′-ends (R package affy: AffyRNAdeg) (5, 11, 49). This score is based on the principle that 5′-ends of mRNA molecules are synthesized first and will thus be more susceptible to degradation (e.g., by 5′-3′ exoribonucleases), such that array hybridizations with more degraded samples will yield relatively higher intensities for probe sets targeting the 3′-ends of genes (5, 11, 49). Based on QC metrics, we identified 12 patients for which either the PP or PN sample was problematic (Supplementary Table S1). These individuals were removed, yielding 217 patients from the eight studies. Data samples were then normalized using robust multichip average (RMA) with samples from each study normalized independently (27). For two studies (GSE13355 and GSE30999), samples were collected in batches, and samples from each batch were normalized independently. Samples were then clustered, and we discerned that two samples from the same patient were outliers, mostly likely because labels for lesional and uninvolved biopsies had been reversed (GSE30999; samples GSM768097 and GSM768096). These samples were removed, and thus further analyses were based upon 216 patients (i.e., 432 PP and PN samples; Supplementary Table S1).
The Affymetrix Human Genome U133 Plus 2.0 array includes probe sets designed to detect expression of 20,493 human genes (Ensembl gene models). For some genes, expression could be detected by multiple “sibling probe sets” (57). To limit redundancy, therefore, we a priori identified a single representative probe set for each Ensembl gene. We preferentially selected those probe sets that were most specific, such that they were annotated with the smallest number of Ensembl gene IDs. After applying this criterion, if there remained multiple probe sets for any one gene, we excluded those probe sets expected to hybridize with targets in a non-specific fashion (i.e., those with “_x_” or “_s_” in the Affymetrix identifier) (1). If there still remained multiple probe sets for a given gene, we selected as a representative whichever probe set had the highest median expression across all 432 PP and PN samples. Following these procedures, we had assigned a representative probe set to all 20,493 Ensembl genes, but in some cases the same representative probe set had been assigned to multiple genes. We thus identified genes for which the same representative probe set had been assigned and retained the single gene most heavily annotated with Gene Ontology terms. Following these procedures, we had uniquely assigned representative probe sets to 18,709 Ensembl gene IDs.
Differential expression analyses included only genes expressed significantly above background with respect to at least one-third of samples in a given dataset (PP and PN). Probe set detection was assessed based upon the difference between perfect match and mismatch probe signal intensities, with a probe set considered detected if the P value for this difference was < 0.05 (one-sided Wilcoxon signed-rank test) (41). Based upon these criteria, the number of genes detected in each study ranged from 11,921 (GSE14905) to 14,885 (GSE50790), with 14,186 genes detected in the pooled dataset. These genes were further analyzed to assess evidence for differential expression. For each patient, the difference in RMA-normalized expression values was calculated between the PP and PN sample. Differences were subsequently analyzed by a moderated one-sample t-test to determine for each gene whether the mean PP − PN difference was significantly different from zero. Linear models were fit for each gene using an n × 1 design matrix (all elements equal to one), where n is the number of patients in the analysis (R package limma: lmFit) (55). Moderated t-statistics were calculated with shrinkage of variance estimates toward a common value (R package limma: eBayes) (55). Raw P values were calculated based upon the moderated t-statistic and were subsequently adjusted by the Benjamini-Hochberg procedure (6).
Processing and mapping of RNA-seq reads.
Raw sequence data associated with GEO accession GSE41745 were downloaded from the National Center for Biotechnology Information's sequence read archive (sample identifiers: SRR605000, SRR605001, SRR605002, SRR605003, SRR605004, and SRR605005) (28). Sequence reads had been generated from three patients with moderate to severe plaque psoriasis (Illumina Genome Analyzer IIx platform) (28). cDNA libraries were prepared using the NuGEN-Ovation System (28), with amplification steps at random locations of transcripts as well as at 3′-ends, but without direct rRNA depletion or poly(A) selection (26). Sequence read files were converted to fastq formatted files using the SRA toolkit “fastq-dump” function (69). An initial QC inspection was carried out with FastQC (4), which indicated positional nucleotide bias favoring increased GC content over the first 10 bases. Potentially, this was due to the use of random primers during the reverse transcription step for generating cDNA (22). The 80 bp reads were therefore trimmed to remove the first 10 positions, yielding 70 bp reads upon which further analyses were based. Initially, samples contained an average of 30.2 million reads (range: 28.9–31.4 million). However, reads were subsequently passed through the “fastq_quality_filter” function from the FASTX toolkit (21), which removed reads with PHRED quality scores < 20 at > 50% of bases. Reads were additionally filtered using the “fastx_artifacts_filter” utility (21). These preprocessing steps removed 13–20% of reads per sample, leaving an average of 25.5 million reads per sample (range: 22.0–27.0 million). Following these steps, none of the six samples failed any of the 11 QC tests reported by FastQC (4).
Reads were mapped to the human genome (Ensembl GRCh37) using TopHat2 (version 2.0.8) (32). Mapping was performed using default settings with specification of the transcript annotation file (i.e., the -G option). On average, 82.2% of quality-filtered reads were mapped among the six samples (range: 77.8–85.6%). The resulting bam files were sorted and indexed using SAMtools (38). The RSeQC script “geneBody_coverage.py” was used to quantify read counts over the length of each gene feature (67), which showed that on average twice as many reads mapped to the 3′-ends of genes compared with 5′-ends (range: 1.92–2.21). Read counts for each gene feature were tabulated using HTSeq (3), with a read assigned to a feature only when at least part of the read overlapped the feature uniquely (i.e., the option “-m intersection-strict”). Fragments per kilobase of transcript per million fragments mapped (FPKM) values and FPKM confidence intervals were calculated using Cufflinks (version 2.1.1) (64). We initially tabulated counts for 22,679 Ensembl genes, but subsequent analyses were performed using 13,913 genes with detectable expression in at least two of the six RNA-seq data samples. For any one sample, two conditions were required for a gene to have detectable expression. First, the count per million mapped reads needed to exceed 0.25 (i.e., ∼5–7 mapped reads per sample). Second, the lower bound on the 95% FPKM confidence interval reported by Cufflinks needed to be >0 (29, 64). This latter criterion was used to guard against including low-expressed genes with many isoforms, for which unambiguous read mapping may be difficult due to their low abundance and absence of unique exons (29, 64).
RNA-seq differential expression analysis (lesional vs. uninvolved skin).
RNA-seq differential expression analysis was performed using edgeR (version 3.0.8) with gene counts normalized using the trimmed mean of M-values method and trended dispersions estimated using the “bin.spline” function (default settings) (52). Library size normalization factors were calculated from the total number of reads mapping to the 13,913 protein-coding genes detected in our analysis. Dispersion estimates were used to fit negative binomial generalized log-linear models, where each model was fit using treatment (PP vs. PN) and subject (patient ID) as covariates (edgeR function: glmFit). The effect of treatment (PP vs. PN) was evaluated for each gene with a likelihood ratio test, with significance evaluated based upon the increase in model deviance that results when the coefficient representing treatment is removed (edgeR function: glmLRT). Raw P values from this approach were adjusted using the Benjamini-Hochberg method to control the false discovery rate (FDR) among the 13,913 protein-coding genes (6). This yielded 1,964 DEGs, including 994 PP-increased DEGs (FDR < 0.05 and FC > 1.50) and 970 PP-decreased DEGs (FDR < 0.05 and FC < 0.67). We also report the number of DEGs obtained using edgeR with different normalization and dispersion estimation methods (Supplementary Table S2), as well as the range of results obtained using DESeq (version 1.10.1) as an alternative analysis method (Supplementary Table S3) (2).
Additional microarray datasets (Enbrel and laser capture microdissection).
