Abstract
Variability in myosin phosphatase (MP) subunits may provide specificity in signaling pathways that regulate muscle tone. We utilized public databases and computational algorithms to investigate the phylogenetic diversity of MP regulatory (PPP1R12A-C) and inhibitory (PPP1R14A-D) subunits. The comparison of exonic coding sequences and expression data confirmed or refuted the existence of isoforms and their tissue-specific expression in different model organisms. The comparison of intronic and exonic sequences identified potential expressional regulatory elements. As examples, smooth muscle MP regulatory subunit (PPP1R12A) is highly conserved through evolution. Its alternative exon E24 is present in fish through mammals with two invariant features: 1) a reading frame shift generating a premature termination codon and 2) a hexanucleotide sequence adjacent to the 3′ splice site hypothesized to be a novel suppressor of exon splicing. A characteristic of the striated muscle MP regulatory subunit (PPP1R12B) locus is numerous and phylogenetically variable transcriptional start sites. In fish this locus only codes for the small (M21) subunit, suggesting the primordial function of this gene. Inhibitory subunits show little intragenic variability; their diversity is thought to have arisen by expansion and tissue-specific expression of different gene family members. We demonstrate differences in the regulatory landscape between smooth muscle enriched (PPP1R14A) and more ubiquitously expressed (PPP1R14B) family members and identify deeply conserved intronic sequence and predicted transcriptional cis-regulatory elements. This bioinformatic and computational study has uncovered a number of attributes of MP subunits that supports selection of ideal model organisms and testing of hypotheses regarding their physiological significance and regulated expression.
Keywords: myosin phosphatase, Mypt1, Mypt2, CPI-17
the sequencing of the human genome (44, 104) opened vast new insights into the variability and diversity encoded within the genome. Before this, predictions for the number of human protein-coding genes ranged between ∼35,000 (24) and upwards of 150,000 (14). The count of protein-coding genes in the most recent build of the human genome (GRCh37.p13, 2009) is 20,805, a number that is on par with that of the simple organism Caenorhabditis elegans, 20,532 (WBcel235, 2012). This has led to the premise that much of the complexity of the human transcriptome is derived from the creation of multiple products from a single gene, such that humans have 69,000 different proteins, whereas in comparison the worm has 25,000, according to current database curations (Ensembl, UniProt). Multiple transcripts are generated from a single gene by alternative usage of exons through alternative splicing or alternative transcriptional start sites, each of which are nearly universal features of multiexon genes in mammals and higher vertebrates (67, 79, 108). An example is the human tropomyosin α (TPM1) gene in which the combination of alternative splicing and multiple transcriptional start sites results in 18 distinct protein coding transcripts (reviewed in Ref. 32).
In humans, the DNA of protein-coding genes accounts for less than 3% of the total genome (20); in the past the remaining 97% of the genome was considered “junk DNA.” However, recent evidence suggests that >80% of the genome is biochemically active: either transcribed (protein coding, noncoding, and pseudogenes), chromatin-associated, or regulatory (20). These active regulatory regions may be intra- or intergenic and contain cis elements that regulate gene transcription or removal of introns (splicing of exons) to convert pre-mRNA into mature protein coding mRNA. Phylogenetic conservation of noncoding DNA sequences has been successfully used to identify transcriptional regulatory elements such as enhancers (reviewed in Ref. 37), though there is not a simple relationship in either direction between sequence conservation and regulatory elements (reviewed in Refs. 72 and 113). Similarly, it has been found that intronic regions flanking alternative exons, and the cis regulatory elements within, are under selection pressure and often exhibit broad conservation (67).
Tissue specificity and tissue gene expression signatures are also frequently conserved, particularly among mammals and higher vertebrates, such that the transcriptome of a given tissue is more similar across species than it is to other cell types within the same species (3, 6, 67). The evolutionary divergence of multiple closely related family members from a single ancestral gene creates diversity in higher organisms through specific expression, either tissue specific or during development, and also through specificity of interactions and substrates. The differing strategies of cell conservation and diversification are illustrated in the protein kinases and phosphatases that regulate approximately one-third of all eukaryotic proteins. Approximately 400 serine-threonine kinases are present in the mammalian genome with a steady increase in the number throughout evolution of eukaryotes. In contrast, only ∼25 corresponding phosphatases are present with little evolutionary change in this number (87). This reflects differing strategies of diversification, as diversity in the activity of Type 1 phosphatases is generated by a large number and variability within associated regulatory subunits and signaling pathways controlling their activity (9).
In smooth muscle the myosin light chain kinase (MLCK) and phosphatase (MP) are primary determinants of the state of contraction of the muscle and thereby contribute to the regulation of blood flow and pressure and other vital organ functions (reviewed in Refs. 34, 38, and 40). MLCK and MP enzymes are also present in striated muscle where their function is less understood but are thought to play a more modulatory role in contractile function (reviewed in Refs. 47 and 94), and in nonmuscle cells where they regulate motility and cytokinesis (reviewed in Ref. 65). The MP holoenzyme is composed of three subunits: the protein phosphatase 1 (PP1) catalytic subunit, the MP targeting/regulatory subunit (MYPT), and the small subunit (M21) (reviewed in Ref. 45). A fourth subunit, CPI-17, is an inhibitory subunit that when phosphorylated inhibits MP activity (reviewed in Ref. 21).
Using traditional methods, we and others have made some progress in determining how variability in the MP regulatory subunits in humans and animal models may provide cell-specific functions. For example, smooth muscle, like striated muscle, may be functionally dichotomized into fast (phasic) versus slow (tonic) contractile phenotypes. Each uses MCLK and MP for activation and dactivation of force, yet the force outputs and how they are regulated are very different (reviewed in Refs. 27 and 100). Within the Mypt1 regulatory subunit an alternative exon (exon 24) that is included in phasic smooth muscle and skipped in tonic smooth muscle is thought to determine regulation of MP activity by cGK1α [nitric oxide (NO) signaling pathway] (51, 83, 120; reviewed in Ref. 15). Similarly, variable expression of an inhibitory subunit of MP, CPI-17 (PPP1R14A), is proposed to determine tissue-specific regulation of MP activity by α-adrenergic signaling (53, 117; reviewed in Ref. 21).
We hypothesized that through interrogation of publicly available databases we could 1) uncover much more about the phylogenetic conservation, variability, and tissue-specific expression of MP subunit isoforms; and 2) discover potential transcriptional and splicing cis-regulatory elements that may control the variability in the expression of MP subunits in muscle tissues, regarding which little is currently understood (reviewed in Refs. 32, 46, 78, and 107). This informatics approach provides new insights into the relationship between MP diversity and muscle diversity and predictions regarding how MP diversity may be generated, providing a foundation for further experimentation and selection of appropriate model organisms.
METHODS
Protein Domain
We used several protein databases to identify conserved protein domains in fly and worm: InterPro (43), PANTHER (102), Pfam (89), and SUPFAM (80).
Conservation and Alignment
Conservation of the Mypt1 alternative exon was determined with the PhastCons conservation track in the UCSC Genome Table Browser (genome.ucsc.edu) (49, 50). All genomic and transcript data were gathered from Ensembl (Release 72, June 2013). Nucleic acid sequence alignment was performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) (98). JalView (112) was used to calculate pairwise sequence identity and to perform alignment editing.
Splice Site and cis-Regulatory Element Predictions
Mypt1 + 2 E24 splice sites were predicted using Human Splicing Finder v2.4.1 (www.umd.be/HSF/) (13) and the Alternative Splice Site Predictor (ASSP; wangcomputing.com/assp) (109), using human and chicken Mypt1 E24 as prediction positive controls. Human Splicing Finder identification of splice sites utilizes a positional weight matrix (PWM) of consensus splice signals (13) to identify potential splice sites that conform to consensus splice signals above threshold. The ASSP prediction algorithm was developed using sequences for known constitutive, alternative, and cryptic splice sites to take into account nonconsensus splice sites (109).
Conserved motifs were identified using Gapped Local Alignment of Motifs (GLAM2; meme.nbcr.net) (28). Predicted cis-splicing regulatory elements (SREs) were then identified using Human Splicing Finder v2.4.1 (www.umd.be/HSF/) (13). Human Splicing Finder reports predicted SREs from several experimental and computational-based prediction sets: ESE Finder (7, 99), RESCUE ESE (25), and other published predictions (33, 110, 119, 121). Of the experimentally derived SRE prediction sets, exonic splicing silencer (ESS) sequences were obtained by testing random decanucleotide sequences in splicing reporters (110). SREs from ESE Finder and Human Splicing Finder were predicted using consensus motif PWMs derived from SELEX (systematic evolution of ligands by exponential enrichment) experiments (7, 13). The other SRE prediction sets were computationally derived through different parameters: predicted exonic splicing enhancers (ESEs) based on hexamer enrichment in weak exons [RESCUE ESE, (25)]; predicted exonic splicing regulatory sequences (ESRs) based on enrichment of conserved (human-mouse) hexamers in human-mouse orthologous exons (33); predicted exonic SREs based on enrichment of octamers in constitutive, noncoding exons (121); predicted SREs as asymmetrically enriched in introns (intronic identity elements: IIEs) or exons (exonic identity elements: EIEs) (119).
Expression Data
Affymetrix exon array data (88) and RNA-Seq tissue expression data (85) were obtained from publicly available sources (genome.ucsc.edu; http://www.ebi.ac.uk/gxa).
