Abstract
σ factors in the σ70 family can be classified into the primary and alternative σ factors according to their physiological functions and amino acid sequence similarities. The primary σ factors are composed of four conserved regions, with the conserved region 1 being divided into two subregions. Region 1.1, which is absent from the alternative σ factor, is poor in conservation; however, region 1.2 is well conserved. We investigated the importance of these two subregions to the function of Bacillus subtilis σA, which belongs to a subgroup of the primary σ factor lacking a 254-amino-acid spacer between regions 1 and 2. We found that deletion of not more than 100 amino acid residues from the N terminus of σA, which removed part or all region 1.1, did not affect the overall transcription activity of the truncated σA-RNA polymerase in vitro, indicating that region 1.1 is not required for the functioning of σA in RNA polymerase holoenzyme. This finding is consistent with the complementation data obtained in vivo. However, region 1.1 is able to negatively modulate the promoter DNA-binding activity of the σA-RNA polymerase. Further deletion of the conserved Arg-103 at the N terminus of region 1.2 increased the content of stable secondary structures of the truncated σA and greatly reduced the transcription activity of the truncated σA-RNA polymerase by lowering the efficiency of transcription initiation after core binding of σA. More importantly, the conserved Arg-103 was also demonstrated to be critical for the functioning of the full-length σA in RNA polymerase.
The RNA polymerase holoenzyme in prokaryotic cells is basically composed of α2ββ′ and σ factor. Designated a core enzyme, α2ββ′ possesses catalytic and regulatory functions. Besides being a transcription factor which confers the specificity of recognition of cognate promoter and initiation of transcription (2, 6, 10, 30, 33, 36), σ factor is involved in open complex formation (16, 24, 28), abortive transcription (13, 29), promoter-proximal pausing (26), and regulation of gene expression (14, 18, 25).
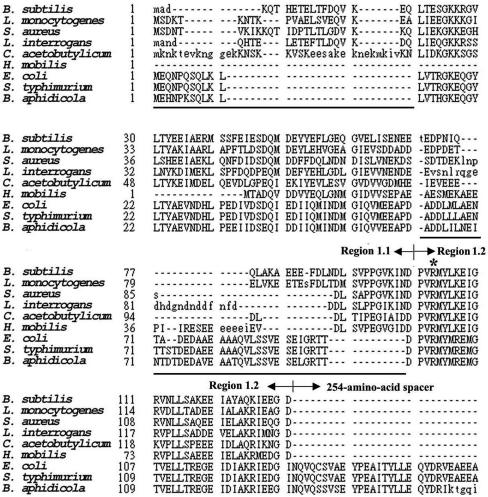
Two families of σ factor (σ70 and σ54) have been reported previously (20, 22). In the σ70 family, σ factors were classified into the primary and alternative σ factors according to their physiological functions and amino acid sequence similarities (20). The primary σ comprises four conserved regions, each containing two to four subregions (12); however, they are poorly conserved in region 1.1 (20) except for the characteristic acidity. Moreover, some of them are devoid of a 254-amino-acid spacer between regions 1 and 2, the function of which is vague (17). On the basis of the presence or absence of the spacer, the primary σ factor can be further separated into two subgroups, with Escherichia coli σ70 and Bacillus subtilis σA referred to as the type members (Fig. 1). However, there is no information about the effect of the above differences on the function of σ factor in the subgroup of σA, which is mostly responsible for transcription of the housekeeping genes. Moreover, it remains unclear whether region 1.1 is dispensable for a primary σ in the σA subgroup, as has been seen for some alternative σ factors (11, 20), or if it is essential for transcription initiation at some promoters as reported previously for E. coli σ70 (1, 32, 35). This study aims to answer whether region 1.1 is essential for the functioning of primary σ factor in the σA subgroup and to determine the essential N-terminal boundary amino acid residue for a truncated σA to function in RNA polymerase holoenzyme.
FIG. 1.
Amino acid sequence alignment of primary σ factors in the σA and σ70 subgroups. Abbreviations denote the following bacterial species: B. subtilis, Listeria monocytogenes, Staphylococcus aureus, Listeria interrogans, Clostridium acetobutylicum, Haemophilus mobilis, E. coli, Salmonella enterica serovar Typhimurium, and Buchnera aphidicola. The primary σ factor can be separated into two subgroups, with those from the top six strains being classified into the σA subgroup and those from the bottom three placed in the σ70 subgroup. Shown in the figure are region 1.1, region 1.2, and part of the 254-amino-acid spacer, all of which are indicated by arrowheads. Region 1.1 of the B. subtilis σA spans amino acid residues 1 through 99, while region 1.2 spans amino acid residues 100 through 131. Amino acid residues which are not conserved between the two subgroups of primary σ factor are underlined. The lowercase letters denote residues that are not considered homologous. The conserved arginine residue at position 103 of σA is indicated by the asterisk.
We have tried to answer the above questions by constructing and characterizing a collection of the N-terminally truncated σA. Here, we present evidences showing that region 1.1, which spans amino acid residues 1 through 99, is not required for the functioning of σA either in vitro or in vivo. However, it is able to negatively modulate the promoter DNA-binding activity of σA-RNA polymerase. We also show that the conserved Arg-103 at the N terminus of region 1.2 is important to ensure efficient functioning of both the truncated and full-length σA in RNA polymerase holoenzyme.
MATERIALS AND METHODS
Construction of truncated sigA and sigA with Arg-103 being replaced.
The truncated sigA genes were constructed by PCR using the wild-type (WT) sigA-containing plasmid pCD2 as template. The forward primers used for N-terminal truncation are designated sndx. They have the sequence 5′-GTCAGAATTCAAGAAGGAGATACAATATG-3′, followed by 18 to 21 nucleotides starting from the codon next to the one being deleted, with the EcoRI site underlined, the putative ribosome-binding site italicized, and the initiation codon (ATG) bolded. The reverse primer MI2 has the sequence 5′-CGTACCTGCAGGGATCCGTATACGCTTACAATAGAAAAT-3′, with the BamHI site underlined. Each of the sigA DNA fragments was digested with EcoRI and BamHI before being cloned into the compatible sites of the transcriptional fusion vector pT7-5 (31).