Gene expression in lesions from patients treated with Enbrel (etanercept) was evaluated using microarray samples available under GEO accession GSE11903 (Affymetrix Human Genome U133A 2.0 array) (76). A moderated t-test was used to identify genes differentially expressed in lesions posttreatment (1 wk and 4 wk) compared with lesions at baseline before treatment (R package limma: lmFit) (55). To localize expression of genes in human skin, we used laser capture microdissection (LCM) data from GEO accession GSE42114 (Affymetrix Human Genome U133 Plus 2.0 array) (19). This study used LCM to dissect out reticular dermis, basal epidermis, and suprabasal epidermis from full-thickness skin sections (n = 3) (19). Genes specifically expressed in a given compartment were identified based upon a two-sample differential expression analysis, in which expression of genes in one compartment was compared with expression in the other two (R package limma: lmFit) (55).
RT-PCR analysis of lesional and uninvolved skin samples.
RT-PCR analysis of selected genes was performed using an independent set of lesional and uninvolved skin samples from eight patients (European Caucasian descent). Patient recruitment and medication washout protocols have been described (43). Biopsies were flash-frozen in liquid nitrogen and stored at −80°C. RNA extractions were performed using RNeasy columns (Qiagen), and RNA quality and quantity were evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA was reverse-transcribed using the High Capacity cDNA Transcription kit (Applied Biosystems, Foster City, CA), and PCR analyses were performed using the 7990HT Fast Real-Time PCR system (Applied Biosystems). TaqMan primers were purchased from Applied Biosystems (cat. no. 4331182 and 4351372; CTNNBIP1, Hs00172016_m1; LOR, Hs01894962_s1; NFASC, Hs00391791_m1; COCH, Hs00990775_m1; ADAMTS2, Hs01029111_m1; ANKRD26, Hs00208680_m1; PSMB10, Hs00988194_g1; TSLP, Hs00263639_m1; MUC1, Hs00159357_m1; PHYHD1, Hs00288878_m1; CIDEA, Hs00154455_m1). Relative expression was quantified using large ribosomal protein P0 (RPLP0) as an internal reference (Applied Biosystems, Hs99999902_m1).
RESULTS
RNA-seq and microarray show similar sensitivity for detection of DEGs in psoriasis lesions.
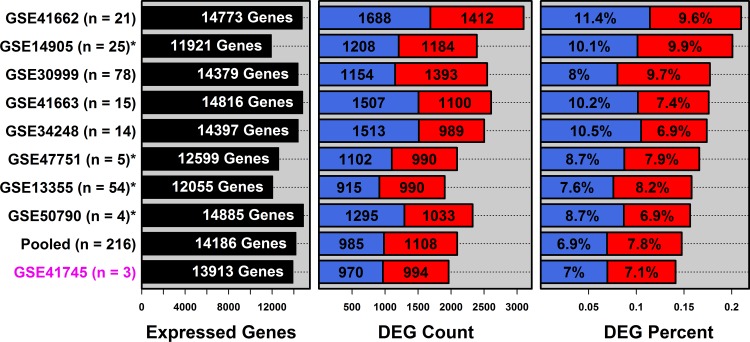
Gene expression differences between lesional (PP) and uninvolved (PN) skin were analyzed using RNA-seq (n = 3 patients) and eight microarray datasets (4 ≤ n ≤ 80). A pooled meta-dataset consisting of all array samples was also analyzed (n = 216 patients). We refer to four array datasets as “low degradation” (GSE13355, GSE47751, GSE50790, and GSE14905), since they included samples with lower posthybridization RNA degradation scores (5, 11, 49) (Supplementary Table S1, Supplementary Fig. S1). We refer to the other array datasets as “high degradation” (GSE41663, GSE34248, GSE41662, and GSE30999), since these included samples with higher RNA degradation scores (Supplementary Table S1, Supplementary Fig. S1).
RNA-seq and microarray identified similar numbers of DEGs, although slightly fewer were identified by RNAs-seq (Fig. 1). RNA-seq identified 1,964 DEGs, including 994 PP-increased DEGs and 970 PP-decreased DEGs (Fig. 1). In contrast, array datasets identified between 2,093 (pooled data) and 3,100 DEGs (GSE41662) (Fig. 1). The percentage of DEGs identified by RNA-seq, relative to the total number of genes tested, was similar or slightly lower than that in the array studies (∼7% for increased and decreased genes, respectively; Fig. 1). For detection of DEGs by RNA-seq (Fig. 1), we utilized default edgeR settings (see materials and methods) (52). When these settings were varied, edgeR identified as few as 1,941 and as many as 2,430 DEGs (Supplementary Table S2). Using DESeq (2), we identified 378 DEGs (280 increased and 98 decreased) with default settings but found that this number varied depending upon normalization and dispersion estimation methods (i.e., between 0 and 3,535 DEGs; Supplementary Table S3). For the remainder of this paper, we focus on edgeR results obtained with default settings (1,964 DEGs, Fig. 1).
Fig. 1.
RNA-seq and microarray show similar sensitivity for detection of differentially expressed genes (DEGs) in psoriasis lesions. Differential expression between psoriasis lesions (PP) and uninvolved skin (PN) was evaluated by RNA-seq (GSE41745) and 8 microarray studies (*low degradation datasets). Left: the number of expressed genes detected in each study and included in differential expression analyses (see materials and methods). Middle: total number of DEGs identified (red = PP-increased, FDR < 0.05 with mean PP/PN fold-change > 1.50; blue = PP-decreased FDR < 0.05 with mean PP/PN fold-change < 0.67). Right: total number of DEGs as a percentage of the number of detected genes in each analysis.
Although their exact number differed, DEG fold-change estimates were usually consistent between technologies. Of 994 RNA-seq PP-increased DEGs, > 93% showed higher expression in PP vs. PN skin in each array study, respectively (Supplementary Figs. S2 and S3). Likewise, of 970 RNA-seq PP-decreased DEGs, > 90% showed lower expression in PP vs. PN skin in each array study, respectively (Supplementary Figs. S2 and S3). In general, therefore, DEGs showed consistent trends, and it was rare for any DEG to show a contrary shift in expression between technologies.
RNA-seq and high degradation microarray data reveal bias of weakly expressed genes toward decreased expression in psoriasis lesions.
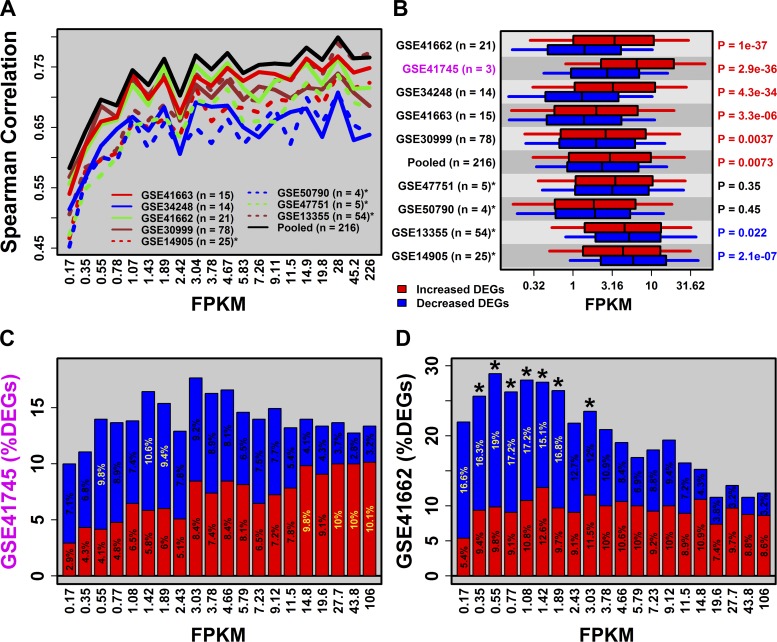
RNA-seq is expected to provide more sensitive detection of low and high abundance mRNAs (28, 37, 59). In partial agreement with this, correspondence of fold-change estimates between microarray and RNA-seq was reduced among genes with weak expression (Fig. 2A). This was observed with respect to each of the array datasets as well as the pooled array data (Fig. 2A). The breakdown point appeared to lie at 1.0 FPKM, with stronger RNA-seq/array correspondence above this threshold (rs ≥ 0.60) and weaker correspondence below (0.45 ≤ rs ≤ 0.60). Notably, there was no decay of the RNA-seq/array correspondence among genes with high expression, suggesting that signal saturation did not limit the dynamic range achieved in microarray studies (Fig. 2A).