Promoter and Enhancer Analysis
Transcriptional start site predictions for human genes were assembled from the SwitchGear Genomics Transcriptional Start Site track in the UCSC Genome Table Browser (genome.ucsc.edu) (49, 50). SwitchGear TSS predictions are based on human GenBank cDNAs. UCSC Genome Browser (human, hg19) ENCODE tracks for H3K4Me1, H3K4Me3, and H3K27Ac from 7 human cell lines (19), ENCODE DNase hypersensitivity clusters from 125 human cell types (20, 103), and ENCODE transcription factor ChIP were used to determine regions of transcriptional activity.
Regions conserved between human and mouse were aligned using the ECR Browser (www.dcode.org) (77). Conserved and aligned transcription factor binding sites (TFBS) were identified using the NCBI dcode package including MultiTF and rVista 2.0 (www.dcode.org) (61, 76). The rVista program utilizes the comprehensive database of TFBS motifs TRANSFAC Pro V10.2 (114) and scores sequence similarity of TFBS to TRANSFAC PWM. We used two cutoffs for TFBS identification: 1) the rVista “optimized for function” method that independently optimizes each TFBS to limit the density of an individual TFBS to ≤3 sites per 10 kb of random sequence (76) and 2) 0.85 fixed cutoff score for sequence similarity to TRANSFAC PWMs.
RESULTS
Phylogenetic Conservation of MP Subunit Families
Catalytic subunit.
The PP1 family (PPP1CA, PPP1CB, PPP1CC) is highly conserved with extensive sequence identity between paralogs (>85%) and among orthologs in other species (Fig. 1A; reviewed in Refs. 9 and 10) (70). Orthologs of this family can be found in the fruit fly, worm, yeast, and plants (58), indicating ancient origins for the PP1 catalytic subunit. The central majority of the PP1 proteins, including the catalytic and binding domains, have nearly identical amino acid sequences. The modest amount of sequence diversity among the paralogs is found in the distal COOH- and NH2-terminal ends of the proteins.
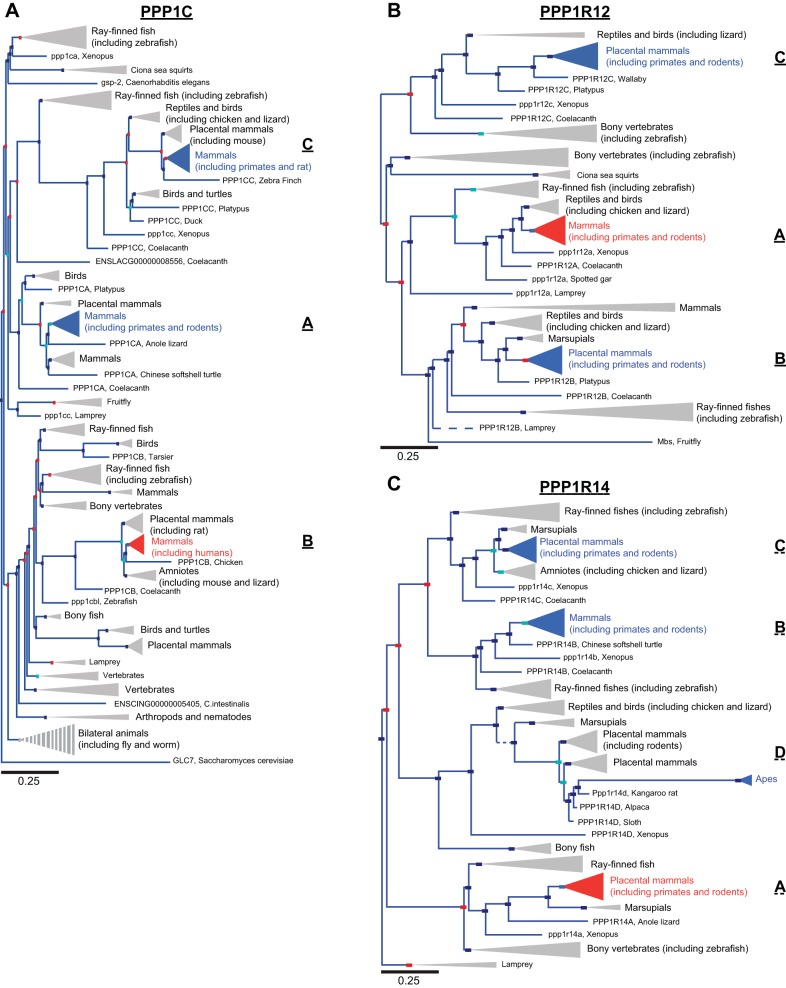
Fig. 1.
Phylogenetic trees of myosin phosphatase subunit families. A: phylogenetic tree of the protein phosphatase 1 (PP1) catalytic subunit family demonstrates high conservation amongst orthologs and paralogs. The node containing the human myosin phosphatase (MP) catalytic subunit, PPP1CB (PP1β), is shown in red, whereas paralogs PPP1CA (PP1α) and PPP1CC (PP1γ) are in blue. B: phylogenetic tree of the MP targeting subunit family is given with the node containing the human smooth muscle MP subunit, protein phosphatase 1 regulatory subunit 12A (PPP1R12A) (MP targeting subunit 1; Mypt1) shown in red and paralogs PPP1R12B (Mypt2) and PPP1R12C (myosin binding subunit 85; MBS85) shown in blue. C: phylogenetic tree of the MP inhibitory subunit family (PP1 regulatory subunit 14; PPP1R14) demonstrates greater variability between paralogs and orthologs compared with the catalytic subunit in A. The node containing the human MP inhibitory subunit PPP1R14A (c-kinase potentiated protein phosphatase 1 inhibitor 17; CPI-17) is shown in red, whereas the paralogs PPP1R14B (phosphatase holoenzyme inhibitor; PHI), PPP1R14C (kinase enhanced phosphatase inhibitor; KEPI), and PPP1R14D (gastric brain phosphatase inhibitor; GBPI) are shown in blue. In the trees the red boxes represents gene duplication and the dark blue boxes speciation events. Light blue boxes are ambiguous nodes. Branch lengths along the horizontal axis correspond to the expected number of changes per nucleotide site in the DNA, as indicated in the corresponding scale bars, which is proportional to evolutionary divergence. The length (horizontal) of solid branches and nodes (triangles) is 1× distance, whereas dashed branches and striped nodes are 10× distance. The width (vertical) of nodes is proportional to the number of orthologs included in the node. Phylogenetic trees were developed by Ensembl, Release 75 (February 2014) (105).
Pairwise analysis of the amino acid and nucleotide sequences indicate that the PPP1CA and PPP1CC paralogs are more similar to each other than to PPP1CB, the gene that codes for the catalytic subunit of MP (also known as PP1β or PP1δ). This is true for the fly as well, suggesting that PPP1CB originated before vertebrates and invertebrates diverged (58). PP1γ (PPP1CC) appears to be a newer isoform, having diverged from PPP1CA, and is less conserved among orthologs than are the PP1α (PPP1CA) and PP1β (PPP1CB). All of the paralogs are ubiquitously expressed.
Thus diversity in the serine-threonine type 1 phosphatases depends on a multitude of PP1 regulatory subunits (reviewed in Ref. 10). The regulatory subunits of the MP catalytic subunit, Mypt1 and CPI-17, have greater evolutionary divergence within their families and greater variability among their orthologs than the catalytic subunit (Fig. 1).
Regulatory subunit.
The Mypt (PPP1R12) regulatory subunit family is part of a very large super family of ankyrin repeat domain proteins composed of two subtrees. Along with the PPP1R12 family, the first subtree contains the PPP1R16 (A-B) family, also known as Mypt3 (PPP1R16A) and TGF-β-inhibited membrane-associated protein (TIMAP; PPP1R16B). The second subtree within this super family contains PPP1R13B, which codes for apoptosis-stimulating protein of p53 1 (ASPP1), PPP1R13L, which codes for inhibitor of apoptosis stimulating protein of p53 (IASPP) protein, and the tumor protein p53 binding protein (TP53BP2). These subfamily members are sometimes classified as Mypts; given their evolutionary distance and lack of cardinal features of Mypt family members (described below) we consider them to be distinct subfamilies.
The myosin phosphatase regulatory subunit family of proteins is composed of Mypt1, Mypt2, and MBS85 (PPP1R12A-C, respectively). The PPP1R12 family is well conserved and there is an orthologous Mypt gene in both the fly (Mbs, Drosophila melanogaster) and worm (Mel-11, C. elegans) (68, 116). Each of the PPP1R12 family members contains an RVxF motif (PP1 binding site) and 7 or 8 ankyrin repeat domains toward the NH2-terminus, 1 or 2 conserved Thr phosphorylation sites, and a leucine zipper motif at the COOH-terminus (reviewed in Ref. 34). Whereas Mypt family members (paralogs) overall have 39–61% amino acid identity, the described conserved domains have much higher sequence identities (50–90%) (reviewed in Ref. 45). These characteristic domains of the Mypt family hold true for the fly and worm orthologs as well (68, 116). The COOH-terminus leucine zipper motif is highly conserved among species and is similar in sequence between family members (88%) (34, 45). BLAST searches of the genome with any of the three PPP1R12 family LZ motifs match only to the other members of the family, suggesting that these specific LZ motif sequences are specific to the PPP1R12 Mypt family.
Inhibitory subunit.
Sequence similarity between inhibitory subunit family member paralogs and orthologs is much less than for the catalytic subunit family (Fig. 1C). However, within the phosphatase inhibitory domain there is >41% sequence similarity (reviewed in Ref. 21). Importantly, the Thr38 phosphorylation site of PPP1R14A (CPI-17), which regulates its inhibitory activity, and the surrounding residues, which are necessary for phosphorylation of Thr38, are conserved in the other three family members (PPP1R14B-D). Orthologs of PPP1R14 are found in lower species, CG17124 in the fly and F55C10.5 in the worm (64, 95) (flybase.org; wormbase.org), and were not previously identified (21). The Thr38 phosphorylation site is conserved in the fly and worm orthologs, while the flanking sequences contain hydrophobic and basic residues but vary from the vertebrate consensus sequence (basic-hydrophobic-Thr-hydrophobic-basic).
