Abstract
Somatic sequencing of cancers has produced new insight into tumorigenesis, tumor heterogeneity, and disease progression, but the vast majority of genetic events identified are of indeterminate clinical significance. Here we describe a NextGen sequencing approach to fully analyze 248 genes, including all those of known clinical significance in melanoma. This strategy features solution capture of DNA followed by multiplexed, high-throughput sequencing, and was evaluated in 31 melanoma cell lines and 18 tumor tissues from patients with metastatic melanoma. Mutations in melanoma cell lines correlated with their sensitivity to corresponding small molecule inhibitors, confirming, for example, lapatinib sensitivity in ERBB4 mutant lines and identifying a novel activating mutation of BRAF. The latter event would not have been identified by clinical sequencing and was associated with responsiveness to a BRAF kinase inhibitor. This approach identified focal copy number changes of PTEN not found by standard methods, such as comparative genomic hybridization (CGH). Actionable mutations were found in 89% of the tumor tissues analyzed, 56% of which would not be identified by standard-of-care approaches. This work shows that targeted sequencing is an attractive approach for clinical use in melanoma.
INTRODUCTION
Clinical sequencing is already an essential part of melanoma treatment. Mutations in BRAF and KIT have both high frequency (45% and 4% respectively (Hodis et al. 2012; Krauthammer et al. 2012)) and clinical implications in melanoma (Flaherty et al. 2010; Kim et al. 2008). Numerous other oncogenic mutations, including NRAS, and PIK3CA (18% and 3% of melanoma) could join the list of clinically actionable mutations, as ongoing clinical trials progress (Ascierto et al. 2013). Additional therapies are also in development for driver mutations that infrequently occur in melanoma (e.g. EZH2, RET, MEK1). Bringing these therapies to bear on melanoma treatment will require rapid identification of tumor genotype across a large, but defined subset of genes.
Recent whole exome sequencing studies have demonstrated substantial power and sensitivity to discover new genes with significantly elevated mutation rates in melanoma. Two such studies recently identified mutations of RAC1 and ARID2 as potential pathogenic events in melanoma, among others, (Hodis et al. 2012; Krauthammer et al. 2012). Studies of melanoma whole exomes, such as The Cancer Genome Atlas project, are powered to identify genes mutated at even lower frequency. Whole exome analysis, however, suffers from several drawbacks. Current methods cost ~$1,000 per sample for sequencing alone, as well as additional costs of the capture methods. As exome sequencing is currently not amenable to a high level of multi-sample pooling (multiplexing), it also incurs high cost in both personnel and sequencing machine time. Perhaps most problematic, whole exome and whole genome studies require computationally intensive bioinformatics analysis and data storage consuming significant resources and specialist interpretation. This problem has been described as “The $1,000 genome, [and] the $100,000 analysis” (Mardis 2010). Lastly, even after a list of high confidence somatic mutations has been generated and annotated from exomic data, the vast majority of such events will be of unknown significance.
More targeted approaches exist that can potentially alleviate the above issues. These options include PCR-based resequencing or in-solution hybrid capture techniques with a more focused target list. These approaches reduce costs and complexity at each stage. In the case of PCR-based resequencing, reduction of the DNA sequence target to a much smaller scale (< 1 megabase, Mb) permits more rapid sequencing and library generation, much higher levels of multiplexing, and more simplified bioinformatics analysis. Hybrid capture technologies, by comparison, retain a slower library generation process, but similarly allow increased multiplexing for reduced sequencing requirements and per-sample costs. Hybrid capture has two principal advantages over PCR-based approaches: it permits analysis of a larger target (several MB), and, with concurrently sequenced matched normal control, can be used to identify copy number events (Chinnaiyan & Palanisamy 2010; J. Li et al. 2012; Sathirapongsasuti et al. 2011; Timmermann et al. 2010). The latter is of particular relevance in melanoma, where deletion of tumor suppressor genes (e.g. CDKN2A, STK11 and PTEN) plays a critical role in tumor pathogenesis.