Overlapping extension PCR was used to construct sigA, with Arg-103 being replaced with Met, Ala, Glu, or Lys. Primers used for the synthesis of the mutant sigA are as follows: A, 5′-CTGCAGAGATCTGAATTCGTTGCAAGCTTTGG-3′ (the underlined bases represent the EcoRI site); D, 5′-CGTACCTGCAGGGATCCGTATACGCTTACAATAGAAAAT-3′ (the underlined bases represent the BamHI site); Bx (x represents R103M, R103A, R103E, or R103K), 5′-ATACATX1X2X3AACTGGGTCATTGATTTTAACG; and Cx, 5′-GACCCAGTTY3Y2Y1ATGTATTTAAAGGAAATCGG-3′. Bx and Cx primers are complementary in the underlined bases. X1X2X3 represents the substituted codon. After restriction enzyme digestion, each of the mutant sigA DNA fragments was cloned into the compatible sites of pT7-5 (31).
Construction of plasmids used for complementation assays.
The plasmids pCF0, pCF100, and pCF104, designed to express WT σA, SND100, and SND104, respectively, were constructed by inserting the corresponding sigA DNA fragment into the SalI and HindIII sites of the pHYP plasmid derived from pHY300PLK. The expression of sigA on the plasmid was under the control of the P1P2 promoter of the B. subtilis sigA operon and the ribosome-binding site of sigA.
Overproduction and purification of σA.
Methods used for overproduction and purification of the mutant σA were similar to those used for the preparation of WT σA (5).
In vitro transcription and abortive transcription assays.
Core enzyme containing the His-tagged β′ subunit was prepared according to the protocol reported previously (15). The method used for the in vitro transcription assay was modified from a previously reported version (5). For in vitro transcription, 10 μl of core enzyme (1 μg) was mixed with an equal volume of purified σA (1.2 μg) before incubation on ice for 10 min. The molar ratio of core to σA was 1:10. Afterward, 20 μl (0.3 μg) of pCT20 or pCT24 plasmid harboring the P1P2 promoter of the B. subtilis sigA operon or the G3b promoter of the B. subtilis φ29 phage (7, 19), respectively, was added to the reconstituted holoenzyme. The mixture was then incubated on ice for 10 min before being transferred to a temperature environment of 37°C. Subsequently, 40 μl of reaction cocktail (40 mM Tris-HCl [pH 7.9]; 10 mM MgCl2; 160 mM KCl; 0.4 mM dithiothreitol; 0.2 mM each UTP, CTP, GTP, and ATP; 3 μCi of [α-32P]ATP; and 5% glycerol) prewarmed at 37°C was added to start the transcription reaction. The reaction was allowed to proceed for 10 min before adding 160 μl of stop buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol). Finally, the RNA products were run on a 6% denaturing polyacrylamide gel, and autoradiography was performed after vacuum drying of the gel.
The method used for abortive transcription was similar to that used for in vitro transcription. In brief, a 10× molar excess of σA was reconstituted with 1 μg of core enzyme. The reconstituted RNA polymerase holoenzyme was then mixed with 0.1 μg of linear G3b promoter DNA (164 bp in length) that had been synthesized by PCR. The template used for the synthesis of linear G3b promoter was the φ29 phage DNA; the two primers were BC1041 (5′-GAGCTCGGATCCAGAGAACGTAGACAACCTC-3′) and BC1046 (5′-CTGCAGAAGCTTGCCATTTCTTCGTCCCACT-3′). The transcription reaction was allowed to proceed for 15 min after the addition of reaction cocktail, and the final transcription products were run on an 18% denaturing polyacrylamide gel.
Core-binding activity of σA.
Equal molar concentrations (5 μM) of core enzyme and σA were mixed and dialyzed against the core-binding buffer (50 mM HEPES [pH 7.9], 0.1 mM EDTA, 100 mM NaCl, 10 mM MgCl2, and 5% glycerol) at 4°C for 8 h before being subjected to a 5-min incubation at 37°C. The reconstituted RNA polymerase holoenzyme was then injected into a Pharmacia Superdex 200 HR 10/30 gel filtration column to separate core-associated and free σA. The fractionated protein samples (1 ml/tube) were then analyzed for the relative contents of β′ and σA by using Western blot analysis.
Preparation and labeling of G3b promoter DNA for gel retardation assay.
The 135-bp linear G3b promoter DNA was synthesized by PCR using the pCoiZA-derived plasmid as template (4). The two primers are BC1041-PstI (5′-GCTGGTCTGCAGAACGTAGACAACAACC-3′) and BC1048-XbaI (5′-GCGTCGTCTAGAATTTGTAGACTCTGTATC-3′). To label the G3b promoter, we suspended 2.5 μg of the DNA fragment recovered by electroelution in 100 μl of Tris-EDTA buffer (10 mM Tris-HCl [pH 8.0] and 0.1 mM EDTA). Next, 2 μl of polynucleotide kinase (10 U/μl), 20 μl of 10× kinase buffer (500 mM Tris-HCl [pH 8.2], 100 mM MgCl2, 1 mM EDTA, 50 mM dithiothreitol, and 1 mM spermidine), 12.5 μl of [γ-32P]ATP (10 μCi/μl), and 65.5 μl of H2O were added to the DNA solution. The mixture was incubated at 37°C for 1 h and then extracted twice with phenol-chloroform. The labeled DNA in the upper aqueous phase was precipitated with 0.3 M sodium acetate, washed with 70% ethanol, dried in vacuum conditions, and dissolved in 100 μl of Tris-EDTA buffer.
Promoter DNA binding of WT and truncated σA-RNA polymerases.
A gel retardation assay was used to analyze the promoter DNA-binding activities of the WT and truncated σA-RNA polymerase holoenzymes. To carry out the experiment, we reconstituted core RNA polymerase with σA in binding buffer (50 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol, 125 mM KCl, and 10% glycerol) on ice for 5 min. Next, the 32P-labeled G3b promoter was added to the reconstituted holoenzyme. The mixture was incubated on ice for 5 min before being transferred to an environment of 37°C for another 5 min of incubation. Afterward, 3 μl of loading buffer (50 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 0.1 mM EDTA, 125 mM KCl, 70% glycerol, and 0.4% bromophenol blue) was added to the binding mixture, and the sample was subjected to electrophoresis on a 5% nondenaturing polyacrylamide gel in 1× TAE buffer (40 mM Tris-acetate and 2 mM EDTA [pH 8.5]).