Interestingly, the chicken genome lacks the PPP1R14A gene, and it has been suggested that chicken smooth muscle MP is inhibited by the CPI-17 paralog PHI (PPP1R14B) (17, 18, 54). However, PPP1R14B is also not found in the completed sequence of the chicken genome (Genome assembly Galgal4, Ensembl Genebuild updated Dec. 2013). Another member of the PPP1R14 family, e.g., the more closely related kinase enhanced phosphatase inhibitor (KEPI) (PPP1R14C), may function as the MP inhibitory subunit in chicken smooth muscle.
Alternative Splicing of Regulatory Subunits and MP Diversity
Mypt1.
The mammalian PPP1R12A (Mypt1) gene consists of 26 exons, 3 of which are alternatively spliced (E13, E14, and E24). Alternative splicing in the central region of Mypt1 is conserved, though the specific alternative exons vary (are not orthologous): E12 in the chicken (16, 96), E9 and E11 in the worm (115) (wormbase.org), and the functional significance of these variants is unknown. Alternative splicing of Mypt1 E24, a 31 nt exon, determines the coding for, or lack of, a COOH-terminal LZ motif in the Mypt1 protein (Fig. 2D). The LZ motif of Mypt1 is thought to be required for LZ-mediated heterodimerization with cGK1α- and cGMP-dependent activation of MP (42, 51, 101) (57). The expression of Mypt1 E24 and central splice variant isoforms has been studied extensively during development (51, 83, 97) and in disease models (16, 29, 35, 62, 84, 97, 120) (reviewed in Refs. 15 and 27).
Fig. 2.
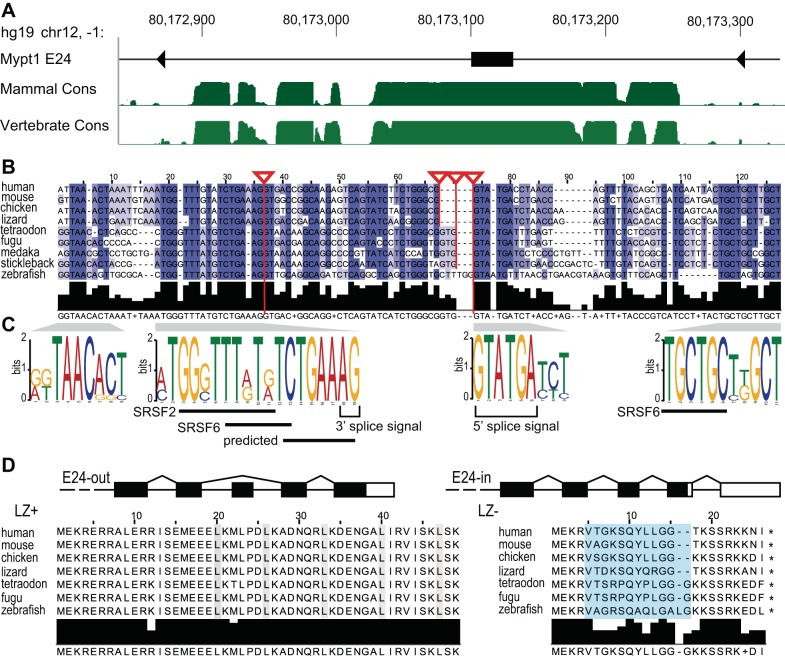
Phylogenetic conservation of isoforms of Mypt1 generated by alternative splicing of Exon 24. A: conservation of the myosin targeting subunit 1 (Mypt1) alternative exon E24 and the flanking intronic region is shown by the phastCons track on the UCSC Genome Browser in the human hg19/GRCh37 genome release. The phastCons track is a multiple alignment of 46 vertebrate species (“Vertebrate Cons”) and a subset of 33 placental mammal (“Mammal Cons”) that estimates the probability of individual nucleotides belonging to a conserved element, by considering both the individual alignment column and its flanking columns. The higher the green bars the more likely the region belongs to a conserved element. B: alignment and coloring based on percent identity of Mypt1 E24 in human, mouse, chicken, and lizard, and the predicted alternative exon in fish demonstrates regions of high conservation immediately flanking the exon. Red lines attached to triangles highlight known and predicted (in fish) splice sites. Conserved sequences were identified and analyzed for splicing cis-regulatory elements as described in methods. C: exon-intron structure of the 3′ end of the Mypt1 gene is shown. Alternative splicing of the 31 nt E24 changes the reading frame. Amino acid sequence alignment of the Mypt1 COOH-terminus for the E24-out/leucine zipper (LZ)+ and the E24-in/LZ− isoforms, demonstrates phylogenetic conservation, with LZ+ more conserved than LZ−. The leucines of the LZ motif for the E24-out isoform (left) are highlighted in gray. The amino acids that are coded by the alternative exon E24 are highlighted in blue.
Mypt1 E24 is located in a region of high conservation spanning approximately 120 bp upstream through 200 bp downstream of E24 (Fig. 2A) (97). E24 is partially conserved in fish genomes (tetraodon, fugu, stickleback, medaka, and zebrafish), though, notably, a homologous sequence is absent in the frog, lamprey, and C. elegans orthologs. Sequence alignment of the alternative exon and 50 bp flanking regions demonstrates that this region is highly conserved in mammals, the chicken, and lizard; in fish there is 67–73% sequence identify of E24 to mammals, whereas the flanking sequence is less well conserved than in the higher vertebrates (Fig. 2B).
The 3′ and 5′ splice sites are computationally predicted in the fish flanking the sequence that is homologous to mammalian Mypt1 E24 alternative exon, resulting in a 34 (most fish) or 37 nt (zebrafish) predicted exon (Fig. 2B). Using RT-PCR, we confirmed that this is indeed a tissue-specific alternative exon in the zebrafish smooth muscle (data not shown). As in higher vertebrates the fish alternative exon also causes a 1-nt shift in the reading frame resulting in a Mypt1 subunit that lacks the COOH-terminal LZ motif (Fig. 2D, right). The LZ motif is highly conserved in the annotated PPP1R12A protein product in tetraodon, fugu, and zebrafish (Fig. 2D, left). The shift in the reading frame also results in a premature termination codon (PTC) in the fish Mypt1, a feature that is also conserved throughout phylogeny (Fig. 2D, right), suggesting functional importance. However, the function and significance of the Mypt1 COOH-terminus alternative (LZ−) amino acid sequence and PTC, respectively, are not known at this time.
The intronic sequence immediately flanking the alternative exon is well conserved (Fig. 2B). The nonconsensus 5′ splice site sequence (guagua) and the lack of an upstream polypyrimidine tract are both characteristic of a weakly spliced (alternative) exon (reviewed in Ref. 2). Two upstream and one downstream blocks of intronic sequence are highly conserved from fish through higher vertebrates suggesting they could function in the regulation of exon splicing (Fig. 1B). Analysis of these sequences using splicing regulatory elements (SRE) prediction algorithms for known cis-splicing regulatory elements (see methods) reveals conserved sites for splicing factors SRSF2 (SRp30b) and SRSF6 (SRp55) in the intronic regions immediately flanking E24 (Fig. 2C). We also identified computationally predicted SREs of unknown function (25, 119) within the highly conserved sequence immediately adjacent to the 3′ splice site: ctgaaa (human-lizard)/ctgaag (fish) and tgaaag (human-lizard)/tgaagG (fish) (Fig. 2C). The proximity of the identified elements to the 3′ splice site suggests that they may function as splicing repressors by blocking recruitment of U2 splicing factor to this site (see discussion). Of note the E24 sequence itself is highly conserved in higher vertebrates but less well conserved in fish. A number of cis-regulators of splicing located within higher vertebrates' E24 (97) are not present within the fish E24 sequence. How this may affect the regulation of E24 splicing is considered further in discussion. A number of other predicted conserved cis-regulatory splicing elements are identified both within the alternative exon and flanking introns (Supplemental Table S1).
Mypt2.
Alternative splicing of exon 24 (numbering based on human gene) of Mypt2 (PPP1R12B) gene product has similarities and differences with that of Mypt1 E24 but has been much less studied. The Mypt2 E24 skipped isoform codes for a highly conserved COOH-terminal LZ motif (Fig. 3C) that is nearly identical in amino acid sequence to the COOH-terminal LZ sequence of family members Mypt1 and p85. In contrast to Mypt1 E24, Mypt2 E24 inclusion codes for an alternative COOH-terminal LZ sequence and contains the PTC (Fig. 3A). Thus MBS85 (PPP1R12C) is the only Mypt family member with an invariant COOH-terminal (LZ) sequence. Like Mypt1 E24, splicing of Mypt2 E24 is highly restricted (described in detail under Complexity of the Mypt2 Locus).
Fig. 3.