To develop a more cost effective sequencing pipeline we designed a targeted hybrid capture approach for use in solid tumors including melanoma. This panel was designed to find all known genetic events of clinical relevance for these cancers. The approach employed is flexible and customizable to addition of new genes as their roles in cancer are better characterized and clinical correlates are defined, and can make use of germline sequencing information when available. Our captured list increased from 75 genes in version 1 to 248 genes in version 6. We first employed this assay on a panel of melanoma cell lines, and identified novel genetic events that correlated with increased sensitivity to corresponding small molecule inhibitors. We subsequently performed targeted exon capture sequencing in 18 tumor tissues obtained from patients with metastatic melanoma, including 4 tumor tissues that were prospectively collected along with matched peripheral blood. Actionable events detected in the prospective samples were disclosed to treating physicians after validation in a clinical laboratory improvement amendment (CLIA) assay.
RESULTS
Targeted Capture and Sequencing Genotyping of Melanoma Cell Lines and Tumors
We performed target resequencing of genes with a reported role in melanoma and other solid tumors by employing a multiplexed, in-solution capture in 31 melanoma cell lines and 18 patients. Variants were identified by comparison to matched normal specimens for the prospective samples, or with the Unified Genotyper single sample mutation calling pipeline (DePristo et al. 2011; McKenna et al. 2010) for the retrospectively collected tumor tissue from patients with advanced melanoma and various melanoma cell lines (see methods). Calls from retrospective samples without a matched normal were subjected to further filtering to remove likely germline variants. This informatics approach was employed to identify possible somatic variants with clinical relevance in melanoma treatment, biology, and prognosis.
By applying version 1 (V1) of the assay in 31 melanoma cell lines, we identified 237 single nucleotide variants (SNV) in coding regions of 59 genes (Table 1 and Supplementary Tables 1 and 2) and 15 insertion-deletion (indel) variants in 10 genes. Of the SNV calls, 64 were synonymous (S) and 173 nonsynonymous (NS), including 9 resulting in a stop codon gain, for a nonsynonymous-to-synonymous (NS:S) ratio of 2.70. This was higher than that seen in previously reported whole-exome study of melanoma cell lines (Stark et al. 2012), likely due to a higher degree of selection on this gene subset. Given our target sequence size of 446 kb in 31 lines, this is an overall somatic mutation rate of 18 somatic mutations per Mb per line (inner quartile range 8.97 to 26.9), consistent with the high mutation rate previously described in melanoma (Hodis et al. 2012; Pleasance et al. 2010; Stark et al. 2012).
Table 1. Selected Mutations in Melanoma Cell Lines and Patients.
Selected genes from melanoma cell lines and patient tumors with potential activating mutations.
| Sample | Gene | Mutation | Previously Observed? | |
|---|---|---|---|---|
| Cell Line | SKMEL100 | KIT | Y362F | No |
| SBC12 | PIK3CA | K678E | No | |
| RPMI8322 | KRAS | G12V | Yes | |
| SKMEL78/88 | HRAS | G12V | Yes | |
| Patient Tumor | RAM99-0621B | KIT | L255Q | No |
| RAM00-0594B | PIK3CA | R88Q | Yes | |
| RAM99-0621B | PIK3CA | E545K | Yes |
To demonstrate the clinical utility of our sequencing approach we subsequently used V2 of the assay (Supplementary Table 1) to analyze 14 retrospectively collected melanoma tumor tissues. For these samples, matched germ line DNA was not available, and therefore likely germline events were filtered using bioinformatic approaches (see methods). By employing this approach we identified 68 SNV and 4 indel variants in coding regions of 52 genes. Of the SNVs, 50 were coding and 18 noncoding for a ratio of 2.78 and an average 25 somatic mutations per Mb per tumor (inner quartile range 12.7 to 42.4).
Finally, by employing V6 of the assay (Supplementary Table 1), we analyzed 4 tumors prospectively collected from patients with metastatic melanoma for which a matched normal tissue was available. SNVs were called by a more robust approach, relying on sequencing of the matched normal to control for germline variants. We identified 105 coding variants, with a range from 2 to 64 somatic mutations per patient.