Fig. 2.
RNA-seq and high degradation microarray data reveal bias of weakly expressed genes toward decreased expression in psoriasis lesions. A: correlation between PP/PN fold-change estimates (RNA-seq vs. microarray). Genes were ordered according to their average expression in the RNA-seq experiment [mean fragments per kilobase of transcript per million fragments mapped (FPKM) in PP and PN samples] and placed into equally sized bins. The Spearman rank correlation was calculated for each bin. B: average FPKM (PP and PN samples) for increased and decreased DEGs. P values (right margin) assess the difference between increased and decreased DEGs (Wilcoxon signed rank test). C and D evaluate differential expression trends for GSE41745 and GSE41662, respectively. Genes were grouped into equally sized bins according to mean FPKM and the percentage of DEGs was calculated for each bin (red = PP-increased; blue = PP-decreased). Yellow percentage values denote bins for which there is significant overabundance of increased or decreased DEGs [false discovery rate (FDR) < 0.05, Fisher's exact test]. *Bins for which the total number of DEGs (increased + decreased) is significantly large (FDR < 0.05, Fisher's exact test).
Weakly expressed genes were biased toward decreased expression in PP skin (Fig. 2, B–D). This trend, which was observed only for RNA-seq (GSE41745) and high-degradation array studies (Fig. 2B), was illustrated by the fact that PP-decreased DEGs were, on average, expressed at lower levels than PP-increased DEGs (Fig. 2B). The trend could also be discerned by evaluating the proportion of PP-increased and PP-decreased DEGs across ordered gene sets with progressively higher FPKM (Fig. 2, C and D). Among genes in the lowest FPKM category, 2.9% were PP-increased DEGs, while 7.1% were PP-decreased DEGs (RNA-seq, Fig. 2C). This reversed, however, among genes in the highest FPKM category, with 10.1% PP-increased DEGs and only 3.2% PP-decreased DEGs (RNA-seq, Fig. 2C). A similar pattern was observed in high-degradation array datasets such as GSE41662 (Fig. 2D), but not in the low degradation array datasets (Fig. 2B).
RNA-seq and low-degradation microarray data reveal bias of high-GC-content genes toward increased expression in psoriasis lesions.
The expression of genes with extreme GC content may be poorly quantified by RNA-seq (7, 65). Consistent with this, correspondence between RNA-seq and microarray fold-change estimates was strongest for GC-neutral genes, with declining correspondence for genes with extreme GC content (Fig. 3A). For high-GC-content genes, this was evident for all array datasets (Fig. 3A). For low-GC-content genes, however, decline in correspondence was more pronounced in low-degradation array datasets (Fig. 3A). RNA-seq/array correspondence is therefore sensitive to GC content, but patterns differ depending upon degradation status (Fig. 3A).
Fig. 3.
RNA-seq and low degradation microarray data reveal bias of high-GC-content genes toward increased expression in psoriasis lesions. A–C: analyses shown in Fig. 2 were repeated, except genes were ordered and grouped into bins according to GC content. D: we identified 1,147 genes with increased expression in lesional skin of patients after 1 wk of Enbrel therapy [P < 0.05 and fold-change (FC) > 1.50], along with 738 genes with decreased expression (P < 0.05 and FC < 0.67) (GSE11903). The proportion of Enbrel-increased (red) and Enbrel-decreased (blue) genes was evaluated for groups of genes binned according to GC content.
The balance of PP-increased to PP-decreased DEGs was also distorted by GC content, and as above, this varied between low- and high-degradation array datasets (Fig. 3). For RNA-seq, PP-increased DEGs had significantly higher GC content than PP-decreased DEGs (Fig. 3B), and consistent with this, the proportion of PP-increased DEGs was greater among genes with higher GC content (Fig. 3C). This trend was weaker for microarray data but was observed in three of four low-degradation array datasets (Fig. 3B). Interestingly, there was an opposite trend in lesions from patients following onset of etanercept (Enbrel) therapy (76). After 1 wk of treatment, high-GC-content genes were disproportionately decreased in lesions from treated patients (as compared with baseline lesions, Fig. 3D). The same pattern was observed in patients after 4 wk of Enbrel treatment (data not shown).
At a genome-wide level, expression was correlated with GC content, although such correlations often differed in strength between PP and PN samples (Supplementary Figs. S4 and S5). For RNA-seq, there were strong negative correlations between expression and GC content (−0.44 ≤ rs ≤ −0.24), with stronger (more negative) correlations in PN samples (Supplementary Fig. S4). A similar difference was noted in low degradation array datasets (Supplementary Fig. S5C). These systemic differences likely contributed to the higher GC content of PP-increased DEGs compared with PP-decreased DEGs (Fig. 3B).
For high-degradation array data, in contrast, genome-wide correlations between GC content and expression were stronger (more negative) but of similar magnitude in PP and PN samples (Supplementary Fig. S5, A–C). Negative correlations between GC content and expression were stronger in samples with high degradation scores (Supplementary Fig. S5, E and F), with smaller PP vs. PN differences in correlation estimates (Supplementary Fig. S5D).
Long genes are biased toward decreased expression in psoriasis lesions regardless of expression profiling technology.
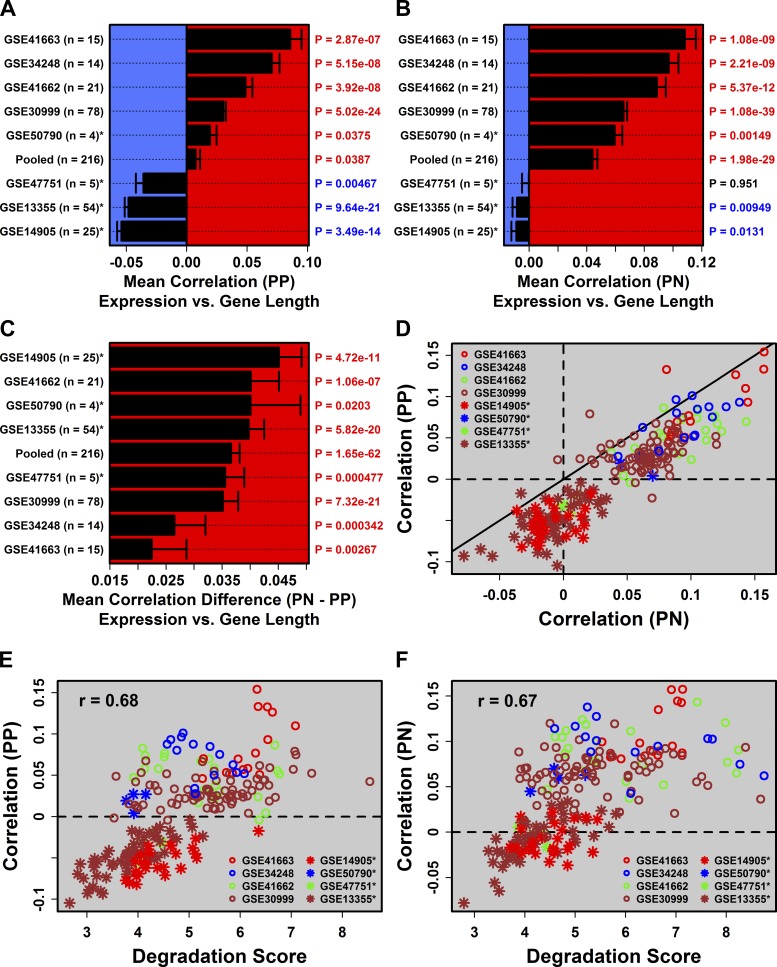
Long genes engender higher read counts in RNA-seq and benefit from increased statistical power in differential expression analyses (46). We thus evaluated the RNA-seq/array correspondence between fold-change estimates across gene groups differing with respect to length (Fig. 4A). This revealed disparities among array datasets, with correspondence breaking down among long genes (>120 kb) for low-degradation array data but not for high-degradation array data (Fig. 4A). RNA-seq/array correspondence, therefore, is also sensitive to gene length, but patterns differ depending upon degradation status (Fig. 4A).