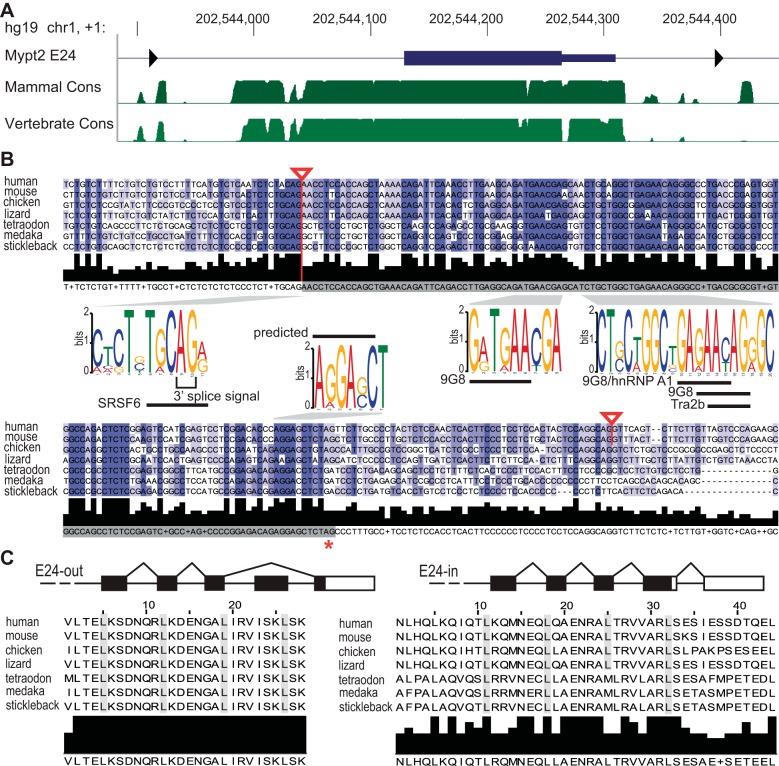
Phylogenetic conservation of isoforms of Mypt2 generated by alternative splicing of Exon 24. A: conservation of the myosin phosphatase targeting subunit 2 (Mypt2) alternative exon and flanking region is shown by the phastCons track on the UCSC Genome Browser in the human hg19/GRCh37 genome release, as described in Fig. 1. B: mammalian (human, mouse) Mypt2 E24 and flanking regions are aligned to the homologous sequences in chicken, lizard, and fish PPP1R12B. Red lines and triangles highlight the known (mammalian) and predicted splice sites for E24. The red asterisk denotes the known (mammalian) and aligned stop codon. Conserved motifs were identified and analyzed for splicing cis-regulatory elements (see methods). C: diagrammed gene structure of the 3′ end of the Mypt2 gene indicates alternative splicing of E24 and the change of the open reading frame, in black. Amino acid sequence alignment of the COOH-terminus of Mypt2 demonstrates high conservation of the E24-out leucine zipper (LZ) motif (left). The E24-in LZ motif (right) is more variable in fish. Leucine residues of the LZ motifs are highlighted in gray.
Like Mypt1 E24, Mypt2 E24 sequence is highly conserved in the genomes of mammals (94% sequence identity to mouse), chicken (76%), and lizard (74%) and also absent in frog. There is 58–63% conservation of the coding portion of the exon of the human Mypt2 E24 in fish (medaka, stickleback, tetraodon) (Fig. 3B). Unlike Mypt1, there is a polypyrimidine-rich tract upstream of Mypt2 E24 and high 3′ splice site consensus conformity (85–91%) in all of the species examined, giving a robust 3′ splice site prediction conforming to the known mammalian splice site (Fig. 3B, red line). The stop codon is also in alignment, though it is a TGA in fish as opposed to a TAG in mammals, chicken, and lizard (Fig. 3B, red asterisk). There is minimal homology in the exonic sequence immediately downstream of the PTC, and the downstream intronic flanking region is not conserved. The annotated mammalian 5′ splice site (Fig. 3B, red line) was not predicted by either the Human Splicing Finder (13) or the Alternative Splice Site Predictor (109). Neither program predicted a 5′ splice site for the other species investigated either, suggesting a very weak 5′ splice site for Mypt2 E24. This along with other features described below, likely accounts for the extremely tissue-restricted and phylogenetically limited splicing of this exon. Alternatively, given the internal PTC in Mypt2 E24, it is conceivable that it could function as a terminal exon, in which case there would be no need for the 5′ splice site, though there is no data to support this scenario at this time.
The COOH-terminal LZ motif coded for by skipping of Mypt2 E24 is highly conserved through fish (Fig. 3C, left). The alternative LZ motif encoded by E24 is 48–52% conserved from human to fish and retains three of the four leucines of the alternative LZ motif (Fig. 3C, right).
Interestingly, a distinct 67 nt alternative exon has been identified in the chicken Mypt2 (63) located in the same intron and nearly 4.5 kb upstream of the sequence with homology to mammalian E24. This chicken Mypt2 alternative E24 has a PTC in the fourth codon, resulting in a COOH-terminal LZ− variant (63). We could not identify with confidence sequences homologous to the distinct chicken Mypt2 E24 in the other species investigated. The chicken genomic sequence homologous to mammalian Mypt2 E24 (Fig. 3B) has not been demonstrated to be a functional exon in chicken, though it could be that the proper tissues have not yet been examined, e.g., chicken skeletal muscle.
Sequence that is conserved between human and fish within the coding portion of the sequence of the Mypt2 E24 contains predicted binding sites for splicing factors 9G8 and Tra2β (Fig. 3B) and an hnRNP A1 site which may act as a splicing silencer (12). Immediately upstream of the PTC is a conserved predicted exonic identity element: AGGAGC (human-lizard)/AGGACC (fish) (Fig. 3B, Supplemental Table S2). In contrast to Mypt1 E24, there is generally a lack of conserved SREs in the intronic flanking regions of Mypt2 E24: upstream of the 3′ splice site is the pyrimidine-rich tract that is conserved as a feature but not at the level of individual nucleotides, while downstream of the 5′ splice site there is a lack of conservation (Fig. 3B, Supplemental Table S2). An exception is the intronic region immediately flanking and including the 3′ splice site, which is highly conserved and contains a predicted binding site for SRSF6 (SRp55) that spans the 3′ splice site (Fig. 3B) and may inhibit recruitment of the U2 splicing factor. Additional conserved SREs were identified (Supplemental Table S2). The absence of conserved SREs near the 5′ splice site and the nonconsensus 5′ splice site itself is consistent with default skipping of the Mypt2 E24.
Complexity of the Mypt2 Locus
Mypt2, hsM21, smM21.
PPP1R12B is a highly polymorphic gene locus where a number of unique transcripts are generated by alternative splicing of exons (described above) and alternative transcriptional start sites (TSS). Unique TSS generate first exons (transcripts) unique to skeletal (Mypt2), cardiac (hsM21) and smooth (smM21) muscle (Fig. 4A) (1, 11, 30). Each of the annotated first exons of human Mypt2, hsM21, and smM21 are associated with indicators of transcriptional activity (H3K4Me1, H3K4Me3, and H3K27Ac), DNase hypersensitivity, and transcription factor binding as well as with TSS predictions (Fig. 4, B, C, E, red). This suggests a relatively unusual situation in which three loci within a single gene are under separate transcriptional control by the three muscle types. Interestingly, the TSS and first exon of hsM21 appears to differ among mammals. The first exon of human hsM21 is conserved in some species (e.g., rhesus, dog, elephant have 96–83% identity) but is missing completely in the mouse and rat (Fig. 4C). Conversely, the sequence of the annotated first exon of mouse hsM21 is conserved in humans (87.5% identity) and is located ∼11 kb downstream of the human hsM21 first exon (Fig. 4A, gray box). There is extensive conservation (>80%) between the human and mouse in the sequence immediately upstream (400 bases) of the sequence for the mouse hsM21 first exon. However, in the human this region lacks a predicted TSS based on human GenBank cDNAs (49, 50). There is also a lack of H3 modifications, DNase hypersensitivity, and TF binding associated with transcriptional activity (Fig. 4D), which, in total, is consistent with different transcriptional start sites and first exons between mouse and human hsM21 with the caveat that these data were obtained from human cell lines. The mouse hsM21 first exon is highly conserved in the rat (95.4%), but only the 3′ half of the exon is conserved in the other mammals (rhesus, dog, elephant, and opossum). Neither the human nor mouse hsM21 first exon sequence is present in chicken or lower species raising the question of whether hsM21 is generated in these species.
Fig. 4.
Complexity of the Mypt2 (PPP1R12B) locus. A: protein phosphatase 1 regulatory subunit 12B (PPP1R12B) locus hosts independent transcriptional start sites (TSS) for Mypt2 (982 aa), heart-specific M21 (hsM21) (208 aa), and smooth muscle M21 (smM21) (186 aa). The first exons and start codons (ATG) for each transcript are highlighted and color coded in this diagram that uses the human PPP1R12B as the template (red: Mypt2; blue: hsM21; green: smM21). The alternate stop codons (TAG) are also diagrammed. The region corresponding to the first exon of the mouse hsM21 is depicted by a gray box. Experimental evidence for independent transcriptional regulation (regions of histone 3 modification, DNase hypersensitivity, TF ChIP) is shown for the first exons of human Mypt2(B) hsM21 (C), and smM21 (E). SwitchGear predicted TSSs are depicted as red lines. The mammalian and vertebrate conservation is also shown. D: experimental evidence for transcriptional regulation of the first exon of mouse hsM21 and conservation are shown.
In contrast the TSS and unique first exon of smM21 (11), located between exons 18 and 19 of Mypt2 are highly conserved in humans, chickens, and fishes (Fig. 4, A and E). Interestingly, the PPP1R12B ortholog in fish (tetraodon, medaka, and stickleback) appears to generate only the smM21 transcript; no genomic sequence with homology to mammalian Mypt2 spanning exons 1–18 is identified in the fish. This suggests that the original PPP1R12B gene may have only coded for the small M21 subunit, and that the NH2-terminal ankyrin repeats and PP1c binding domains were later acquired through a recombination with another Mypt gene, most likely PPP1R12C given its closer phylogenetic relationship (Fig. 1B). Alternatively, the 5′ end of the PPP1R12B gene could have been lost during fish evolution. Whereas the cause of this variability in the PPP1R12B gene structure during evolution is not defined, the variability itself is consistent with the difficulty in defining the function of MP (Mypt2) in striated muscle.