Internal and External Validation
We previously sequenced NRAS, KIT, BRAF, MET and CDKN2A/B in the panel of 31 melanoma cell lines ((Shields et al. 2007) and not shown). These results were confirmed using the V1 of our next generation sequencing (NGS) assay. A normal primary fibroblast cell line processed in parallel with the melanoma cell lines resulted in only one synonymous variant called as an SNV, validating that our approach has a low rate of false positives. In the four melanomas with a matched normal, we identified seven mutations, out of which five were successfully validated using a CLIA-certified assay. Although it is possible that our failure to confirm these two mutations may reflect sequencing errors of our NGS assay, more likely, we believe the observed discordance reflects lower sensitivity of the CLIA-certified assay (e.g. Sanger sequencing), especially in the presence of stromal contamination (Nollau & Wagener 1997). These results suggest the assay is highly accurate with regard to the detection of SNVs in cell lines and primary tumors.
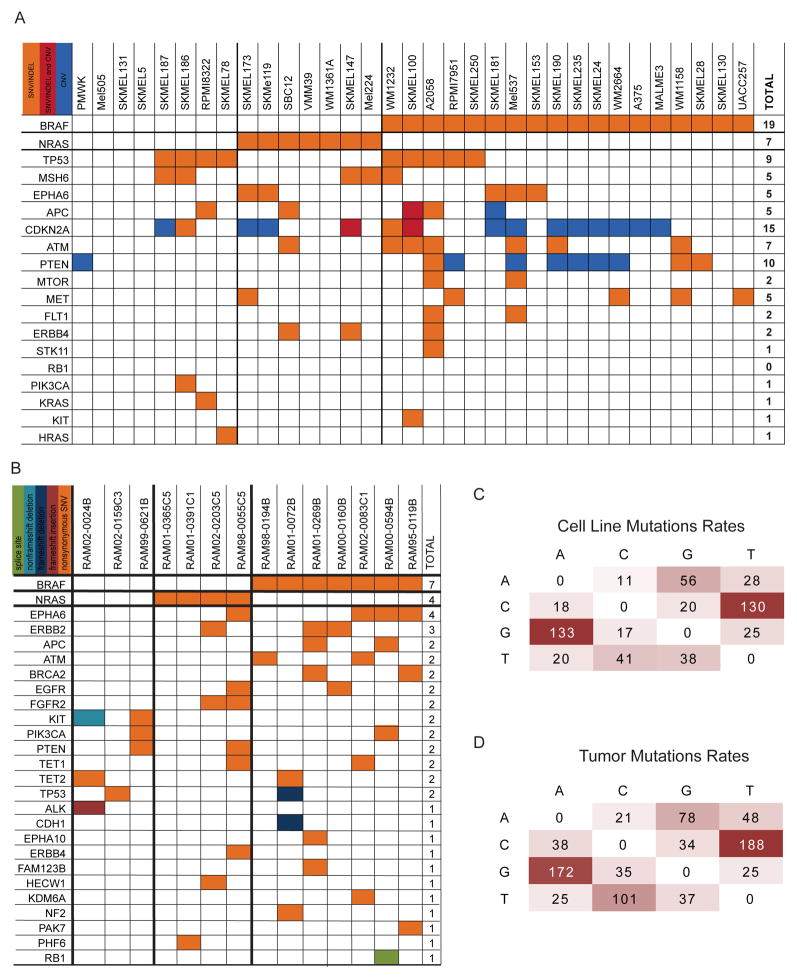
In comparison with published melanoma datasets (COSMIC (Forbes et al. 2011) and two studies (Hodis et al. 2012; Krauthammer et al. 2012)), somatic single nucleotide mutation frequencies and profiles were similar for both melanoma cell lines and tumors (Fig. 1A,B). For example, BRAF and NRAS mutations were noted in 52% and 21% respectively of all samples (cell lines and patient samples). Canonical BRAF V600E and NRAS Q61R mutations were present at a range of estimated allele frequencies in patient derived samples (from 16% to 91%). This suggests robust detection of mutations in the presence of substantial aneuploidy or stromal contamination. In addition, single base pair mutations in cell lines and unmatched tumor samples exhibited C → T mutation bias (Fig. 1C,D). This mutation profile has been reported in whole genome (Pleasance et al. 2010) and exome sequencing (Hodis et al. 2012; Krauthammer et al. 2012) in melanoma, suggesting common ultraviolet light-induced mutagenesis. These data suggest targeted NextGen sequencing yields mutation frequencies and spectrum that are similar to large-scale whole genome and exome sequencing strategies.