Partial proteolysis of N-terminally truncated σA.
Basically, 15 μg of truncated σA in TEDG buffer (10 mM Tris-HCl [pH 7.9], 0.1 mM EDTA, 0.1 mM dithiothreitol, and 5% glycerol) supplemented with 0.2 M NaCl was mixed with 15 ng of trypsin or chymotrypsin, incubated at 37°C (final volume, 20 μl), and quenched at various time points (0, 5, 10, 20, or 30 min) by adding phenylmethylsulfonyl fluoride to a final concentration of 1 mM. The digested protein sample was mixed with 2× sample buffer (0.125 M Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate, 20% glycerol, 0.002% bromophenol blue, and 10% β-mercaptoethanol) and was then run on a sodium dodecyl sulfate-12% polyacrylamide gel. The proteolytic patterns of the truncated σA were compared after we stained the gels with Coomassie brilliant blue.
Structural analysis of σA protein.
The circular dichroism (CD) spectra of the truncated σA were measured by using a Jasco 710 CD spectrophotometer. Each of the purified σA samples was dialyzed against TEDG buffer supplemented with 0.2 M NaCl and adjusted to a final concentration of 0.2 mg/ml before CD analysis. The spectral data were analyzed by using Jasco software.
RESULTS
Effect of N-terminally truncated σA on transcription activity of σA-RNA polymerase.
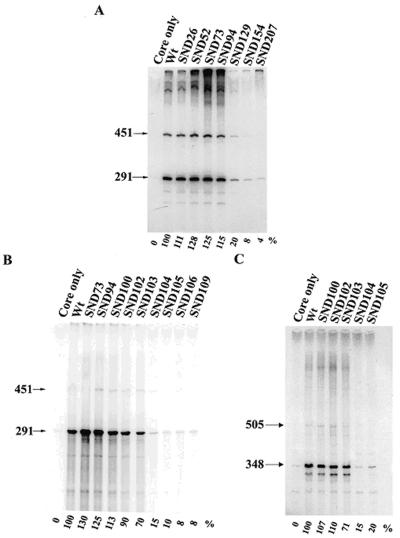
The B. subtilis housekeeping σA factor belongs to a subgroup of primary σ factors which are devoid of a 254-amino-acid spacer between regions 1 and 2 (Fig. 1). In order to assess whether region 1.1 of this σ factor is dispensable for its function as a known alternative σ factor which lacks region 1.1, we first constructed a series of N-terminally truncated σA factors. To avoid the possible effect of extra amino acid residues fused at the N terminus of the truncated σA on its function, the pT7-5 vector (31), which is suitable for transcriptional fusion, was adopted so as to overproduce the truncated σA. In this way, the most N-terminal amino acid residue of the truncated σA would be methionine or the amino acid residue directly C terminal to that designed for deletion, depending on whether N-terminal processing of the truncated σA occurred. After purification of the overproduced σA, each of them was analyzed for in vitro activity on the G3b promoter after reconstitution with core RNA polymerase. As shown in Fig. 2A, SND26, SND52, SND73, and SND94, with part of region 1.1 deleted, increased the transcription activity of the truncated σA; however, further deletion of region 1.2 or region 2 (such as SND129, SND154, and SND207) drastically reduced the activity, indicating that elimination of amino acid residues from the N terminus to a certain position within amino acid residues 94 to 129 of σA is detrimental to σA function.
FIG. 2.
In vitro transcription assays of the N-terminally truncated σA-RNA polymerase holoenzymes. (A and B) In vitro transcription activities of the truncated σA-RNA polymerase holoenzymes on the G3b promoter of B. subtilis φ29 phage. (C) In vitro transcription activities of the truncated σA-RNA polymerase holoenzymes on the P1P2 promoter of the B. subtilis sigA operon. The method used for the assay is described in Materials and Methods. The truncated σA-RNA polymerases assayed are shown at the top of each panel. The arrowheads indicate the mRNA transcripts (451 and 291 bases for the G3b promoter; 505 and 348 bases for the P1P2 promoter) terminated at the T1T2 tandem terminator (19). The value below each lane is the relative transcription activity of the assayed RNA polymerase, with that of the WT σA-RNA polymerase referred to as 100%. Each value is the average of at least three individual assays with a standard deviation of less than 10%.
To precisely map the position, more N-terminally truncated σA variants (SND100 to SND109) were created, overproduced, N-terminally sequenced (Table 1), and analyzed for the activities on both the G3b promoter of B. subtilis φ29 phage (Fig. 2B) and the P1P2 promoter of the B. subtilis sigA operon (Fig. 2C). Our data showed that the truncated σA-RNA polymerases can be divided into three activity groups (Fig. 2B). SND73, SND94, SND100, and SND102, from which most or all of region 1.1 was deleted, belonged to the high-activity group and were at least as active as the WT. Furthermore, SND103, with the conserved Arg-103 (Fig. 1) replaced with methionine (Table 1), had a moderate activity which was about 70% of that of the WT. SND104 to SND109, with Arg-103 removed, were in the low-activity group. Less than 15% of WT activity was retained by RNA polymerases containing these truncated σA variants. Similar results were obtained for the truncated σA-RNA polymerases on the P1P2 promoter (Fig. 2C). Since the nucleotide sequences of the G3b and P1P2 promoters are different, especially at the −35 region (7, 19), it was assumed that the transcription defect caused by deletion of Arg-103 in SND104 is promoter independent. Moreover, our results clearly manifested that the conserved Arg-103 is an essential N-terminal boundary amino acid residue for the truncated σA; the loss of this arginine, rather than its preceding amino acid sequence, is detrimental to σA function.
TABLE 1.
N-terminal amino acid sequencesa of truncated σA factors
| σA derivative | Amino acid residue at the N terminus of region 1.2b at position no.:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | |
| WT | D | P | V | R | M | Y | L | K |
| SND100 | −c | P | V | R | M | Y | L | K |
| SND102 | − | M | R | M | Y | L | K | |
| SND103 | − | M | M | Y | L | K | ||
| SND104 | − | M | Y | L | K | |||
| SND105 | − | M | L | K | ||||
The N-terminal sequence of each truncated σA factor was determined by using an amino acid sequencer (ABI model 494) at National Chung-Hsing University.