The complexity of the locus is compounded by the tissue-specific expression of these independent transcripts. Mypt2 is transcribed in the skeletal muscle, heart, and, to a lesser extent, brain (30); hsM21, as indicated by the name, has tissue-specific expression in cardiac muscle (1); and smM21 is expressed in smooth muscle (69, 88). Additionally, splicing of PPP1R12B E24 is highly regulated (1, 88). Inclusion of mammalian E24 in Mypt2 is highly restricted to skeletal muscle (unspecified type) in the context of the full-length Mypt2 transcript, with about half of the total transcript levels composed of the alternative isoform (1). Additionally, both the E24-in and E24-out isoforms of hsM21 were cloned from human cardiac tissue (1), though there is no report of their relative proportions. Inclusion of the distinct avian PPP1R12B 67 nt E24 occurs in the context of smM21 and is also tissue specific, representing the primary isoform in the fast (phasic) gizzard smooth muscle (51, 63). In contrast inclusion of E24 (181 or 67 nt) in mammalian smooth muscle smM21 transcripts is not identified in published studies (63) nor public gene expression datasets. More thorough investigations of the pattern of splicing of Mypt2 E24 are required to define its tissue specificity, potential coupling to gene transcription, and phylogenetic conservation.
MP Inhibitory Subunits
Four family members of the inhibitory subunit of myosin phosphatase (PPP1R14A-D) have been identified with little variation within each transcript (21). Exon 2 of CPI-17 (PPP1R14A) has been reported as alternative in human aorta (118), but our surveys did not uncover an exon skipped variant in a survey of rat smooth muscle tissue (data not shown) and there are no other reports of this variant in the databases. Alternative translational start sites have been proposed for human and mouse PHI (−1,2) (PPP1R14B) (22), but using the current mouse and human genome sequence the proposed PHI-2 AUG sequence 167 nt and 154 nt upstream of the well-validated start codons of mouse and human PHI-1, respectively, 1) lack Kozak consensus sequence for translational initiation and 2) would change the reading frame and cause premature termination after 24 amino acids, casting doubt on this as a bona fide translational variant.
The diversity in the inhibitory subunits arises by their regulated transcription, yet the expressional and functional relationships between the inhibitory subunit family members are not well defined. The initial reports of CPI-17 (PPP1R14A), the first identified family member, demonstrated high levels of expression specific to smooth muscle (23). This finding is confirmed in public databases of human and mouse RNA-Seq and microarray tissue surveys (85, 88), with high levels of CPI-17 transcript in the aorta, bladder, lung, and prostate and much lower levels in striated muscle and nonmuscle cells, mirroring Mypt1 expression. In contrast, PHI (PPP1R14B) expression is fairly ubiquitous with published immunoblot evidence of protein expression (22) and ubiquitous expression in RNA-Seq data in general agreement (85, 88). The expression of the other inhibitory subunit family members KEPI (PPP1R14C) and gastric brain phosphatase inhibitor (GBPI) (PPP1R14D) is less pervasive and less robust. Published studies have characterized KEPI expression as most robust in cardiac tissue (60) while GBPI expression is primarily limited to the colon (59).
There is evidence both for (18) and against (81) redundancy between the two main inhibitory subunits expressed in smooth muscle, PPP1R14A and B, yet there are no mouse knockout experiments to define the roles of the subunits. Our unpublished data from Deep RNA sequencing indicates that CPI-17 (PPP1R14A) transcripts are approximately two- to ninefold higher than PHI-1 in vascular smooth muscle (unpublished data, RP Dippold and SA Fisher). Consistent with restricted expression and specialized function of PPP1R14A, the region upstream of the PPP1R14A TSS lacks TATA and CAAT boxes, has little phylogenetic conservation, and lacks indicators of transcriptional activity in cell lines (Fig. 5A). A limited region of 188 bases ∼300–500 bases upstream of the TSS could be aligned between human and mouse and putative TFBS identified, including a GC box that was previously identified and shown to function as a minimal promoter in vitro [(52) and Supplemental Table S3].
Fig. 5.
Analysis of CPI-17 (PPP1R14A) and PHI-1 (PPP1R14B) noncoding sequence for potential transcriptional regulatory activity. A: protein phosphatase 1 regulatory subunit 14A (PPP1R14A). Portions of noncoding sequence in introns 1 and 3 (red boxes 1 and 2) are well conserved between human and mouse and have other indicators suggestive of transcriptional enhancer activity (H3 modification, DNase hypersensitivity, and TF ChIP). The transcription factor binding sites (TFBS) predicted in each of these blocks of sequence are shown. The TFBS in black are predicted using the rVista “optimized for function” approach to reduce false positives (see methods). The TFBS in gray have a singular cutoff of 0.85 similarity to the TRANSFAC positional weighted matrix (PWM) and are of interest to muscle gene regulation. B: PPP1R14B. noncoding sequence is well conserved immediately upstream of the TSS and in intron 1 (red boxes 1 and 2) and have other features suggestive of transcriptional promoter and enhancer activity, respectively. The TFBS predicted in each of these blocks of sequence are shown. The full compilation of TFBS for the two cutoffs can be found in Supplemental Tables S3 and S4.
We used a number of criteria in an attempt to computationally identify potential PPP1R14A (CPI-17) transcriptional enhancers (see methods) (36, 111). Regions within the first intron immediately upstream of exon 2 and within the third intron are well conserved suggesting possible regulatory functions (Fig. 5A). These regions were hypersensitive to DNAse and contained histone modifications consistent with enhancer activity. Algorithms predicted a number of binding motifs for TFs involved in smooth muscle phenotypic determination (reviewed in Ref. 55), including Enhancer box (E-box), cAMP response element-binding protein (CREB), nuclear factor of activated T-cells (NFAT), and peroxisome proliferator-activated receptor-γ (PPARγ) but notably no CARG motifs for the binding of serum response factor (SRF) and myocardin, a well-validated smooth muscle cis-regulatory motif (reviewed in Ref. 82). The absence of indicators of transcriptional activity near the TSS likely reflects the tissue-specific transcription of PPP1R14A and its low expression in the various cell lines in which these assays are performed.
In contrast to PPP1R14A, PPP1R14B appears to have considerably more transcriptional activity in cell lines as is suggested by the histone 3 modifications, DNase hypersensitivity, and TF ChIP (Fig. 5B), likely indicative of its un-restricted expression. The intronic and upstream regions of PPP1R14B are more highly conserved, allowing for a more extensive report of conserved TFBS. Potential conserved regulatory elements include a muscle-specific TATA (mTATA) box (gctggcccTTTAAGggg)-SRF combination 44 bp upstream of the TSS, an AP2 site within 20 bp of the SRF site, multiple SP1 sites, and an Ebox site in the conserved region proximal to the TSS (Fig. 5B). The first intron of PPP1R14B contains many conserved, high-scoring putative TFBS (Fig. 5, B2, Supplemental Table S4). As with the upstream promoter region, several of the putative TFBS in the first intron are involved with muscle-specific gene expression, muscle differentiation, homeostasis, and growth such as Ebox, NFAT, CREB, and Forkhead box protein O1 (FOXO1) (reviewed in Ref. 5). This computational prediction of transcriptional control of the MP inhibitory subunits provides a foundation for experimental testing of their functional importance in different cell types.
DISCUSSION
The control of muscle function by protein phosphorylation reflecting the regulated activities of Ser-Thr Type 1 phosphatases and kinases is pervasive and also muscle type specific. Yet there remains limited understanding of the diversity within the components of the signaling pathway and its effect on the control of muscle function. Here we focused on the myosin phosphatase and analyzed public databases to define modes of diversity and the regulation of the variability of the MP subunits. Considerable diversity is present within its regulatory subunits, which reflects evolutionary genomic diversification as a whole and includes increases in the number of regulatory (inhibitory) gene family members multiplied by greatly increasing combinations of alternative transcriptional start sites and exon splicing that vastly increase the number of unique transcripts generated from each gene locus.
The completion of sequencing of many genomes facilitated a phylogenetic analysis of Mypt1 E24 splice variants. The skipping of Mypt1 E24, the evolutionary and tissue default, codes for a COOH-terminal LZ motif, a highly conserved feature of Mypt family members (Mypt1, Mypt2, and MBS85; reviewed in Ref. 34), consistent with its proposed role in LZ-mediated heterodimerization with cGK1α and NO/cGMP-dependent activation of MP (42, 51, 101) (57). The “LZ,” in which a leucine (or isoleucine) residue is present at every seventh amino acid in the context of an a-helical coiled-coil domain, was originally described in what is now termed the B-ZIP family of transcription factors (56). In this large family, high throughput and computational studies suggest that specificity in LZ-mediated hetero- and homodimerizations produce great diversity and specificity in the transcriptional output (90, 106). There is some evidence for similar specificity and diversity in LZ-mediated interactions in the regulation of myosin phosphatase and other contractile proteins controlling muscle function (reviewed in Refs. 15 and 39). Mypt1 and Mypt2 have alternative COOH-termini generated by alternative splicing of E24 that abolishes or creates an alternative LZ motif, respectively. The Mypt1 E24 sequence is highly conserved as an alternative exon from fish to mammals, but not present in the ancestral ortholog in flies and worms, and is also absent from some vertebrate classes such as amphibians. This suggests that this alternative exon emerged during evolution and was under strong selection pressure to be retained. We propose that it arose as a mechanism to suppress NO/cGMP regulation of MP activity in phasic smooth muscle tissues. A number of studies have shown that NO or its second messenger cGMP are unable to activate MP as a means for relaxation of prototypical phasic smooth muscle such as the rat portal vein (26, 83) and chicken gizzard (51, 86). MP expression and activity is severalfold higher in phasic versus tonic smooth muscle (31), all supporting the hypothesis that higher basal, yet unregulated MP activity, at least with respect to NO/cGMP signaling, is required for cycling of phasic smooth muscle contraction and relaxation. This hypothesis could be tested by deletion of E24 in the mouse converting all smooth muscle tissues to the E24− (LZ+) isoform.