Figure 1. Aggregate Analysis of Mutation Calls.
(A) Co-occurrence of mutations in melanoma cell lines including SNVs, CNV calls from SNP array, and both. (B) Co-occurrence of mutations in metastatic cutaneous melanoma showing KIT mutations in BRAF and NRAS wild type lines. (C) Putative somatic mutation rates in melanoma cell lines and (D) putative somatic mutations in tumors.
Exemplary Events not Found by Standard Analyses
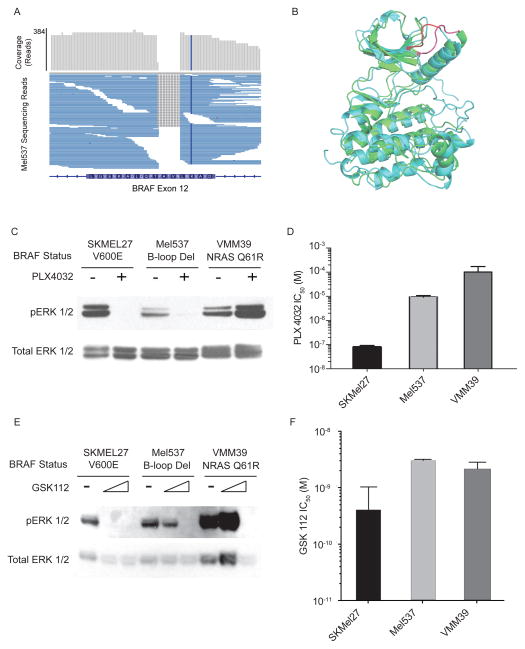
Apart from hot-spot mutations of RAS/RAF/KIT detected by standard molecular pathology approaches, the assay was able to identify a diverse set of genetic aberrations not found using standard clinical sequencing. For example a 5 codon in-frame deletion mutation of BRAF in the Mel537 cell line was discovered (BRAF del486-491) (Fig. 2A). This mutation exhibited remarkable structural similarity (Fig. 2B) to activating in-frame deletions of the EGFR kinase found in non-small cell lung cancer (Jackman et al. 2006). Consistent with this mutation being an activating event, this line co-clustered with NRAS and BRAF mutant tumors, but not with RAS/RAF wildtype tumors, in an RNA expression analysis (Supplementary Fig. 1). Additionally this line exhibited increased levels of phospho-ERK that decreased with treatment with a BRAF inhibitor, PLX-4032 (Fig. 2C,D), in contrast to the paradoxical increase in phospho-ERK that occurs in NRAS mutant tumors (Fig. 2C, VMM39, see also (Carnahan et al. 2010; Halaban et al. 2010; Heidorn et al. 2010)). Compared to lines harboring the classical V600E BRAF mutation, Mel537 was comparatively less sensitive to both BRAF (PLX-4032) and MEK inhibitors (trametinib or GSK1120212 (Stones et al. 2013)) (Fig. 2C-F). This observation may be driven by altered downstream signaling in the BRAF del486-491 mutant, altered binding of drug substrates, or by additional mutations in the Mel537 cell line not related to the RAS-RAF-ERK signaling cascade. These results suggest that the BRAF del486-491 mutation is a non-canonical mechanism to activate ERK and downstream targets in melanoma, and is associated with moderate sensitivity to BRAF and MEK inhibitors.
Figure 2. Non-Frameshift BRAF Mutation Sensitive to PLX4032.
(A) Mel537 cell line aggregate read depth and mapped reads show homozygous non-frameshift deletion mutation in exon 12 of BRAF. (B) Structural alignment of inhibitor bound BRAF structure (CYAN) to inhibitor bound EGFR (GREEN) showing structural homology of the BRAF exon 12 deletion (RED) and know activating EGFR exon 19 deletions (PINK). (C) ERK and activated ERK (pERK) blots in serum-starved lines with and without PLX4032 treatment. (D) PLX4032 sensitivity of SKMEL27 (BRAF V600E), Mel537, and VMM39 (NRAS Q61R). (E) ERK and activated ERK (pERK) blots in serum-starved lines with and without MEK inhibition by GSK112. (F) GSK112 sensitivity of SKMEL27, Mel537 and VMM39.