Region 1.2 spans residues 100 through 131 of the B. subtilis σA factor.
−, the specific amino acid residue and those N terminal to it have been deleted.
With region 1.1 deleted, σA is functional in vivo.
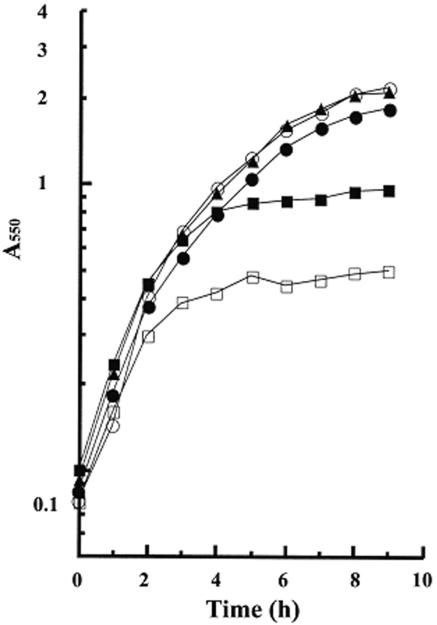
The dispensability of region 1.1 for σA to function in vitro suggested that the truncated σA with region 1.1 deleted might be also functional in vivo. To test this idea, we used the plasmids pCF0, pCF100, and pCF104, which were able to express WT σA, SND100, and SND104, respectively, in B. subtilis (data not shown), to complement the growth defect of B. subtilis DB3200, in which the WT sigA is under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible spac promoter (19). As expected, B. subtilis DB3200 was low in growth yield in minimal medium lacking IPTG (Fig. 3). Introduction of pCF0 or pCF100 increased the growth yield of B. subtilis DB3200 to about the same level as that observed in the presence of IPTG. However, pCF104 only partially restored the growth of B. subtilis DB3200. The difference in capability of pCF100 and pCF104 to complement the growth defect of B. subtilis DB3200 in vivo correlated well with the difference in transcription activity of the corresponding truncated σA-RNA polymerases in vitro (Fig. 2). These results suggest that the truncated σA with region 1.1 deleted is functional in vivo and that further deletion of the conserved Arg-103 in region 1.2 is somehow detrimental to the functioning of the truncated σA in vivo.
FIG. 3.
Complementation of the growth defect of B. subtilis DB3200 by the N-terminally truncated σA in vivo. The plasmids pCF0, pCF100, and pCF104 (see Materials and Methods) were introduced separately into B. subtilis DB3200. The growth curve of each strain of B. subtilis in minimal medium was then measured. ○ and □ indicate the growth of B. subtilis DB3200 in the presence and absence of IPTG, respectively. •, ▴, and ▪ represent the growth of B. subtilis DB3200/pCF0, DB3200/pCF100, and DB3200/pCF104, respectively, in the absence of IPTG. A550, absorbance at 550 nm.
Importance of conserved Arg-103 at the N terminus of region 1.2 to functional properties of N-terminally truncated σA.
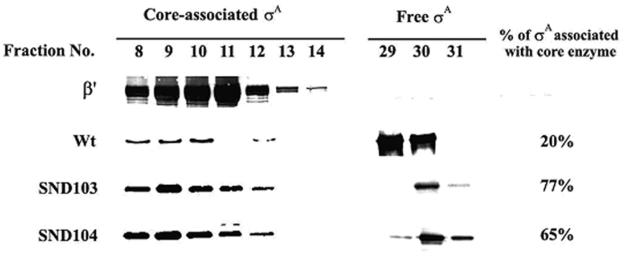
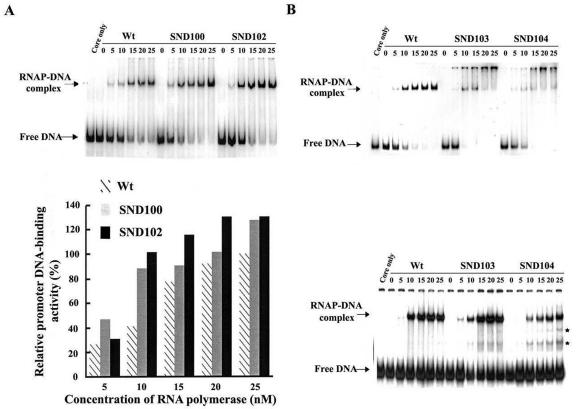
The drastic reduction in transcription activity of SND104 σA-RNA polymerase suggests that SND104 must be defective at a certain step of transcription initiation. To clarify this point, we first analyzed the core-binding efficiency of the truncated σA. In the analyses, equal numbers of moles of core enzyme and truncated σA were mixed and then subjected to molecular sieving, during which σA can be separated into the free and core-associated states. As shown in Fig. 4, about 20% of the WT σA was found to associate with core enzyme, as assessed by its coelution with the β′ subunit of RNA polymerase; however, much higher levels of core binding were observed for SND103 (77%) and SND104 (65%) (Fig. 4). These results indicate that the drastic reduction in the activity of SND104 σA-RNA polymerase (Fig. 2) is not attributed to the core binding of SND104. The reduction should be due to a certain defect arising after core binding. Thus, the activities of promoter DNA binding of WT, SND100, SND102, SND103, and SND104 σA-RNA polymerases were further analyzed. To diminish the effect of possible differences in core binding of the truncated σA on promoter DNA binding of the corresponding σA-RNA polymerases, we used RNA polymerase holoenzymes reconstituted from a 10× molar excess of σA for gel retardation assays. As shown in Fig. 5, a slight increase in the promoter DNA-binding activity was observed for both SND100 and SND102 σA-RNA polymerases (Fig. 5A). However, it was difficult to assess the promoter DNA-binding activities of SND103 and SND104 σA-RNA polymerases since the retarded RNA polymerase-DNA complexes started to disappear when the concentration of RNA polymerase was above 10 nM, especially for SND104 σA-RNA polymerase (Fig. 5B, upper panel). This phenomenon might be attributed to aggregation of the binary complexes, which was eliminated as heparin was incorporated into the binding buffer. With the improvement, a clearly lower promoter DNA-binding activity was observed for SND104 σA-RNA polymerase than that of the WT or SND103 σA-RNA polymerase (Fig. 5B), indicating that removal of Arg-103 from the truncated σA will affect promoter DNA binding of the truncated σA-RNA polymerase and/or the stability of the binary complexes thus formed.