There has been less study of the expression pattern of the Mypt2 E24 splice variants coding for COOH-terminal LZ variants. The few studies that have examined this describe the sample as skeletal muscle, leaving open the possibility that the expression of the variants may vary by striated muscle type (fast, slow), species, or developmental age. Interestingly, the fish Mypt2 homologue only codes for the small (M21) subunit, suggesting that either 1) MP activity is not required for striated muscle function in fish or 2) this function is served by another Mypt family member. LZ-mediated dimerization partners of each Mypt2 LZ variant have not been determined, though given the strong similarity of the E24− encoded LZ to Mypt1 (and MBS85) it seems likely that it would also bind PKG1α. Both E24− and E24+-encoded Mypt2 LZ motifs are slightly basic and have similar charge profiles. However, the E24+ encoded LZ motif is uniquely followed by a COOH-terminal acidic tail of 11 residues that could modulate interactions with regulatory proteins. Entirely missing from the models of LZ motifs and MP protein interactions is consideration of the small (M21) subunit of MP, which has yet to be well characterized in terms of tissue-specific expression of isoforms and function. Finally, it is worth noting that the upstream regulatory kinase PKG1 also has two evolutionarily conserved isoforms containing alternative NH2-terminal LZ motifs (α, β) generated from alternative transcriptional start sites (75). The individual LZ motifs of the two isoforms are thought to create specificity in their substrates though there remains limited data in this regard (reviewed in Refs. 8 and 39). The current study describing the tissue-specific and phylogenetic patterns of expression of the MP LZ containing subunits provides a foundation for experimental testing of the role of PKG1 activation of MP in muscle-type specific control of function in appropriate model organisms.
Portions of the intronic sequence flanking Mypt1 E24 are also highly conserved, consistent with a prior genome-wide evolutionary study of alternative splicing that found evolutionary conservation of alternative exons was associated with conservation of flanking intronic regulatory sequences (67). Aside from the splice site junction sequences, the most invariant sequence is the hexanucleotide 5′-TCTGAA-3′ located just upstream of the AG of the 3′ splice site. This sequence was computationally predicted to be an exonic splicing enhancer and exonic identity element (25, 119). Exonic elements of this nature are thought to be necessary for proper exon identification and splicing (4). However, when found in intronic regions flanking exons, they can act as splicing repressors (48, 66, 119). The proximity of this element to the E24 3′ splice site would predict that it would suppress splicing of the exon by blocking recruitment of U2 to this site. Its function as a cis-regulator of E24 splicing remains to be tested, which given its conservation could be performed in any model system and most expeditiously in the zebrafish. The high conservation of Mypt1 E24 sequence among higher vertebrates (mammals, birds, reptiles: 26/31 nt identity) is less present in the fish. The exonic cis-element (GCAAGAGU) that binds the splicing factor Tra2β and enhances splicing of Mypt1 E24 (29, 97) is absent. Whether this results in less efficient tissue-specific splicing of E24 in fish or reflects different control mechanisms requires further study. Predicted binding sites for other classic Ser/Arg-rich (SR) splicing factors [SRSF2 (SRp30b) and SRSF6 (SRp55) (92)] are also highly evolutionarily conserved and could be involved in the regulated splicing of this exon. Overall, there remains limited understanding of the regulation of alternative splicing in the generation of smooth muscle phenotypic diversity and somewhat more understanding of this topic in striated muscle (reviewed in Refs. 27, 32, and 46). The highly programmed and tissue-specific nature of Mypt1 and Mypt2 E24 splicing make them good candidates for model exon approaches to this problem.
The four PPP1R14 family members have much less complex gene structures with little variability within individual genes, not surprising given their small size and few (4–5) exons. There is a single and well-validated report of a splice variant of PPP1R14A (CPI-17) in human aortic tissue in which skipping of exon 2 deletes a segment of the PP1 inhibitory domain (118). However, alternative splicing of this highly conserved exon in other species is not present in the queried databases such that this variation may be unique to humans. Rather, the diversity in the inhibitory subunit seems to derive from the variability among the family members and the highly regulated and tissue-specific expression of these genes, though it should be acknowledged that the function and relationships between the family members in vivo have not yet been defined through gene knock-outs. Expression of CPI-17 mRNA and protein is highly tissue-specific, being greatly enriched in smooth muscle with some variation between muscle types (117), and dynamically altered in disease (41, 71, 73, 74, 93). Little is known about its transcriptional regulation in these contexts. An upstream minimal promoter was identified (52), but there is little conservation of this upstream region between species, and no CAAT or TATA boxes, all consistent with a highly regulated transcript. We found highly conserved noncoding sequence in introns 2 and 3 that are predicted to contain a number of transcriptional cis-regulatory elements. Some transcription factors that bind these predicted elements, such as NFAT and PPARγ (5, 91), mediate the slow muscle gene program and regulate responses to external signals, as do CREB, Stat, SMAD, and Ets (reviewed in Ref. 5). Definition of these conserved and predicted transcriptional enhancer domains will require functional testing; not all regulatory sequences are conserved, and conservation of noncoding and putative regulatory sequence provides an increased likelihood but not certainty that they would function as such. In contrast PHI has the features of a less tightly regulated gene, with a highly conserved upstream sequence that in cell lines has histone marks and DNAse hypersensitivity indicative of an active promoter.
Perspectives and Signficance
The phosphorylation and dephosphorylation of myosin is the primary means by which force is controlled in smooth muscle and is thought to provide a modulatory role in striated muscle. In this study we have used computational analyses of large publicly available databases to describe the diversity in MP subunits. This more comprehensive analysis has revealed a number of features of the MP subunits that are predicted to underlie the functional significance and expressional regulation of this diversity. Of particular note are 1) the deeply conserved alternative splicing of Mypt1 E24, putative upstream splicing regulatory element, and PTC, 2) the substantial phylogenetic variability at the Mypt2 (PPP1R12B) locus with a variety of tissue-specific transcripts and alternative splicing that matches its uncertain role in striated muscle function, and 3) the phylogenetic expansion of the inhibitory subunit (PPP1R14A-D) gene family with little intragenic diversity, and difference in structure of putative promoter-enhancers between the highly (CPI-17) and less highly (PHI-1) regulated family members. This comprehensive phylogenetic analysis of MP subunit diversity will enable the optimal selection of model organisms for testing hypotheses as to the regulation and function of subunit isoforms in determining specificity in the control of signaling pathways that regulate muscle function.
GRANTS
This work was supported by National Institutes of Health Grants HL-66171 to S. A. Fisher and T32 AR007592 to R. P. Dippold.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: R.P.D. and S.A.F. conception and design of research; R.P.D. performed experiments; R.P.D. and S.A.F. analyzed data; R.P.D. and S.A.F. interpreted results of experiments; R.P.D. prepared figures; R.P.D. drafted manuscript; R.P.D. and S.A.F. edited and revised manuscript; R.P.D. and S.A.F. approved final version of manuscript.