Fig. 4.
Long genes are biased toward decreased expression in psoriasis lesions regardless of expression profiling technology. Analyses shown in Fig. 3 were repeated, except genes were ordered and grouped into bins according to length. In F, we analyzed 1,770 genes with increased expression in lesional skin of patients after 4 wk of Enbrel therapy (FDR < 0.05 and FC > 1.50; red), along with 1,532 genes with decreased expression (FDR < 0.05 and FC < 0.67; blue) (GSE11903).
We observed platform-independent differences between PP-increased and PP-decreased DEGs with respect to gene length, with PP-decreased DEGs significantly longer in all datasets (Fig. 4B). The trend was observed in all datasets (RNA-seq and microarray) but was strongest and most evident with RNA-seq (Fig. 4, B and C). Curiously, for RNA-seq, the total proportion of DEGs increased among the longest genes (Fig. 4C), as expected (46), but we also noted an overabundance of DEGs among the shortest genes, such that DEG abundance had a U-shaped relationship with gene length (Fig. 4C). In contrast, for array data, there tended to be an overabundance of DEGs among longer genes but not shorter genes (Fig. 4D). In all cases, however, the proportion of PP-decreased DEGs increased with gene length (Fig. 4B). The opposite pattern was observed in patients as early as 1 wk following Enbrel therapy, with long genes biased toward increased expression in lesions from treated patients (Fig. 4, E and F).
These trends reflected PP vs. PN differences in the genome-wide correlation between gene length and gene expression (Fig. 5 and Supplementary Fig. S6). For RNA-seq, length-expression correlations were weak when expression was quantified by FPKM (0.08 ≤ rs ≤ 0.17, Supplementary Fig. S6), although as expected, stronger correlations were observed when expression was measured in terms of log-transformed counts (0.21 ≤ rs ≤ 0.30). Regardless of the quantification method, however, such correlations were always stronger in PN samples, consistent with bias of long genes towards PP-decreased expression (Fig. 4). Inspection of the array data revealed similar trends, except high degradation samples were associated with stronger (more positive) genome-wide length-expression correlations (Fig. 5, E and F). Regardless of the degradation scores, however, length-expression correlations were almost always higher in PN samples (Fig. 5, C and D), consistent with bias of long genes toward PP-decreased expression (Fig. 4).
Fig. 5.
Genome-wide correlations between gene length and expression are stronger in degraded samples and consistently higher in PN samples. A: average Spearman correlation between gene length and expression in PP samples. P values evaluate whether the median correlation coefficient was significantly different from 0 (2-tailed Wilcoxon signed rank test). B: the same analysis was repeated for PN samples. C: the same analysis was repeated for the difference (PN − PP) in correlation coefficients. D: scatterplot comparing genome-wide length-expression correlations in paired PP and PN samples. Diagonal denotes equal correlation estimates in PP and PN samples. Association between degradation scores and length-expression correlations in PP samples (E) and PN samples (F).
Gene length is the dominant factor determining differential expression and predicts DEGs as well as anatomical localization.
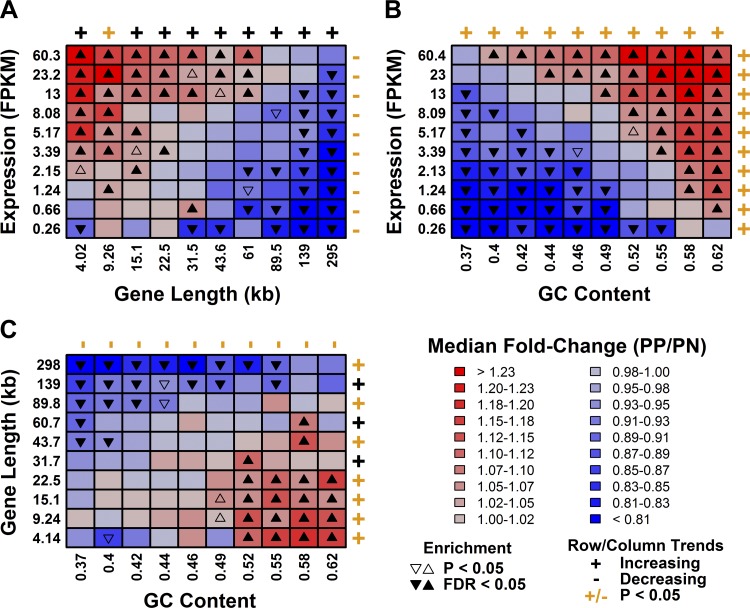
We identified three gene characteristics that influence differential expression in psoriasis: mRNA abundance, GC content, and gene length. Focusing on the RNA-seq data (GSE41745), we inspected bivariate combinations of these factors and could discern mostly additive relationships, with each factor having similar effects on fold-change estimates (PP/PN) irrespective of the other two (Fig. 6). Gene length, however, appeared to be dominant: long genes were biased toward decreased expression even if they were expressed at high levels or if they had high GC content (Fig. 6, A and C).
Fig. 6.
mRNA abundance, GC content, and gene length have additive effects on differential expression in psoriasis lesions. A: genes were placed into 100 equally sized bins based upon their expression level and gene length. Median fold-change (PP/PN) was calculated for genes in each bin. B: the analysis was repeated for GC content and gene expression. C: the analysis was repeated for GC content and gene length.
Psoriasis lesions contain of a mixture of cell types and the anatomical localization of mRNAs within lesions appears to be a factor influencing differential expression (37, 60, 62). Genes specifically expressed in epidermis or KCs, for instance, tend to show PP-increased expression, while genes specifically expressed in dermis tend to show PP-decreased expression (37, 60, 62). To assess the influence of anatomical localization, we used LCM data to localize expression of genes to the reticular dermis, basal epidermis, or suprabasal epidermis (19). As expected, genes specifically expressed in the reticular dermis were biased toward PP-decreased expression, while genes specifically expressed in the suprabasal epidermis were biased toward PP-increased expression (RNA-seq, Supplementary Fig. S7). Notably, weakly expressed genes were not PP-decreased unless they were also specifically expressed in the dermis (RNA-seq, Supplementary Fig. S7A). Gene length, however, was the dominant factor, with long genes biased toward PP-decreased expression, regardless of their anatomical localization (RNA-seq, Supplementary Fig. S7).
We next used regression modeling to determine which gene characteristics best predicted differential expression in psoriasis, by screening models that include mRNA abundance, GC content, gene length, as well as variables indicating the degree to which genes are specifically expressed in an anatomical compartment (reticular dermis, basal epidermis, and suprabasal epidermis). PP-increased DEGs (RNA-seq) were equally well predicted by gene length and suprabasal epidermis localization [area under the curve (AUC) = 0.606, Supplementary Fig. S8A], and these two factors also provided the best performing bivariate model (AUC = 0.651, Supplementary Fig. S8B). For prediction of PP-decreased DEGs (RNA-seq), gene length was clearly the dominant factor and provided the best univariate model (AUC = 0.633, Supplementary Fig. S9A).