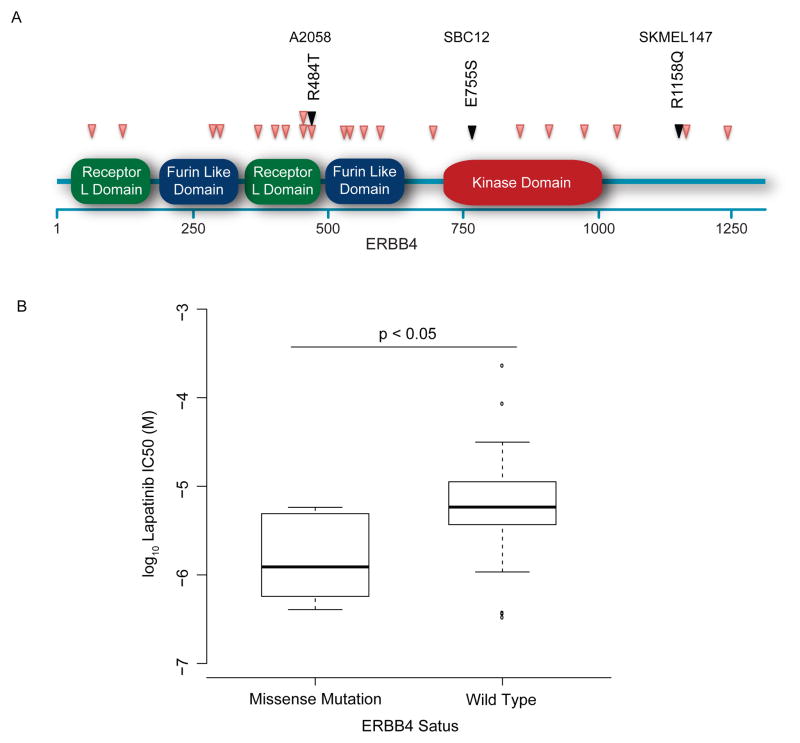
Consistent with a prior report (Prickett et al. 2009; Stark et al. 2012), we noted several mutations in diverse regions (Fig. 3A) of the ERBB4 gene in melanoma cell lines and tumors. Melanoma cell lines harboring mutations of this kinase exhibited a trend (p=0.1) towards enhanced sensitivity to the EGFR-inhibitor, lapatinib, in accordance with prior findings (Fig. 3B) (Prickett et al. 2009). While these mutations appear common (observed in 10% of melanoma primaries and cell lines assessed in this sample), the dispersion of these mutations throughout the gene makes their identification difficult by classical (Sanger) sequencing. Other potential clinically actionable events were observed in both melanoma cell lines (Supplementary Table 2) and clinical samples (Table 2) and included previously described as well as novel mutations in PIK3CA and KIT.
Figure 3. ERBB4 Mutation Predicts Lapatinib Sensitivity.
(A) Previously reported mutations in ERBB4 (PINK) and in three cell lines (BLACK) span multiple domains of the gene. (B) Lapatinib sensitivity in three ERBB4 mutant lines was increased compared to ERBB3 wild type lines (p = 0.1).
Table 2. Targeted Resequencing Identifies Clinically Actionable Mutations.
Clinical characteristics of 14 patients with advanced melanoma whose tumors underwent NGS. Mutation calls and mutational profiles, with potentially actionable mutations are highlighted in red. Somatic mutations validated by CLIA-certified testing and reported to physicians are underlined.