Supplementary Material
REFERENCES
- 1.Arimura T, Suematsu N, Zhou YB, Nishimura J, Satoh S, Takeshita A, Kanaide H, Kimura A. Identification, characterization, and functional analysis of heart-specific myosin light chain phosphatase small subunit. J Biol Chem 276: 6073–6082, 2001. [DOI] [PubMed] [Google Scholar]
- 2.Ast G. How did alternative splicing evolve? Nat Rev Genet 5: 773–782, 2004. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, Kim T, Misquitta-Ali CM, Wilson MD, Kim PM, Odom DT, Frey BJ, Blencowe BJ. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593, 2012. [DOI] [PubMed] [Google Scholar]
- 4.Bourgeois CF, Popielarz M, Hildwein G, Stevenin J. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol Cell Biol 19: 7347–7356, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nature Rev 12: 349–361, 2011. [DOI] [PubMed] [Google Scholar]
- 6.Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, Albert FW, Zeller U, Khaitovich P, Grutzner F, Bergmann S, Nielsen R, Paabo S, Kaessmann H. The evolution of gene expression levels in mammalian organs. Nature 478: 343–348, 2011. [DOI] [PubMed] [Google Scholar]
- 7.Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESE finder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res 31: 3568–3571, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casteel DE, Smith-Nguyen EV, Sankaran B, Roh SH, Pilz RB, Kim C. A crystal structure of the cyclic GMP-dependent protein kinase Iβ dimerization/docking domain reveals molecular details of isoform-specific anchoring. J Biol Chem 285: 32684–32688, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceulemans H, Stalmans W, Bollen M. Regulator-driven functional diversification of protein phosphatase-1 in eukaryotic evolution. Bioessays 24: 371–381, 2002. [DOI] [PubMed] [Google Scholar]
- 10.Ceulemans H, Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol Rev 84: 1–39, 2004. [DOI] [PubMed] [Google Scholar]
- 11.Chen YH, Chen MX, Alessi D, Campbell DG, Shanahan C, Cohen P, Cohen PT. Molecular cloning of cDNA encoding the 110 kDa and 21 kDa regulatory subunits of smooth muscle protein phosphatase 1. FEBS Lett 356: 51–55, 1994. [DOI] [PubMed] [Google Scholar]
- 12.Del Gatto-Konczak F, Olive M, Gesnel MC, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol 19: 251–260, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson D. Gene estimate rises as US and UK discuss freedom of access. Nature 401: 311, 1999. [DOI] [PubMed] [Google Scholar]
- 15.Dippold RP, Fisher SA. Myosin phosphatase isoforms as determinants of smooth muscle contractile function and calcium sensitivity of force production. Microcirculation 21: 239–248, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dirksen WP, Vladic F, Fisher SA. A myosin phosphatase targeting subunit isoform transition defines a smooth muscle developmental phenotypic switch. Am J Physiol Cell Physiol 278: C589–C600, 2000. [DOI] [PubMed] [Google Scholar]
- 17.El-Touhky A, Given AM, Cochard A, Brozovich FV. PHI-1 induced enhancement of myosin phosphorylation in chicken smooth muscle. FEBS Lett 579: 4271–4277, 2005. [DOI] [PubMed] [Google Scholar]
- 18.El-Toukhy A, Given AM, Ogut O, Brozovich FV. PHI-1 interacts with the catalytic subunit of myosin light chain phosphatase to produce a Ca(2+) independent increase in MLC(20) phosphorylation and force in avian smooth muscle. FEBS Lett 580: 5779–5784, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eto M. Regulation of cellular protein phosphatase-1 (PP1) by phosphorylation of the CPI-17 family, C-kinase-activated PP1 inhibitors. J Biol Chem 284: 35273–35277, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eto M, Karginov A, Brautigan DL. A novel phosphoprotein inhibitor of protein type-1 phosphatase holoenzymes. Biochemistry 38: 16952–16957, 1999. [DOI] [PubMed] [Google Scholar]
- 23.Eto M, Senba S, Morita F, Yazawa M. Molecular cloning of a novel phosphorylation-dependent inhibitory protein of protein phosphatase-1 (CPI17) in smooth muscle: its specific localization in smooth muscle. FEBS 410: 356–360, 1997. [DOI] [PubMed] [Google Scholar]
- 24.Ewing B, Green P. Analysis of expressed sequence tags indicates 35,000 human genes. Nat Genet 25: 232–234, 2000. [DOI] [PubMed] [Google Scholar]
- 25.Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science 297: 1007–1013, 2002. [DOI] [PubMed] [Google Scholar]
- 26.Feletou M, Hoeffner U, Vanhoutte PM. Endothelium-dependent relaxing factors do not affect the smooth muscle of portal vein. Blood Vessels 26: 21–32, 1989. [PubMed] [Google Scholar]
- 27.Fisher SA. Vascular smooth muscle phenotypic diversity and function. Physiol Genomics 42A: 169–187, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frith MC, Saunders NF, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol 4: e1000071, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu K, Mende Y, Bhetwal BP, Baker S, Perrino BA, Wirth B, Fisher SA. Tra2beta is required for tissue-specific splicing of a smooth muscle myosin phosphatase targeting subunit alternative exon. J Biol Chem 287: 16575–16585, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujioka M, Takahashi N, Odai H, Araki S, Ichikawa K, Feng J, Nakamura M, Kaibuchi K, Hartshorne DJ, Nakano T, Ito M. A new isoform of human myosin phosphatase targeting/regulatory subunit (MYPT2): cDNA cloning, tissue expression, and chromosomal mapping. Genomics 49: 59–68, 1998. [DOI] [PubMed] [Google Scholar]
- 31.Gong MC, Cohen P, Kitazawa T, Ikebe M, Masuo M, Somlyo AP, Somlyo AV. Myosin light chain phosphatase activites and the effects of phosphatase inhibitors in tonic and phasic smooth muscle. J Biol Chem 267: 14662–14668, 1992. [PubMed] [Google Scholar]
- 32.Gooding C, Smith CW. Tropomyosin exons as models for alternative splicing. Adv Exp Med Biol 644: 27–42, 2008. [DOI] [PubMed] [Google Scholar]
- 33.Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G. Comparative analysis identifies exonic splicing regulatory sequences–The complex definition of enhancers and silencers. Mol Cell 22: 769–781, 2006. [DOI] [PubMed] [Google Scholar]
- 34.Grassie ME, Moffat LD, Walsh MP, MacDonald JA. The myosin phosphatase targeting protein (MYPT) family: a regulated mechanism for achieving substrate specificity of the catalytic subunit of protein phosphatase type 1delta. Arch Biochem Biophys 510: 147–159, 2011. [DOI] [PubMed] [Google Scholar]
- 35.Han YS, Brozovich FV. Altered reactivity of tertiary mesenteric arteries following acute myocardial ischemia. J Vasc Res 50: 100–108, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet 16: 369–372, 2000. [DOI] [PubMed] [Google Scholar]
- 37.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet 13: 469–483, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartshorne DJ, Ito M, Erdodi F. Role of protein phosphatase type 1 in contractile functions: myosin phosphatase. J Biol Chem 279: 37211–37214, 2004. [DOI] [PubMed] [Google Scholar]
- 39.Hofmann F, Bernhard D, Lukowski R, Weinmeister P. cGMP regulated protein kinases (cGK). Handbk Exp Pharmacol 137–162, 2009. [DOI] [PubMed] [Google Scholar]
- 40.Hong F, Haldeman BD, Jackson D, Carter M, Baker JE, Cremo CR. Biochemistry of smooth muscle myosin light chain kinase. Arch Biochem Biophys 510: 135–146, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu W, Mahavadi S, Li F, Murthy KS. Upregulation of RGS4 and downregulation of CPI-17 mediate inhibition of colonic muscle contraction by interleukin-1β. Am J Physiol Cell Physiol 293: C1991–C2000, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang QQ, Fisher SA, Brozovich FV. Unzipping the role of myosin light chain phosphatase in smooth muscle cell relaxation. J Biol Chem 279: 597–603, 2004. [DOI] [PubMed] [Google Scholar]
- 43.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJ, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res 40: D306–D312, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Human Genome Sequencing Consortium. Lander E.S, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature 409: 860–921, 2001. [DOI] [PubMed] [Google Scholar]
- 45.Ito M, Nakano T, Erdodi F, Hartshorne DJ. Myosin phosphatase: structure, regulation and function. Mol Cell Biochem 259: 197–209, 2004. [DOI] [PubMed] [Google Scholar]
- 46.Kalsotra A, Cooper TA. Functional consequences of developmentally regulated alternative splicing. Nat Rev Genet 12: 715–729, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamm KE, Stull JT. Signaling to myosin regulatory light chain in sarcomeres. J Biol Chem 286: 9941–9947, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381: 535–538, 1996. [DOI] [PubMed] [Google Scholar]
- 49.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 12: 996–1006, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khatri JJ, Joyce KM, Brozovich FV, Fisher SA. Role of myosin phosphatase isoforms in cGMP-mediated smooth muscle relaxation. J Biol Chem 276: 37250–37257, 2001. [DOI] [PubMed] [Google Scholar]
- 52.Kim JI, Urban M, Young GD, Eto M. Reciprocal regulation controlling the expression of CPI-17, a specific inhibitor protein for the myosin light chain phosphatase in vascular smooth muscle cells. Am J Physiol Cell Physiol 303: C58–C68, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitazawa T, Kitazawa K. Size-dependent heterogeneity of contractile Ca2+-sensitization in rat arterial smooth muscle. J Physiol 590: 5401–5423, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitazawa T, Polzin AN, Eto M. CPI-17-deficient smooth muscle of chicken. J Physiol 557: 515–528, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kudryavtseva O, Aalkjaer C, Matchkov VV. Vascular smooth muscle cell phenotype is defined by Ca2+-dependent transcription factors. FEBS J 280: 5488–5499, 2013. [DOI] [PubMed] [Google Scholar]
- 56.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240: 1759–1764, 1988. [DOI] [PubMed] [Google Scholar]
- 57.Lee E, Hayes DB, Langsetmo K, Sundberg EJ, Tao TC. Interactions between the leucine-zipper motif of cGMP-dependent protein kinase and the C-terminal region of the targeting subunit of myosin light chain phosphatase. J Mol Biol 373: 1198–1212, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Q, Buckler ESt Muse SV, Walker JC. Molecular evolution of type 1 serine/threonine protein phosphatases. Mol Phylogenet Evol 12: 57–66, 1999. [DOI] [PubMed] [Google Scholar]
- 59.Liu QR, Zhang PW, Lin Z, Li QF, Woods AS, Troncoso J, Uhl GR. GBPI, a novel gastrointestinal- and brain-specific PP1-inhibitory protein, is activated by PKC and inactivated by PKA. Biochem J 377: 171–181, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu QR, Zhang PW, Zhen Q, Walther D, Wang XB, Uhl GR. KEPI, a PKC-dependent protein phosphatase 1 inhibitor regulated by morphine. J Biol Chem 277: 13312–13320, 2002. [DOI] [PubMed] [Google Scholar]