Both RNA-seq and microarray uniquely identify genes as differentially expressed in psoriasis lesions.
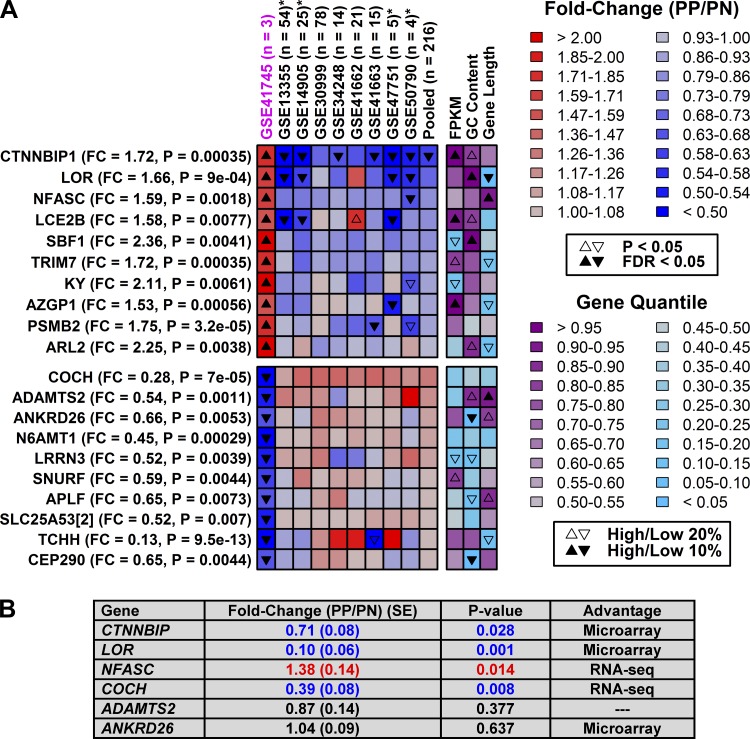
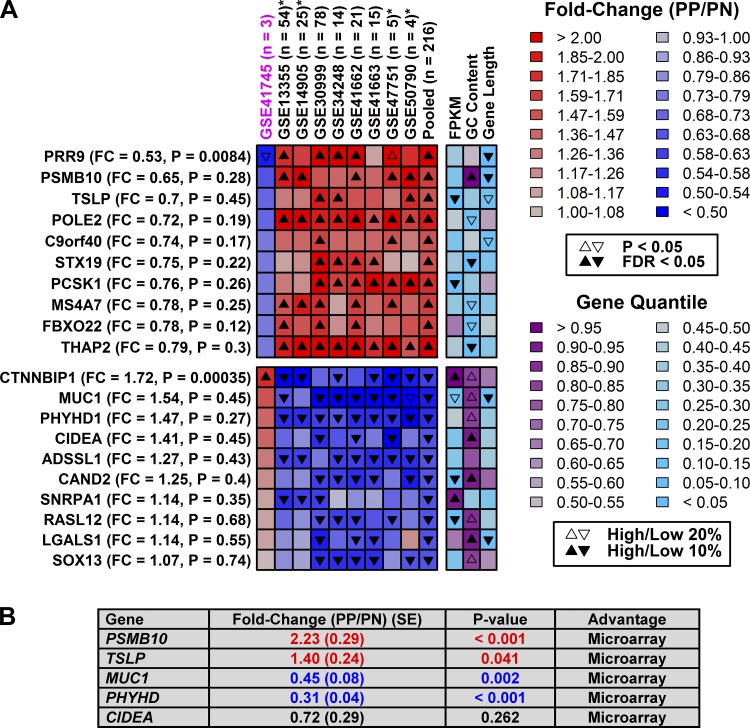
Most genes showed similar trends between expression profiling technologies (Supplementary Figs. S2 and S3), although we could identify some genes for which shifts in expression were platform-specific (Figs. 7 and 8). Genes identified as increased by RNA-seq (but not array) often had high GC content (e.g., LOR and SPF1), while those identified as decreased by RNA-seq (not array) tended to have low GC content (e.g., ANKRD26 and CEP290) (Fig. 7). Those identified as increased by array (not RNA-seq) often had low GC content (e.g., STX19 and THAP2), while those identified as decreased by array (not RNA-seq) often had higher GC content (e.g., CIDEA and CAND2) (Fig. 8). We attempted to validate six RNA-seq-specific DEGs using RT-PCR and independent samples from eight patients (Fig. 7B). The RNA-seq result was confirmed for two of six genes, but for three others the array result was confirmed (Fig. 7B). Likewise, we attempted to validate five array-specific DEGs with RT-PCR, and in all five cases the array result was verified, while the RNA-seq result was not supported (Fig. 8B). Potentially, the higher confirmation rate for array-specific DEGs may be attributed to the larger sample size of the pooled array data (n = 216 patients) compared with the RNA-seq data (n = 3).
Fig. 7.
Genes uniquely identified as differentially expressed by RNA-seq. A, top: genes significantly elevated in psoriasis lesions according to RNA-seq (GSE41745) but decreased in microarray studies (pooled data, n = 216 patients). Bottom: genes significantly decreased in psoriasis lesions according to RNA-seq but increased in microarray studies. Genes are listed in the left margin along with FC (PP/PN) and FDR values (RNA-seq). Final heat-map columns indicate percentile rankings of genes with respect to their mean expression in PP and PN samples (FPKM), GC content, and length. B: RT-PCR analysis of 6 genes based upon independent samples (n = 8 patients).
Fig. 8.
Genes uniquely identified as differentially expressed by microarray. A, top: genes significantly elevated in psoriasis lesions in microarray studies (pooled data, n = 216 patients) but not according to RNA-seq (GSE41745). Bottom: genes significantly decreased in psoriasis lesions in microarray studies but not RNA-seq. Genes are listed in the left margin along with fold-change (PP/PN) and FDR values (RNA-seq). Final heat-map columns indicate percentile rankings of genes with respect to their mean expression in PP and PN samples (FPKM), GC content, and length. B: RT-PCR analysis of 5 genes based upon independent samples (n = 8 patients).
DISCUSSION
RNA-seq is a revolutionary technology for expression profiling and has continued to generate insights in basic research and translational medicine. RNA-seq findings, however, may not be fully consistent with those obtained by microarray, and synthesizing results from both technologies should provide a stronger basis for discovery than strict reliance on one platform alone. We thus evaluated correspondence between RNA-seq and microarray findings in psoriasis research, a context that is rich in data resources due to high-quality studies carried out over the last decade (9, 30, 37, 28, 43, 58, 62, 75). Our results show that RNA-seq and microarrays identify similar numbers of DEGs, with each technology identifying some DEGs uniquely. We further showed that correspondence between platforms is influenced by mRNA abundance, GC content, and gene length and that these factors distorted differential expression trends to alter the balance of increased to decreased DEGs. Gene length had the strongest impact on differential expression, regardless of the expression profiling technology. This finding has implications for future work in psoriasis, since it demonstrates that differential expression cannot be explained on the basis of gene function alone but is at least partly dependent on structural features of the mRNA molecule itself. A second implication is that, compared with de novo transcription, mRNA stability may be equally important for determining steady-state transcript levels in psoriasis lesions.