| Pt No | Age | Race | Sex | Non Synonymous Single Nucleotide and Indel Variations | Survival (Months) |
|---|---|---|---|---|---|
| 1 | 87 | W | M |
|
0.8 |
| 2 | 38 | W | M |
|
11+ |
| 3 | 69 | W | M |
|
71 |
| 4 | 57 | W | M |
|
37 |
| 5 | 28 | W | M |
|
12 |
| 6 | 31 | W | M |
|
17 |
| 7 | 70 | W | M |
|
74 |
| 8 | 41 | W | M |
|
9 |
| 9 | 70 | Afr | F |
|
17 |
| 10 | 37 | As | F | KDM6AT726K PTCH1A741V TP53V31I | 21 |
| 11 | 73 | W | F |
|
16 |
| 12 | 24 | W | F |
|
204 |
| 13 | 38 | W | M |
|
31 |
| 14 | 72 | W | F |
|
4 |
| M1 | - | - | - | EPHB6P165PS TSC2FS | N/A |
| M2 | - | - | - |
|
N/A |
| M3 | - | - | - |
|
N/A |
| M4 | - | - | - |
|
N/A |
Tumor Suppressor Loss
In addition to detecting mutations in oncogenes, we employed our NextGen assay to detect genetic aberration in tumor suppressor genes that play important roles in melanoma progression (Dankort et al. 2009; Liu et al. 2012), overall prognosis (Nathanson et al. 2011), and response to small molecule inhibitors (Trunzer et al. 2013). To estimate tumor suppressor inactivation in the melanoma cell line panel, data from NextGen sequencing were analyzed to identify complete deletion of targeted exons. For this analysis, we identified exons with no sequencing coverage in a subset of melanoma cell lines with otherwise substantial coverage (>30x) in other lines. These calls were validated by CGH array data where available.
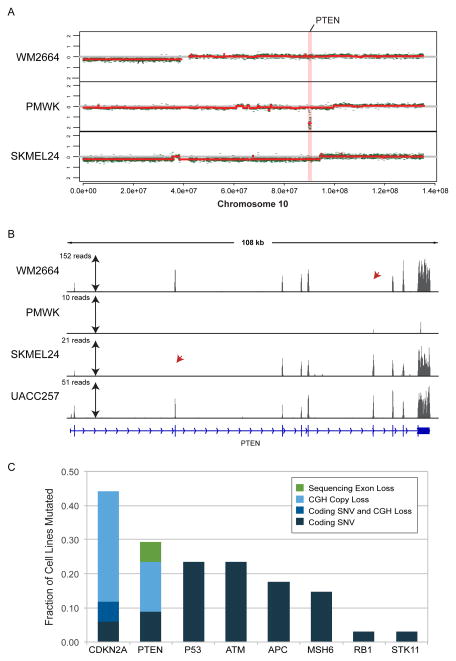
All NextGen-based deletion calls were consistent with CGH arrays, and vice-versa, with the exception of highly focal deletions of PTEN noted in two melanoma cell lines that were called in sequencing but were undetectable by CGH (Fig. 4A). These were regions with complete loss of coverage of individual exons of PTEN in the WM2664 and SKMEL24 lines (Fig. 4B). These findings were in keeping with the total absence of PTEN protein observed in these lines by ourselves and others ((Stahl et al. 2003) and not shown). Melanoma cell lines with larger deletions spanning the PTEN gene were apparent as loss of sequencing coverage of all exons, while the majority of lines showed intact PTEN genes with coverage of all exons. When these genetic aberrations were combined with SNV/indel calls, the observed rates of CDKN2A, PTEN and TP53 mutation were 44%, 29%, and 24% respectively (Fig. 4C), consistent with prior analyses of melanoma cell lines (Forbes et al. 2011; Stark et al. 2012) and TCGA (see e.g. cBioPortal (Cerami et al. 2012)).
Figure 4. Sequencing Detects Focal Deletions at High Resolution.
(A) CNV calls using SNP microarrays. (B) Sequencing coverage of PTEN exons in WM2664, PMWK, SKMEL24 and UACC257 with very low coverage in a PTEN deletion line (PMWK) as well as isolated exons in other lines (red arrowheads). (C) Aggregated mutation rates in selected tumor suppressor genes in cell lines using CGH array and sequencing.