FIG. 4.
The core-binding efficiency of the WT and truncated σA. The method used for measuring the core-binding efficiency of σA is described in Materials and Methods. Sample collection was started after elution of the molecular sieving column with buffer equal to the void volume. The fractionated samples (1 ml/tube) were analyzed for the contents of β′ and truncated σA by Western blotting. The core-associated σA was assessed by its coelution with β′. The percentage of core-associated σA was calculated by dividing the band density of core-associated σA by the total band densities of core-associated and free σA.
FIG. 5.
The promoter DNA-binding activities of the WT and N-terminally truncated σA-RNA polymerase holoenzymes. (A) Promoter DNA-binding activity of the WT, SND100, or SND102 σA-RNA polymerase in the absence of heparin. In the assay, a 1 nM concentration of 32P-labeled G3b promoter DNA was mixed with various concentrations (0, 5, 10, 15, 20, and 25 nM) of the reconstituted RNA polymerase holoenzyme. The molar ratio of σA to core was 10:1 for enzyme reconstitution. The gel retardation assay used for measuring the promoter DNA-binding activity is described in Materials and Methods. The relative promoter-DNA binding activity shown in the bottom panel was calculated by dividing the band density of the binary complexes formed at the indicated concentration of the truncated σA-RNA polymerase by that formed by a 25 nM concentration of the WT σA-RNA polymerase. Each value is the average of three individual tests. (B) Promoter DNA-binding activities of the WT, SND103, and SND104 σA-RNA polymerase holoenzymes in the absence (top panel) or presence (bottom panel) of heparin. The method used for the assay was the same as that used for panel A, except that a 0.1 nM (0.6 μg/ml) concentration of heparin was incorporated in the binding buffer when needed. The two bands indicated by asterisks are putative complexes formed by the G3b promoter and free SND103 or SND104. RNAP, RNA polymerase.
Interestingly, two extra bands with electrophoretic mobilities faster than those of the binary complexes but slower than those of the free promoter DNA were observed in binding mixture containing SND103 or SND104 σA-RNA polymerase (Fig. 5B, bottom panel). These two bands seem to be the complexes formed by the G3b promoter and free SND103 or SND104, since they were also detectable in the absence of core enzyme (data not shown).
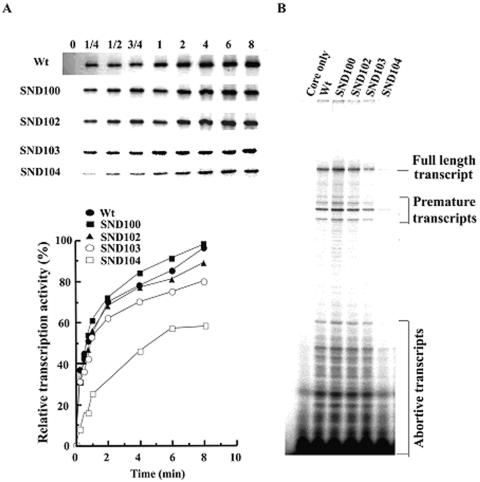
Besides promoter DNA binding, other transcription properties such as open complex formation and abortive transcription were also analyzed for the truncated σA-RNA polymerases. The efficiency of open complex formation was estimated by single-cycle transcription, while the activity of abortive transcription was estimated by measuring the radioactivity of abortive transcripts. Parallel to the decrease in promoter DNA binding (Fig. 5B, bottom panel), a significant reduction in the activity of single-cycle transcription (Fig. 6A) and abortive transcription (Fig. 6B) was detected for SND104 σA-RNA polymerase. The efficiency of open complex formation of SND104 σA-RNA polymerase ranged from about 25 to 60% of that of the WT depending on the time of incubation prior to heparin addition (Fig. 6A, bottom panel). Moreover, the activity of abortive transcription of SND104 σA-RNA polymerase, as assessed by measuring the total radioactivity of abortive transcripts, was only 20% of that of the WT (Fig. 6B). The above results indicate that the RNA polymerase containing SND104 is defective in transcription initiation after core binding.
FIG. 6.
Single-cycle transcription and abortive transcription assays of the N-terminally truncated σA-RNA polymerases. (A) Single-cycle transcription activities of the truncated σA-RNA polymerases. The method used for the single-cycle transcription assay was similar to that used for in vitro transcription. Briefly, 2 μg of σA-RNA polymerase holoenzyme was mixed with 0.8 μg of pCT24 plasmid, which harbors the G3b promoter. The reaction mixture (40 μl) was then incubated at 37°C for 0, 1/4, 1/2, 3/4, 1, 2, 4, or 8 min before 0.3 μg of heparin (0.1 μg/μl) was added. The molar ratio of heparin to G3b promoter was 200:1. Subsequently, 37 μl of reaction cocktail was added, and the reaction was allowed to proceed for 5 min at 37°C. The top panel shows the band densities of in vitro transcript (291 bases) generated by each of the σA-RNA polymerase holoenzyme. The bottom panel shows the relative transcription activities of the tested RNA polymerases, with that of SND100 σA-RNA polymerase obtained at 8 min referred to as 100%. Each value is the average of three individual tests, and the deviation is less than 10%. (B) Abortive transcription activities of the truncated σA-RNA polymerases. The method used for abortive transcription is the same as that described in Materials and Methods. The full-length transcript is 61 bases in length. The RNA polymerase tested is as indicated at the top of each lane.
Importance of conserved Arg-103 at the N terminus of region 1.2 to structure of N-terminally truncated σA.