Previous RNA-seq studies have described a purely technical gene length bias in which longer genes benefit from increased statistical power due to the larger number of reads mapping to such genes (46). There was some indication of this in our study (Fig. 4C), but the trend we identified appeared nontechnical and attributable to psoriasis biology. Long genes were heavily biased toward decreased expression in psoriasis lesions, while short genes were biased toward increased expression (Fig. 4). This trend was platform independent, and, importantly, the opposite pattern was observed in lesions from patients as early as 1 wk after Enbrel therapy (Fig. 4E), which further indicated that the effect is nontechnical and due to mechanisms at work in active and resolving lesions. Although we restricted our analyses to protein-coding mRNAs, we expect that this gene length bias would broadly influence coding and noncoding mRNA species. A previous microarray study focusing on miRNAs, for instance, identified 42 psoriasis-increased miRNAs but only five psoriasis-decreased miRNAs (78). Likewise, an RNA-seq study identified 71 psoriasis-increased miRNAs compared with only 27 psoriasis-decreased miRNAs (31). As predicted based upon our findings, therefore, short miRNA species appear biased toward increased expression in psoriasis (31, 78).
Potentially, gene length bias in psoriasis is due to length-dependent differences in mRNA stability between lesions and uninvolved skin, which would not necessarily result from mRNA degradation taking place after lesions are sampled but may rather be attributable to the lesional skin microenvironment. For both lesional and uninvolved skin, mRNA samples with greater degradation scores show increased expression of long genes and decreased expression of short genes (Fig. 5, E and F), consistent with previous studies of ex situ mRNA degradation in clinical samples (45). Independent of this process, however, in situ mRNAs from psoriasis lesions are subject to a harsh microenvironment in which endoribonuclease activity may preferentially target longer mRNAs (18, 42, 53). Short mRNAs, in contrast, may be less often targeted and may indeed be elevated since they can be transcribed more rapidly as part of a coordinated defense response (56, 66). These mechanisms, in combination, may lead to lower genome-wide correlations between gene length and expression in lesional skin (Fig. 5D), resulting in decreased expression of long mRNAs and increased expression of short mRNAs (Fig. 4, B–D).
Heightened endoribonuclease activity in psoriasis lesions may result from two cytokine-driven mechanisms, including 1) induction of RNASE7 by cytokines such as IFN-γ (24, 25, 54) and 2) activation of RNase L mediated by IFN-α and IFN-β and induction of 2′-5′-oligoadenylate synthetases (39, 61, 75). The first mechanism has been well characterized in psoriasis lesions, which feature heightened production of antimicrobial peptides (AMPs), such as β-defensins, psoriasin (S100A7), cathelicidin (LL-37), dermcidin, and RNase 7 (23). Of these, RNase 7 has especially strong antimicrobial activity and is a potent ribonuclease, with one study indicating that RNase 7 digested yeast tRNA with 50-fold greater efficiency than recombinant eosinophil cationic protein (RNase 3) (24). The second mechanism is based upon activation of pathways connecting viral RNA sensors to RNase L activation (e.g., RIG-I/MDA-5 → IFN-α/IFN-β → OAS → RNase L) (33, 50, 75). Stimulation of this pathway leads to synthesis of 2′,5′-oligoadenylates (2–5As) and activation of the latent endoribonuclease RNase L, which in turn cleaves self and nonself ssRNA species (39). Both RNase 7 and RNase L may promote elevated endoribonuclease activity in psoriasis lesions. This may decrease abundance of long mRNAs while increasing abundance of shorter mRNAs (5, 53), contributing to the gene length bias observed in this study (Fig. 4).
RNA-seq is expected to improve dynamic range for detection of genes with weak or strong expression (28, 37, 59). In partial agreement with this idea, correspondence between RNA-seq- and array-based fold-change estimates declined among low-expressed genes (< 1.0 FPKM), although this did not occur among genes with high expression (Fig. 2A). Interestingly, for both RNA-seq and microarray, weakly expressed genes were biased toward decreased expression in psoriasis lesions. This pattern may be attributable to differences in the architecture of psoriasis lesions and normal skin (37). Low-abundance mRNAs, for instance, may be predominantly derived from dermal cells and thus driven toward decreased expression in psoriasis lesions, since in relative terms the quantity of dermal cells declines with epidermal expansion (37). It remains unclear, however, why such a “dermis dilution” effect would be more pronounced in samples with higher degradation scores, as observed in this study (Fig. 2B). A possible explanation is that RNA degradation leads to a paradoxical increase in the ability of both technologies to detect signals arising from low-expressed genes, such as those associated with dermal fibroblasts (49). A previous study of postmortem brain tissue, for instance, revealed that RNA degradation had differential effects on low- vs. high-expressed genes, with degradation favoring increased expression of low abundance mRNAs (49). Further work is needed to understand the significance of RNA degradation and how it affects bias of weakly expressed genes. We expect that such biases will be magnified for studies in which nonstringent thresholds are used to filter out genes with weak or undetectable expression.
GC content bias has been recognized as an important contributor to technical variation in RNA-seq and microarray studies of gene expression (7, 17, 65). Our analyses suggest a mixed pattern concerning GC content bias, which may be explained by both technical and biological considerations. Correlations between GC content and expression were stronger for RNA-seq compared with microarray, and we noted that genes with discrepant trends between technologies were often those with extreme GC content (Figs. 7 and 8). These results appear consistent with PCR amplification bias favoring GC-neutral content as previously documented for RNA-seq data (7, 17, 65). At the same time, in the absence of RNA degradation, lesional and uninvolved skin differed with respect to the global correlation between GC content and gene expression (Supplementary Fig. S5C). This led to bias of high GC content genes toward increased expression in psoriasis lesions (Fig. 3, B and C). The biological relevance of this trend was supported by our analysis of (resolving) lesions from patients following Enbrel treatment (76), which showed the opposite pattern, with decreased expression of high GC content genes (Fig. 3, E and F). These results suggest mechanisms favoring high-GC-content mRNAs in psoriasis lesions, potentially due to effects of GC content on mRNA stability. As noted above, for instance, heightened ribonuclease activity in lesions may be due to RNase L, which is known to target UA and UU dinucleotides (20, 68, 71).
Previous studies of gene expression in psoriasis reported that RNA-seq identified more DEGs than microarrays (28, 37). This contrasts with our findings, however, since we found that microarrays identified more DEGs than RNA-seq (Fig. 1). The number of DEGs identified does not necessarily provide a good basis for comparison of expression profiling technologies. This is because DEGs include both false-positives and true-positives (Figs. 7 and 8), while the number of DEGs identified by RNA-seq varies considerably depending upon the statistical method used to test for differential expression (Supplementary Tables S2 and S3). RNA-seq and microarray also generate data with different distributions (i.e., negative binomial vs. log-normal) and thus require different statistical tests that may not be equally powered. Nevertheless, three factors likely explain why microarrays identified more DEGs in our study. First, there was greater statistical power for detection of DEGs by microarray, since microarray datasets included more patients (4 ≤ n ≤ 80) than the RNA-seq study (n = 3). Second, we identified DEGs based upon modest fold-change thresholds (FC > 1.5 or FC < 0.67). More stringent thresholds (e.g., FC > 2 or FC < 0.50) would have more strongly reduced the number of DEGs identified by microarray, since microarray fold-change estimates are compressed relative to RNA-seq (10, 13). Third, microarray platforms may offer a genuine advantage over RNA-seq with respect to reduction of noise levels, particularly among low-expressed genes, leading to increased signal-to-noise ratios and improved DEG detection (10). For determining which technology to utilize, investigators will need to balance the relative strengths of each approach, although our findings caution against the idea that RNA-seq will yield more true-positive DEGs at a given FDR.