- 61.Loots GG, Ovcharenko I. rVISTA 2.0 evolutionary analysis of transcription factor binding sites. Nucleic Acids Res 32: W217–W221, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, Zhang H, Gokina N, Mandala M, Sato O, Ikebe M, Osol G, Fisher SA. Uterine artery myosin phosphatase isoform switching and increased sensitivity to SNP in a rat l-NAME model of hypertension of pregnancy. Am J Physiol Cell Physiol 294: C564–C571, 2008. [DOI] [PubMed] [Google Scholar]
- 63.Mabuchi K, Gong BJ, Langsetmo K, Ito M, Nakano T, Tao T. Isoforms of the small non-catalytic subunit of smooth muscle myosin light chain phosphatase. Biochim Biophys Acta 1434: 296–303, 1999. [DOI] [PubMed] [Google Scholar]
- 64.Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ. FlyBase: improvements to the bibliography. Nucleic Acids Res 41: D751–D757, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matsumura F, Hartshorne DJ. Myosin phosphatase target subunit: many roles in cell function. Biochem Biophys Res Commun 369: 149–156, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McNally LM, McNally MT. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol Cell Biol 18: 3103–3111, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338: 1593–1599, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mizuno T, Tsutsui K, Nishida Y. Drosophila myosin phosphatase and its role in dorsal closure. Development 129: 1215–1223, 2002. [DOI] [PubMed] [Google Scholar]
- 69.Moorhead G, Johnson D, Morrice N, Cohen P. The major myosin phosphatase in skeletal muscle is a complex between the beta-isoform of protein phosphatase 1 and the MYPT2 gene product. FEBS Lett 438: 141–144, 1998. [DOI] [PubMed] [Google Scholar]
- 70.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J 417: 401–409, 2009. [DOI] [PubMed] [Google Scholar]
- 71.Mueed I, Zhang L, MacLeod KM. Role of the PKC/CPI-17 pathway in enhanced contractile responses of mesenteric arteries from diabetic rats to alpha-adrenoceptor stimulation. Br J Pharmacol 146: 972–982, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson AC, Wardle FC. Conserved non-coding elements and cis regulation: actions speak louder than words. Development 140: 1385–1395, 2013. [DOI] [PubMed] [Google Scholar]
- 73.Ohama T, Hori M, Momotani E, Iwakura Y, Guo F, Kishi H, Kobayashi S, Ozaki H. Intestinal inflammation downregulates smooth muscle CPI-17 through induction of TNF-α and causes motility disorders. Am J Physiol Gastrointest Liver Physiol 292: G1429–G1438, 2007. [DOI] [PubMed] [Google Scholar]
- 74.Ohama T, Hori M, Sato K, Ozaki H, Karaki H. Chronic treatment with interleukin-1β attenuates contractions by decreasing the activities of CPI-17 and MYPT-1 in intestinal smooth muscle. J Biol Chem 278: 48794–48804, 2003. [DOI] [PubMed] [Google Scholar]
- 75.Orstavik S, Natarajan V, Tasken K, Jahnsen T, Sandberg M. Characterization of the human gene encoding the Type Ia and Type Ib cGMP-dependent protein kinase (prkg1). Genomics 42: 311–318, 1997. [DOI] [PubMed] [Google Scholar]
- 76.Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res 15: 184–194, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res 32: W28-W286, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84: 767–801, 2004. [DOI] [PubMed] [Google Scholar]
- 79.Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, Dahmane N, Davuluri RV. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res 21: 1260–1272, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pandit SB, Gosar D, Abhiman S, Sujatha S, Dixit SS, Mhatre NS, Sowdhamini R, Srinivasan N. SUPFAM–a database of potential protein superfamily relationships derived by comparing sequence-based and structure-based families: implications for structural genomics and function annotation in genomes. Nucleic Acids Res 30: 289–293, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pang H, Guo Z, Xie Z, Su W, Gong MC. Divergent kinase signaling mediates agonist-induced phosphorylation of phosphatase inhibitory proteins PHI-1 and CPI-17 in vascular smooth muscle cells. Am J Physiol Cell Physiol 290: C892–C899, 2006. [DOI] [PubMed] [Google Scholar]
- 82.Parmacek MS. Myocardin–not quite MyoD. Arterioscler Thromb Vasc Biol 24: 1535–1537, 2004. [DOI] [PubMed] [Google Scholar]
- 83.Payne MC, Zhang HY, Prosdocimo T, Joyce KM, Koga Y, Ikebe M, Fisher SA. Myosin phosphatase isoform switching in vascular smooth muscle development. J Mol Cell Cardiol 40: 274–282, 2006. [DOI] [PubMed] [Google Scholar]
- 84.Payne MC, Zhang HY, Shirasawa Y, Koga Y, Ikebe M, Benoit JN, Fisher SA. Dynamic changes in expression of myosin phosphatase in a model of portal hypertension. Am J Physiol Heart Circ Physiol 286: H1801–H1810, 2004. [DOI] [PubMed] [Google Scholar]
- 85.Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, McMurry J, Marioni JC, Malone J, Megy K, Rustici G, Tang AY, Taubert J, Williams E, Mannion O, Parkinson HE, Brazma A. Expression Atlas update–a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res 42: D926–D932, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfitzer G, Merkel L, Ruegg JC, Hofmann F. Cyclic GMP-dependent protein kinase relaxes skinned fibers from guinea pig taenia coli but not from chicken gizzard. Pflügers Arch 407: 87–91, 1986. [DOI] [PubMed] [Google Scholar]
- 87.Plowman GD, Sudarsanam S, Bingham J, Whyte D, Hunter T. The protein kinases of caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc Natl Acad Sci USA 96: 13603–13610, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pohl AA, Sugnet CW, Clark TA, Smith K, Fujita PA, Cline MS. Affy exon tissues: exon levels in normal tissues in human, mouse and rat. Bioinformatics 25: 2442–2443, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res 40: D29-D301, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reinke AW, Baek J, Ashenberg O, Keating AE. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 340: 730–734, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, Deriaz O. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-gamma coactivator-1 and peroxisome proliferator-activated receptor-alpha in skeletal muscle. Diabetes 52: 2874–2881, 2003. [DOI] [PubMed] [Google Scholar]
- 92.Sanford JR, Ellis J, Caceres JF. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem Soc Trans 33: 443–446, 2005. [DOI] [PubMed] [Google Scholar]
- 93.Sato K, Ohkura S, Kitahara Y, Ohama T, Hori M, Sato M, Kobayashi S, Sasaki Y, Hayashi T, Nasu T, Ozaki H. Involvement of CPI-17 downregulation in the dysmotility of the colon from dextran sodium sulphate-induced experimental colitis in a mouse model. Neurogastroenterol Motil 19: 504–514, 2007. [DOI] [PubMed] [Google Scholar]
- 94.Scruggs SB, Solaro RJ. The significance of regulatory light chain phosphorylation in cardiac physiology. Arch Biochem Biophys 510: 129–134, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shaye DD, Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLos One 6: e20085, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shimizu H, Ito M, Miyahara M, Ichikawa K, Okubo S, Konishi T, Naka M, Tanaka T, Hirano K, Hartshorne DJ, Nakano T. Characterization of the myosin-binding subunit of smooth muscle myosin phosphatase. J Biol Chem 269: 30407–30411, 1994. [PubMed] [Google Scholar]
- 97.Shukla S, Fisher SA. Tra2beta as a novel mediator of vascular smooth muscle diversification. Circ Res 103: 485–492, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith PJ, Zhang C, Wang J, Chew SL, Zhang MQ, Krainer AR. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum Mol Genet 15: 2490–2508, 2006. [DOI] [PubMed] [Google Scholar]
- 100.Somlyo AP, Somlyo AV. Signal transduction and regulation in smooth muscle. Nature 372: 231–236, 1994. [DOI] [PubMed] [Google Scholar]
- 101.Surks HK, Mochizuki N, Kasai Y, Georgescu SP, Tang KM, Ito M, Lincoln TM, Mendelsohn ME. Regulation of myosin phosphatase by a specific interaction with cGMP- dependent protein kinase Ialpha. Science 286: 1583–1587, 1999. [DOI] [PubMed] [Google Scholar]
- 102.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature 489: 75–82, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science 291: 1304–1351, 2001. [DOI] [PubMed] [Google Scholar]
- 105.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19: 327–335, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta 1759: 4–12, 2006. [DOI] [PubMed] [Google Scholar]
- 107.Wang DZ, Olson EN. Control of smooth muscle development by the myocardin family of transcriptional coactivators. Curr Opin Genet Dev 14: 558–566, 2004. [DOI] [PubMed] [Google Scholar]
- 108.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang M, Marin A. Characterization and prediction of alternative splice sites. Gene 366: 219–227, 2006. [DOI] [PubMed] [Google Scholar]
- 110.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845, 2004. [DOI] [PubMed] [Google Scholar]
- 111.Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE. Human-mouse genome comparisons to locate regulatory sites. Nat Genet 26: 225–228, 2000. [DOI] [PubMed] [Google Scholar]
- 112.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet 26: 66–74, 2010. [DOI] [PubMed] [Google Scholar]
- 114.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 24: 238–241, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wissmann A, Ingles J, Mains PE. The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol 209: 111–127, 1999. [DOI] [PubMed] [Google Scholar]
- 116.Wissmann A, Ingles J, McGhee JD, Mains PE. Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Develop 11: 409–422, 1997. [DOI] [PubMed] [Google Scholar]
- 117.Woodsome TP, Eto M, Everett A, Brautigan DL, Kitazawa T. Expression of CPI-17 and myosin phosphatase correlates with Ca(2+) sensitivity of protein kinase C-induced contraction in rabbit smooth muscle. J Physiol 535: 553–564, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yamawaki K, Ito M, Machida H, Moriki N, Okamoto R, Isaka N, Shimpo H, Kohda A, Okumura K, Hartshorne DJ, Nakano T. Identification of human CPI-17, an inhibitory phosphoprotein for myosin phosphatase. Biochem Biophys Res Commun 285: 1040–1045, 2001. [DOI] [PubMed] [Google Scholar]
- 119.Zhang C, Li WH, Krainer AR, Zhang MQ. RNA landscape of evolution for optimal exon and intron discrimination. Proc Natl Acad Sci USA 105: 5797–5802, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang H, Fisher SA. Conditioning effect of blood flow on resistance artery smooth muscle myosin phosphatase. Circ Res 100: 730–737, 2007. [DOI] [PubMed] [Google Scholar]
- 121.Zhang XHF, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Develop 18: 1241–1250, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.