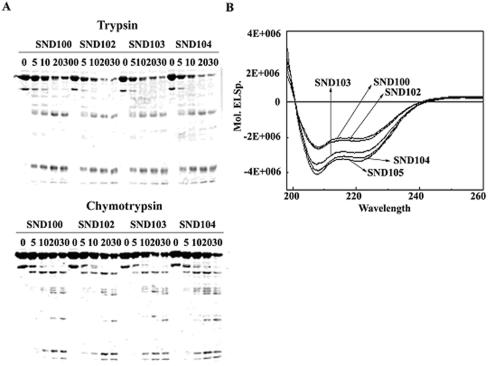
To understand the reasons for the drastic change in transcription properties of SND104 σA-RNA polymerase, we analyzed the overall conformation of the truncated σA by using limited proteolysis. No significant difference in trypsin or chymotrypsin digestion pattern was observed among SND100, SND102, SND103, and SND104 (Fig. 7A), suggesting that the gross structures of the truncated σA were similar despite the differences at their N termini (Table 1). Since the data obtained from limited proteolysis could not rule out the possibility of a minor but detrimental structural change in SND104, CD was adopted for further analysis. As shown in Fig. 7B, the CD spectra of the truncated σA were separated into three groups according to their superimposability. SND100 and SND102, with high transcription activity, had very similar CD spectra. SND104 and SND105, with low transcription activity, were also superimposable in CD spectra. SND103, with moderate transcription activity, had an absorption curve falling just between those for the two above-mentioned groups. The relatively higher negative absorptions of SND103, SND104, and SND105 (also SND106 and SND109 [data not shown]) in CD spectra demonstrate that the substitution (SND103) or deletion (SND104) of Arg-103 has increased the content of stable secondary structures of the truncated σA with moderate and low activities.
FIG. 7.
The structural properties of the N-terminally truncated σA. (A) Partial proteolysis analyses of the N-terminally truncated σA. The truncated σA subunits were partially digested with trypsin (top panel) or chymotrypsin (bottom panel) (see Materials and Methods). The time points (in minutes) to stop the proteolytic reaction are shown on top of each panel. (B) CD spectra of the N-terminally truncated σA. Each of the N-terminally truncated σA (0.2 mg/ml) subunits was scanned with UV light from 190 to 260 nm. The absorption curves for the truncated σA are as indicated.
Arg-103 is critical for full-length σA to function.
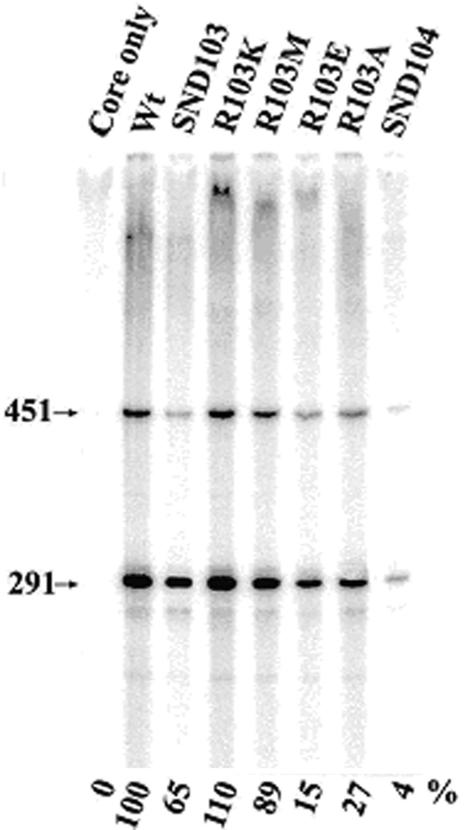
To rule out the possibility that the effect of Arg-103 deletion on transcription of SND104 σA-RNA polymerase is due to the creation of an unusual N terminus of the truncated σA—and to see whether Arg-103 is also important for the functioning of full-length σA in an RNA polymerase holoenzyme—we constructed, overproduced, and compared the transcription activities of a set of full-length σA variants in which Arg-103 was replaced with Lys (K), Met (M), Ala (A), or Glu (E). Here, R103E was designed to enable analysis of the importance of the positive charge of the arginine side chain, and R103A allowed analysis of both the positive charge and size of the side chain. As shown in Fig. 8, the transcription activity of R103E or R103A σA-RNA polymerase on the G3b promoter was about 15 or 27%, respectively, of that of the WT, indicating that both the positive charge and bulky side chain of Arg-103 are important for σA to function in a holoenzyme. This notion was supported by the facts that the R103K σA-RNA polymerase, the Arg-103 of which was replaced with a positively charged and comparably sized lysine, was 10% more active than the WT and that the R103M σA-RNA polymerase, of which the positive charge but not the size of the amino acid side chain was removed, retained about 89% of the activity of the WT (Fig. 8). Taken together, these results demonstrate that the conserved Arg-103 at the N terminus of region 1.2 is also important for the full-length σA to function in an RNA polymerase holoenzyme.
FIG. 8.
In vitro transcription activity of mutant σA-RNA polymerase holoenzyme, with Arg-103 of σA replaced with Lys (K), Met (M), Ala (A), or Glu (E). The method used for in vitro transcription is the same as that described for the truncated σA. The mutant σA-RNA polymerases assayed are shown at the top. The arrowheads indicate the two mRNA transcripts (291 and 451 bases long) generated from the G3b promoter. The value below each lane indicates the relative transcription activity of the assayed RNA polymerase, with that of the WT σA-RNA polymerase referred to as 100%. The standard deviation for three individual tests was less than 10%.
DISCUSSION
We have herein characterized a collection of the B. subtilis N-terminally truncated σA factors. Our results demonstrate that region 1.1 is not required for σA to function either in vitro or (probably) in vivo. Furthermore, the conserved Arg-103 at the N terminus of region 1.2 is critical for the N-terminally truncated σA to function in vitro. Deletion of this arginine residue will increase the content of stable secondary structures of the truncated σA and greatly reduce the transcription activity of the truncated σA-RNA polymerase by lowering the efficiency of transcription initiation after σA and core association. Moreover, the conserved Arg-103 is also important for the functioning of full-length σA in an RNA polymerase holoenzyme.
Three functions have been ascribed to region 1.1 of E. coli σ70, which include autoinhibition of promoter binding by free σ70 (8, 9), modulation of formation of stable polymerase-promoter complexes (32), and enhancement of efficiency of transcription initiation (35). Similar to that reported for E. coli σ70, autoinhibition of promoter binding by region 1.1 was observed for free σA (Fig. 5B, bottom panel). However, region 1.1 of the B. subtilis σA is not essential for transcription initiation of σA-RNA polymerase in vitro, including promoter DNA binding (SND100 in Fig. 5A), open complex formation (SND100 in Fig. 6A), and abortive transcription (SND100 in Fig. 6B). The fact that region 1.1 is dispensable for the functioning of σA in vitro is consistent with the finding that pCF100 is able to fully complement the growth defect of the WT σA-deficient mutant, B. subtilis DB3200 (Fig. 3), and the temperature-sensitive sigA mutant, B. subtilis DB1001, in vivo (data not shown). The dispensability of region 1.1 for σA to function is similar to that happening to the alternative σ, which is devoid of region 1.1 and a long spacer (20), and also to that recently reported for the E. coli σS, which has region 1.1 but no spacer (11).