Gene expression in psoriasis has been extensively studied to generate insights into disease mechanisms, and promising ideas have been advanced for clinical applications based upon expression profiling (48, 60). It is likely that RNA-seq will increasingly be the technology of choice for both research and clinical applications, but in agreement with other studies (34), our findings illustrate the complementary nature of these technologies and show how integrative analyses of both data types can be used to distinguish 1) technical trends arising from platform-specific artifacts and 2) nontechnical trends potentially related to the biological processes. Our findings also highlight directions for future progress. It has often been assumed, for example, that most gene expression shifts in psoriasis lesion can be attributed to de novo transcription and DNA/protein regulatory interactions (40, 61). Length-dependence of expression patterns in psoriasis, however, suggests that mRNA stability and degradation may play an equally important role. Further work is therefore needed to define transcription-associated expression shifts, apart from those driven by posttranscriptional mechanisms (8, 47, 70). This should lead to clearer signals in the data and facilitate identification of mechanisms that control gene transcript levels.
ENDNOTE
At the request of the author(s), readers are herein alerted to the fact that additional materials related to this manuscript may be found at the institutional website of one of the authors, which at the time of publication they indicate is: http://www.derm.med.umich.edu/PhysGen_00022_2014.pdf. These materials are not a part of this manuscript and have not undergone peer review by the American Physiological Society (APS). APS and the journal editors take no responsibility for these materials, for the website address, or for any links to or from it.
GRANTS
This work was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grants AR-054966, AR-042742, AR-050511 (J. T. Elder), K08 AR-060802 (J. E. Gudjonsson), and K01 AR-064765 (A. Johnston). Additional support was provided by the Babcock Endowment Fund (A. Johnston and J. E. Gudjonsson), the A. Alfred Taubman Medical Research Institute Kenneth and Frances Eisenberg Emerging Scholar Award (J. E. Gudjonsson), and the Doris Duke Foundation (J. E. Gudjonsson). J. T. Elder is supported by the Ann Arbor VA Hospital. W. R. Swindell is funded in part by the American Skin Association Carson Family Research Scholar Award in Psoriasis.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: W.R.S., A.J., and J.E.G. conception and design of research; W.R.S. analyzed data; W.R.S., A.J., and J.E.G. interpreted results of experiments; W.R.S. prepared figures; W.R.S. drafted manuscript; W.R.S., J.J.V., J.T.E., A.J., and J.E.G. edited and revised manuscript; X.X. performed experiments; J.J.V., J.T.E., A.J., and J.E.G. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Sean Caron (University of Michigan, School of Public Health and Biostatistics) for providing us with assistance and computational support for this project.
REFERENCES
- 1.Affymetrix Inc. Design and Performance of the GeneChip® Human Genome U133 Plus 2.0 and Human Genome U133A 2.0 Arrays. Affymetrix. http://media.affymetrix.com/support/technical/technotes/hgu133_p2_technote.pdf (27 Feb. 2014).
- 2.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 11: R106, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8: 1765–1786, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (27 Feb. 2014).
- 5.Atz M, Walsh D, Cartagena P, Li J, Evans S, Choudary P, Overman K, Stein R, Tomita H, Potkin S, Myers R, Watson SJ, Jones EG, Akil H, Bunney WE, Jr, Vawter MP, Members of National Institute of Mental Health Conte Center, Pritzker Neuropsychiatric Disorders Research Consortium. Methodological considerations for gene expression profiling of human brain. J Neurosci Methods 163: 295–309, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a powerful and practical approach to multiple testing. J Roy Stat Soc B 57: 289–300, 1995 [Google Scholar]
- 7.Benjamini Y, Speed TP. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Res 40: e72, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 15: 163–175, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bigler J, Rand HA, Kerkof K, Timour M, Russell CB. Cross-study homogeneity of psoriasis gene expression in skin across a large expression range. PLoS One 8: e52242, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black MB, Parks BB, Pluta L, Chu TM, Allen BC, Wolfinger RD, Thomas RS. Comparison of microarrays and RNA-seq for gene expression analyses of dose-response experiments. Toxicol Sci 137: 385–403, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Bolstad BM, Collin F, Brettschneider J, Simpson K, Cope L, Irizarry RA, Speed TP. Quality assessment of Affymetrix GeneChip data. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor, edited by Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. New York: Springer, 2005 [Google Scholar]
- 12.Bottomly D, Walter NA, Hunter JE, Darakjian P, Kawane S, Buck KJ, Searles RP, Mooney M, McWeeney SK, Hitzemann R. Evaluating gene expression in C57BL/6J and DBA/2J mouse striatum using RNA-Seq and microarrays. PLoS One 6: e17820, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradford JR, Hey Y, Yates T, Li Y, Pepper SD, Miller CJ. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics 11: 282, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11: 94, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chavan SS, Bauer MA, Peterson EA, Heuck CJ, Johann DJ. Towards the integration, annotation and association of historical microarray experiments with RNA-seq. BMC Bioinformatics 14, Suppl 14: S4, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung LM, Ferguson JP, Zheng W, Qian F, Bruno V, Montgomery RR, Zhao H. Differential expression analysis for paired RNA-Seq data. BMC Bioinformatics 14: 110, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res 36: e105, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Förster FJ, Neufahrt A, Stockum G, Bauer K, Frenkel S, Fertig U, Leonhardi G. Subcellular distribution of phosphatases, proteinases, and ribonucleases in normal human stratum corneum and psoriatic scales. Arch Dermatol Res 254: 23–28, 1975 [DOI] [PubMed] [Google Scholar]
- 19.Gulati N, Krueger JG, Suárez-Fariñas M, Mitsui H. Creation of differentiation-specific genomic maps of human epidermis through laser capture microdissection. J Invest Dermatol 133: 2640–4642, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han JQ, Wroblewski G, Xu Z, Silverman RH, Barton DJ. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J Interferon Cytokine Res 24: 664–676, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Hannon GJ. FASTX-Toolkit. Cold Spring Harbor Laboratory. http://hannonlab.cshl.edu/fastx_toolkit//(27 Feb. 2014).