Combined SNV and copy number analysis of NextGen sequencing yielded a high total rate of less common tumor suppressor gene (TSG) mutations in our clinical samples as well (Table 2). Mutations in APC, ATM, BRCA1, BRCA2, PTEN, RB1 or TET1 were seen in 10 out of 14 completed patients (71%) lacking matched normal. We additionally identified copy number changes in both target and off-target regions by comparing tumor coverage to that of the matched normal, where available. When restricting the analysis to captured genes, two tumors had copy number changes reaching significance (adjusted p < 0.10). These were in CCND2 (5.4 fold copy number vs. normal) and PTEN (0.3 fold copy number vs. normal), and both are commonly mutated in melanoma (Forbes et al. 2011), TCGA (cBioPortal (Cerami et al. 2012)). Collectively, our results show the ability of targeted capture exome NGS to detect various genetic aberrations, both mutation and copy number changes, in common as well as infrequently mutated genes, which may provide further insights about their role in prognosis and treatment decisions.
Assessment of Clinical Actionability and Result Reporting
To assess the potential clinical applicability of this targeted sequencing approach, we identified patients with somatic mutations that might influence clinical care (Table 2). These mutations were associated with either a potential treatment (small molecule inhibitors) or with use in prognosis and diagnosis (Table 3). Therapeutic indications were mutations associated with sensitivity to agents i) approved for use in melanoma, ii) approved for use in a different tumor type or iii) in human clinical trials for melanoma. Of the 18 patient-derived tumors, 16 patients (89%) carried mutations that could influence treatment decisions, including possible enrollment into a clinical trial. These included BRAF activating mutations (V600E and D594E) as well as mutations in ALK, ERBB4, KIT, and PIK3CA. Even excluding BRAF mutations found by standard clinical testing, 9 patients exhibited mutations that could influence therapy. This ‘actionability’ rate is comparable to that recently reported by another group in a large study of multiple cancer types including melanoma (Frampton et al. 2013).
Table 3. Genes and Associated Clinical Action.
Genes discovered in patient tumors with clinical actionability, along with the associated action and therapy.
| Gene | Category | Action |
|---|---|---|
| ABL | Approved Therapy | Imatinib, Desatinib, Nilotinib, Ponatinib |
| ALK | Approved Therapy | Crizotinib |
| BRAF | Approved Therapy | Vemurafenib |
| ERBB4 | Clinical Trials | Lapatinib |
| KDR | Approved Therapy and Clinical Trials | Nonspecific kinase inhibition: Sunitinib, Sorafenib, Pazopenib |
| KIT | Approved Therapy | Desatinib |
| MET | Approved Therapy | Crizotinib, Carbozantinib |
| NOTCH4 | Clinical Trials | Gamma Secretase Inhibitors |
| NRAS | Future Clinical Trial | MEK/ERK inhibitors |
| PDGFRA | Clinical Trials | PDGFR inhibitors (e.g. Crenolanib) |
| PDGFRB | Clinical Trials | PDGFR inhibitors (e.g. Crenolanib) |
| PIK3CA | Clinical Trials | PI3K Inhibitors |
| PTEN | Future Clinical Trial | PI3K Inhibitors |
Patients with metastatic melanoma who consented to have their tumor sequenced for actionable somatic mutations underwent NextGen sequencing using our assay as part of an ongoing clinical trial (NCT01457196). Somatic mutations deemed clinically actionable by the above criteria could be reported to a physician after validation using a CLIA-certified assay. In the four melanomas analyzed, we found between 0 and 6 actionable mutations per tumor (Table 3). One tumor had the BRAF V600E mutation alone. Two other tumors had several mutations, including one in MAP3K1, recently reported to have tumor suppressor activity in melanoma (Stark et al. 2012), as well as ERBB4 and PIK31R. Mutations in five genes (ABL, BRAF, MET, PDGFRA, and PDGFRB, Table 2 underlined) from 3 patients were confirmed by a CLIA-certified method and reported in the clinical record. These results indicates that this low-cost, capture-based sequencing can be performed in clinical real-time to provide actionable information beyond currently performed clinical sequencing.