Regions 1.1 of the primary σ factor are poorly conserved in amino acid sequence, albeit with a preservation of the characteristic acidity. Moreover, this region is assumed to form a disordered structure since it is undetectable in the electron density maps of the cocrystal of Taq RNA polymerase and promoter DNA (23). Actually, region 1.1 of the B. subtilis σA is also largely disordered, as predicted by a protein disorder predictor (data not shown). The dispensability of this region for B. subtilis σA to function (Fig. 2, 3, 5, and 6) implies that it can be independent and separated from other domains of σA. Support for this idea came from in situ proteolysis of the purified Taq σA in crystal trials, in which region 1.1 was shown to separate completely from other fragments consisting of the conserved regions 1.2 to 2.4, 3.0 to 3.1, and 4.1 to 4.2 (3); further support also comes from our previous study, in which Arg-103 was shown to be more accessible to trypsin digestion (34). It has been proposed that the acidic region 1.1 of Taq σA localizes directly inside the main channel and interacts with the basic surface of the channel wall of the RNA polymerase core enzyme (23). If this is true, there would be extra positive charges on the channel surface of the N-terminally truncated σA-RNA polymerase, which would lead in turn to an enhanced interaction of the truncated σA-RNA polymerase with the negatively charged promoter DNA. Our observation of an enhanced promoter DNA-binding activity of SND100 or SND102 σA-RNA polymerase comparing with that of the WT (Fig. 5A) strongly supports this notion. Thus, region 1.1 of σA is able to modulate the promoter DNA-binding activity of σA-RNA polymerase.
The disappearance of binary complexes at relatively high concentrations of SND103 or SND104 σA-RNA polymerase is interesting (Fig. 5B). It is unlikely that this event is attributed to the loss of region 1.1, as no such phenomenon was observed for SND100 or SND102 σA-RNA polymerase. In other words, this event does not result simply from the exposure of extra positive charges on the channel wall of SND103 or SND104 σA-RNA polymerase as mentioned previously. It is most probably triggered by a combination of the extra positive charges on the channel wall of SND103 or SND104 σA-RNA polymerase and the conformation of each of the two truncated σA factors in RNA polymerase holoenzyme.
Our results demonstrate that the conserved Arg-103 in region 1.2 is important for both the N-terminally truncated (Fig. 2) and full-length (Fig. 8) σA forms to function in an RNA polymerase holoenzyme. The high conservation of this arginine in known primary σ factors (Fig. 1) suggests that it may play similar roles for all of the primary σ factors. The relatively high efficiency of core binding of SND104 (Fig. 4) but low promoter binding of SND104 σA-RNA polymerase (Fig. 5B, bottom panel) implies that a certain transcription defect(s) arises in the SND104 σA-RNA polymerase after core and σA association. Most likely, the SND104 σA-RNA polymerase does not assume a conformation suitable for recognition and binding of the promoter DNA. Since SND103 σA-RNA polymerase remains efficient in promoter DNA binding (Fig. 5B), we propose that the conserved Arg-103, rather than its preceding amino acid residues, is critical for σA to undergo a proper conformational change in the core enzyme. Moreover, the differential effect of substitution of Arg-103 for Lys, Met, Glu, or Ala in σA on the transcription activity of σA-RNA polymerase (Fig. 8) suggests that both the positive charge and bulky side chain of the conserved Arg-103 are important for ensuring a proper conformational change of the RNA polymerase holoenzyme.
Although many structural and functional aspects of σ factors have been investigated, some important issues, including the binding of free σ factor to the promoter, the conformational change of σ factor upon core binding, and the DNA melting by σ factor, remain to be solved. The mutant σA obtained in the present study may enable us to study the conformational change of σA during core binding. The direct binding of SND103 or SND104 to the G3b promoter in the presence of heparin (Fig. 5B) implies that the binding strength may be strong enough for future footprinting study of σA. Moreover, the heparin resistance of the binary complexes containing the G3b promoter and free SND103 or SND104 (Fig. 5B) implies that the truncated σA alone may be able to melt the double-stranded DNA (21, 27). Detailed analyses of the above properties are now being undertaken.
Acknowledgments
This research was supported by the National Science Council, Taiwan, Republic of China (NSC 90-2311-B-005-008).