- 22.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res 38: e131, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harder J, Dressel S, Wittersheim M, Cordes J, Meyer-Hoffert U, Mrowietz U, Fölster-Holst R, Proksch E, Schröder JM, Schwarz T, Gläser R. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J Invest Dermatol 130: 1355–1364, 2010 [DOI] [PubMed] [Google Scholar]
- 24.Harder J, Schroder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. J Biol Chem 277: 46779–46784, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Harder J, Schröder JM. Psoriatic scales: a promising source for the isolation of human skin-derived antimicrobial proteins. J Leukoc Biol 77: 476–486, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Head SR, Komori HK, Hart GT, Shimashita J, Schaffer L, Salomon DR, Ordoukhanian PT. Method for improved Illumina sequencing library preparation using NuGEN Ovation RNA-Seq System. Biotechniques 50: 177–180, 2011 [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jabbari A, Suárez-Fariñas M, Dewell S, Krueger JG. Transcriptional profiling of psoriasis using RNA-seq reveals previously unidentified differentially expressed genes. J Invest Dermatol 132: 246–249, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25: 1026–1032, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston A, Guzman AM, Swindell WR, Wang F, Kang S, Gudjonsson JE. Early tissue responses in psoriasis to the anti-TNF-α biologic etanercept suggest reduced IL-17R expression and signalling. Br J Dermatol [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, Zhang W, Bowcock AM. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet 20: 4025–4040, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitamura H, Matsuzaki Y, Kimura K, Nakano H, Imaizumi T, Satoh K, Hanada K. Cytokine modulation of retinoic acid-inducible gene-I (RIG-I) expression in human epidermal keratinocytes. J Dermatol Sci 45: 127–134, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Kogenaru S, Qing Y, Guo Y, Wang N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics 13: 629, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kvam VM, Liu P, Si Y. A comparison of statistical methods for detecting differentially expressed genes from RNA-seq data. Am J Bot 99: 248–256, 2012 [DOI] [PubMed] [Google Scholar]
- 36.Landau WM, Liu P. Dispersion estimation and its effect on test performance in RNA-seq data analysis: a simulation-based comparison of methods. PLoS One 8: e81415, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B, Tsoi LC, Swindell WR, Gudjonsson JE, Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, Kochkodan JJ, Voorhees JJ, Kang HM, Nair RP, Abecasis GR, Elder JT. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J Invest Dermatol [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; Subgroup, 1000 Genome Project Data Processing. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li XL, Ezelle HJ, Hsi TY, Hassel BA. A central role for RNA in the induction and biological activities of type 1 interferons. Wiley Interdiscip Rev RNA 2: 58–78, 2011 [DOI] [PubMed] [Google Scholar]
- 40.Lu X, Du J, Liang J, Zhu X, Yang Y, Xu J. Transcriptional regulatory network for psoriasis. J Dermatol 40: 48–53, 2013 [DOI] [PubMed] [Google Scholar]
- 41.McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics 7: 49, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melbye SW, Brant BA, Freedberg IW. Epidermal nucleases. III. The ribonucleases of human epidermis. Br J Dermatol 97: 355–364, 1977 [DOI] [PubMed] [Google Scholar]
- 43.Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, Ruether A, Schreiber S, Weichenthal M, Gladman D, Rahman P, Schrodi SJ, Prahalad S, Guthery SL, Fischer J, Liao W, Kwok PY, Menter A, Lathrop GM, Wise CA, Begovich AB, Voorhees JJ, Elder JT, Krueger GG, Bowcock AM, Abecasis GR; Collaborative Association Study of Psoriasis. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet 41: 199–204, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlén M, Nielsen J. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res 40: 10084–10097, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Opitz L, Salinas-Riester G, Grade M, Jung K, Jo P, Emons G, Ghadimi BM, Beissbarth T, Gaedcke J. Impact of RNA degradation on gene expression profiling. BMC Med Genomics 3: 36, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 4: 14, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulsen MT, Veloso A, Prasad J, Bedi K, Ljungman EA, Magnuson B, Wilson TE, Ljungman M. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods 67: 45–54, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perera GK, Ainali C, Semenova E, Hundhausen C, Barinaga G, Kassen D, Williams AE, Mirza MM, Balazs M, Wang X, Rodriguez RS, Alendar A, Barker J, Tsoka S, Ouyang W, Nestle FO. Integrative biology approach identifies cytokine targeting strategies for psoriasis. Sci Transl Med 6: 223ra22, 2014 [DOI] [PubMed] [Google Scholar]
- 49.Popova T, Mennerich D, Weith A, Quast K. Effect of RNA quality on transcript intensity levels in microarray analysis of human post-mortem brain tissues. BMC Genomics 9: 91, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prens EP, Kant M, van Dijk G, van der Wel LI, Mourits S, van der Fits L. IFN-alpha enhances poly-IC responses in human keratinocytes by inducing expression of cytosolic innate RNA receptors: relevance for psoriasis. J Invest Dermatol 128: 932–938, 2008 [DOI] [PubMed] [Google Scholar]
- 51.Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, Betel D. Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol 14: R95, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santiago TC, Purvis IJ, Bettany AJ, Brown AJ. The relationship between mRNA stability and length in Saccharomyces cerevisiae. Nucleic Acids Res 14: 8347–8360, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simanski M, Rademacher F, Schröder L, Schumacher HM, Gläser R, Harder J. IL-17A and IFN-γ synergistically induce RNase 7 expression via STAT3 in primary keratinocytes. PLoS One 8: e59531, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: 3, 2004 [DOI] [PubMed] [Google Scholar]
- 56.Solier S, Ryan MC, Martin SE, Varma S, Kohn KW, Liu H, Zeeberg BR, Pommier Y. Transcription poisoning by Topoisomerase I is controlled by gene length, splice sites, and miR-142–3p. Cancer Res 73: 4830–4839, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stalteri MA, Harrison AP. Interpretation of multiple probe sets mapping to the same gene in Affymetrix GeneChips. BMC Bioinformatics 8: 13, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suárez-Fariñas M, Li K, Fuentes-Duculan J, Hayden K, Brodmerkel C, Krueger JG. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol 132: 2552–2564, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su Z, Li Z, Chen T, Li QZ, Fang H, Ding D, Ge W, Ning B, Hong H, Perkins RG, Tong W, Shi L. Comparing next-generation sequencing and microarray technologies in a toxicological study of the effects of aristolochic acid on rat kidneys. Chem Res Toxicol 24: 1486–1493, 2011 [DOI] [PubMed] [Google Scholar]
- 60.Swindell WR, Johnston A, Voorhees JJ, Elder JT, Gudjonsson JE. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics 14: 527, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swindell WR, Johnston A, Xing X, Voorhees JJ, Elder JT, Gudjonsson JE. Modulation of epidermal transcription circuits in psoriasis: new links between inflammation and hyperproliferation. PLoS One 8: e79253, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swindell WR, Xing X, Stuart PE, Chen CS, Aphale A, Nair RP, Voorhees JJ, Elder JT, Johnston A, Gudjonsson JE. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One 7: e34594, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.'t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX, Boer JM, van Ommen GJ, den Dunnen JT. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res 36: e141, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Dijk EL, Jaszczyszyn Y, Thermes C. Library preparation methods for next-generation sequencing: Tone down the bias. Exp Cell Res 322: 12–20, 2014 [DOI] [PubMed] [Google Scholar]
- 66.Veloso A, Biewen B, Paulsen MT, Berg N, Carmo de Andrade Lima L, Prasad J, Bedi K, Magnuson B, Wilson TE, Ljungman M. Genome-wide transcriptional effects of the anti-cancer agent camptothecin. PLoS One 8: e78190, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28: 2184–2185, 2012 [DOI] [PubMed] [Google Scholar]
- 68.Washenberger CL, Han JQ, Kechris KJ, Jha BK, Silverman RH, Barton DJ. Hepatitis C virus RNA: dinucleotide frequencies and cleavage by RNase L. Virus Res 130: 85–95, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36: D13–D21, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wongpalee SP, Sharma S. The pre-mRNA splicing reaction. Methods Mol Biol 1126: 3–12, 2014 [DOI] [PubMed] [Google Scholar]
- 71.Wreschner DH, McCauley JW, Skehel JJ, Kerr IM. Interferon action–sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289: 414–417, 1981 [DOI] [PubMed] [Google Scholar]
- 72.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res 40: e133, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia J, Joyce CE, Bowcock AM, Zhang W. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Hum Mol Genet 22: 737–748, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu X, Zhang Y, Williams J, Antoniou E, McCombie WR, Wu S, Zhu W, Davidson NO, Denoya P, Li E. Parallel comparison of Illumina RNA-Seq and Affymetrix microarray platforms on transcriptomic profiles generated from 5-aza-deoxy-cytidine treated HT-29 colon cancer cells and simulated datasets. BMC Bioinformatics 14, Suppl 9: S1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao Y, Richman L, Morehouse C, de los Reyes M, Higgs BW, Boutrin A, White B, Coyle A, Krueger J, Kiener PA, Jallal B. Type I interferon: potential therapeutic target for psoriasis? PLoS One 3: e2737, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaba LC, Suárez-Fariñas M, Fuentes-Duculan J, Nograles KE, Guttman-Yassky E, Cardinale I, Lowes MA, Krueger JG. Effective treatment of psoriasis with etanercept is linked to suppression of IL-17 signaling, not immediate response TNF genes. J Allergy Clin Immunol 124: 1022–1010, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLoS One 9: e78644, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zibert JR, Løvendorf MB, Litman T, Olsen J, Kaczkowski B, Skov L. MicroRNAs and potential target interactions in psoriasis. J Dermatol Sci 58: 177–185, 2010 [DOI] [PubMed] [Google Scholar]