REFERENCES
- 1.Bowers, C. W., and A. J. Dombroski. 1999. A mutation in region 1.1 of σ70 affects promoter DNA-binding by Escherichia coli RNA polymerase holoenzyme. EMBO J. 18:709-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgess, R. R., A. A. Travers, J. J. Dunn, and E. K. F. Bautz. 1969. Factor stimulating transcription by RNA polymerase. Nature 221:43-46. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase specificity σ subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 4.Chang, B. Y., C. T. Liao, Y. D. Wen, and W. H. Wang. 1997. The temperature sensitivity of Bacillus subtilis is due to insufficient activity, rather than insufficient concentration, of the mutant σA factor. Microbiology 143:1299-1308. [DOI] [PubMed] [Google Scholar]
- 5.Chang, B. Y., and R. H. Doi. 1990. Overproduction, purification, and characterization of the Bacillus subtilis RNA polymerase σA factor. J. Bacteriol. 172:3257-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniels, D., P. Zuber, and R. Losick. 1990. Two amino acids in an RNA polymerase σ factor involved in the recognition of adjacent base pairs in the −10 region of cognate promoter. Proc. Natl. Acad. Sci. USA 87:8075-8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison, B. L., C. L. Murray, and J. C. Rabinowitz. 1980. Specificity of promoter site utilization in vitro by bacterial RNA polymerases on Bacillus subtilis φ29 DNA. Transcription mapping with exonuclease III. J. Biol. Chem. 255:8819-8830. [PubMed] [Google Scholar]
- 8.Dombroski, A. J., W. A. Walter, M. T. Record, Jr., D. A. Siegele, and C. A. Gross. 1992. Polypeptide containing highly conserved regions of transcription initiation σ70 factor exhibit specificity of binding to promoter DNA. Cell 70:501-512. [DOI] [PubMed] [Google Scholar]
- 9.Dombroski, A. J., W. A. Walter, and C. A. Gross. 1993. Amino-terminal amino acids modulate σ-factor DNA-binding. Genes Dev. 7:2446-2455. [DOI] [PubMed] [Google Scholar]
- 10.Gardella, T., H. Moyle, and M. M. Susskind. 1989. A mutant Escherichia coli σ70 subunit of RNA polymerase with altered promoter specificity. J. Mol. Biol. 206:579-590. [DOI] [PubMed] [Google Scholar]
- 11.Gowrishankar, J., K. Yamamoto, P. R. Subbarayan, and A. Ishihama. 2003. In vitro properties of RpoS (σS) mutants of Escherichia coli with postulated N-terminal subregion 1.1 or C-terminal region 4 deleted. J. Bacteriol. 185:2673-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmann, J. D., and M. J. Chamberlin. 1988. Structure and function of bacterial sigma factor. Annu. Rev. Biochem. 57:839-872. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez, V. J., L. M. Hsu, and M. Cashel. 1996. Conserved region 3 of Escherichia coli sigma 70 is implicated in the process of abortive transcription. J. Biol. Chem. 271:18775-18779. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez, V. J., and M. Cashel. 1995. Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. J. Mol. Biol. 252:536-549. [DOI] [PubMed] [Google Scholar]
- 15.Huang, W. C. 2000. Identification and characterization of the functional domains of Bacillus subtilis σA factor. Master's thesis. National Chung-Hsing University, Taiwan, Republic of China.
- 16.Juang, Y. L., and J. D. Helmann. 1994. A promoter melting region in the primary σfactor of Bacillus subtilis: identification of functionally important aromatic amino acids. J. Mol. Biol. 235:1470-1488. [DOI] [PubMed] [Google Scholar]
- 17.Kumar, A., H. S. Williamson, N. Fujita, A. Ishihama, and R. S. Hayward. 1995. A partially functional 245-amino-acid internal deletion derivative of Escherichia coli sigma 70. J. Bacteriol. 177:5193-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landini, P., and S. J. Busby. 1999. The Escherichia coli Ada protein can interact with two distinct determinants in the σ70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J. Bacteriol. 181:1524-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao, C. T., Y. D. Wen, W. H. Wang, and B. Y. Chang. 1999. Identification and characterization of a stress-responsive promoter in the macromolecular synthesis operon of Bacillus subtilis. Mol. Microbiol. 33:377-388. [DOI] [PubMed] [Google Scholar]
- 20.Lonetto, M., M. Gribskov, and C. A. Gross. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174:3843-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melancon, P., R. R. Burgess, and M. T. Record, Jr. 1982. Nitrocellulose filter binding studies of the interaction of E. coli RNA polymerase holoenzyme with deoxyribonucleic acid restriction fragments: evidence for multiple classes of non-promoter interactions, some of which display promoter-like properties. Biochemistry 21:4318-4331. [DOI] [PubMed] [Google Scholar]
- 22.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor σ54 (σN). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 23.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 24.Panaghie, G., S. E. Aiyar, K. L. Bobb, R. S. Hayward, and P. L. de Haseth. 2000. Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 299:1217-1230. [DOI] [PubMed] [Google Scholar]
- 25.Rhodius, V. A., and S. J. Busby. 2000. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase sigma 70 subunit: application of suppression genetics. J. Mol. Biol. 299:311-324. [DOI] [PubMed] [Google Scholar]
- 26.Ring, B. Z., W. S. Yarnell, and J. W. Robert. 1996. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell 86:485-493. [DOI] [PubMed] [Google Scholar]
- 27.Roe, J. H., R. R. Burgess, and M. T. Record, Jr. 1984. Kinetics and mechanism of the interaction of Escherichia coli RNA polymerase with the λPR promoter. J. Mol. Biol. 176:495-521. [DOI] [PubMed] [Google Scholar]
- 28.Rong, J. C., and J. D. Helmann. 1994. Genetic and physiological studies of Bacillus subtilis σA mutants defective in promoter melting. J. Bacteriol. 176:5218-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen, R., H. Nagai, V. J. Hernandez, and N. Shimamoto. 1998. Reduction in abortive transcription from the lambda PR promoter by mutations in region 3 of the sigma-70 subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 273:9872-9877. [DOI] [PubMed] [Google Scholar]
- 30.Siegele, D. A., J. C. Hu, W. A. Walter, and C. A. Gross. 1989. Altered promoter recognition by mutant forms of the σ70 subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 206:591-603. [DOI] [PubMed] [Google Scholar]
- 31.Tabor, S. 1990. Expression using the T7 RNA polymerase/promoter system, p. 16.2.1-16.2.11. In F. A. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, N.Y.
- 32.Vuthoori, S., C. W. Bowers, A. McCrachen, A. J. Dombroski, and D. M. Hinton. 2001. Domain 1.1 of the σ70 subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 309:561-572. [DOI] [PubMed] [Google Scholar]
- 33.Waldburger, C., T. Gardella, R. Wong, and M. M. Susskind. 1990. Changes in conserved region 2 of Escherichia coli σ70 affecting promoter recognition. J. Mol. Biol. 215:267-276. [DOI] [PubMed] [Google Scholar]
- 34.Wen, Y. D., C. T. Liao, K. M. Liou, W. H. Wang, W. C. Huang, and B. Y. Chang. 2000. Structural and functional properties of a Bacillus subtilis temperature-sensitive σA factor. Proteins 40:613-622. [DOI] [PubMed] [Google Scholar]
- 35.Wilson, C., and A. J. Dombroski. 1997. Region 1 of sigma 70 is required for efficient isomerization and initiation of transcription by Escherichia coli RNA polymerase. J. Mol. Biol. 267:60-74. [DOI] [PubMed] [Google Scholar]
- 36.Zuber, P., J. Healy, H. L. Carter, S. Cutting, C. P. Moran, and R. Losick. 1989. Mutation changing the specificity of an RNA polymerase sigma factor. J. Mol. Biol. 206:605-614. [DOI] [PubMed] [Google Scholar]