DISCUSSION
In this report, we describe the development and performance of a customized, capture-based NextGen assay to identify somatic genetic aberrations, both mutations and copy number changes, in genes associated with cancer development, progression, prognosis, and response to treatment. Identification of such genetic aberrations may influence clinical care of patients with melanoma and other cancers. This approach can be easily modified to include additional genes whose role in cancer is being better established. Using this approach, we demonstrated in a limited number of tumor tissues obtained from patients with advanced melanoma that such patients harbor a high frequency of actionable genetic aberrations beyond BRAF, NRAS and KIT. We observed actionable events both for oncogenes outside of regions commonly subjected to sequencing in the current clinical setting (e.g. “non-canonical” mutations of BRAF) and of druggable oncogenes not traditionally considered to drive melanoma formation (e.g. PIK3CA and ERBB4). For three cases, events identified using this approach were validated using CLIA-approved methods and reported to patients and caregivers within the context of a clinical trial.
While whole exome/genome analyses have their merit for scientific discovery, we believe that the use of highly targeted sequencing panels is presently the most cost-effective approach for use in clinical decision-making. In addition to cost-efficacy, highly targeted sequencing can produce greater depth of sequencing coverage over the target, allowing the identification of mutations with relatively low minor allele frequency (<10%) within a sample. This is essential for samples with stromal contamination, which is likely to be a common feature of clinical samples, or in the presence of genetic heterogeneity within the tumor. Such an approach will be valuable in upcoming clinical trials that attempt to: identify in real-time patient subsets that may exhibit primary resistance to small molecule inhibitors (Trunzer et al. 2013), understand the genetic patterns of resistance to small molecule inhibitors (Nazarian et al. 2010; Shi et al. 2012), and provide guidance for rational treatment combinations applicable only to select patient subgroups. This approach leverages the information from large discovery-driven projects (Hodis et al. 2012; Krauthammer et al. 2012) and TCGA while allowing significantly reduced costs through multiplexing. Additional multiplexing is likely to be possible as sequencing technologies continue to advance. Of note, we have now extended the assay to formalin-fixed paraffin embedded (FFPE) samples, allowing for greater utility in common clinical care. DNA quality from FFPE in general is adequate for capture based NextGen sequencing, yielding results comparable to those obtained using frozen material (not shown and (Frampton et al. 2013)).
Using this approach, we identified an unusual mutation (del486-491) in BRAF in a cell line that would not be identified by standard-of-care approaches. Although we are not aware of a prior identification of this BRAF mutation in melanoma, the del486-491 mutation has been previously reported in an ovarian cell line (Hanrahan et al. 2012). The ability of this mutation to lead to activation of ERK and its moderate sensitivity to BRAF and MEK inhibitors suggests this mutation merits clinical assessment. We believe this mutation may be more common than appreciated, having escaped prior identification because it is not in a commonly analyzed exon (12 as opposed to 15 and 11) and is a 15 bp deletion, requiring approaches dedicated toward identifying these events. Targeted sequencing successfully identified BRAF and NRAS mutations observed in melanoma, but also discovered more rare events in these genes. This approach also readily identified canonical, activating mutations of other druggable oncogenes, many of which are the target of ongoing clinical trials (e.g. PIK3CA).
The ability to uncover many simultaneous mutations within a melanoma affords new opportunities for multiple targeted therapies. However, the principles of such treatments remain unclear. Which mutations, if any, should take priority for targeting, or should multiple oncogenic mutations always be targeted with multiple therapies? Though it seems likely that multiple therapies will more effectively target resistance pathways, as with dual BRAF/MEK inhibition in melanoma, multiple pathway targeting has also lead to increased toxicity (e.g. as seen with combinations of MEK and PI3K inhibitors). Most difficult is the rarity of any one particular assemblage of mutations, given the number of possible mutations involved. In the near term, the personalization of cancer therapy remains experimental.
Finally, capture offers a platform for determination of genetic aberrations in tumor suppressors genes, allowing simultaneous assay of SNVs, indels, and copy loss in genes such as ERBB4, STK11, RB1, CDKN2A, and PTEN. Inactivation of these genes is common in melanoma, and likely will strongly affect tumor biology. For example, RB1/CDKN2a status correlates with sensitivity to CDK4/6 inhibitors (Wiedemeyer et al. 2010), whereas loss of ERBB4 is associated with lapatinib sensitivity (Figure 4 and ((Prickett et al. 2009)), which is currently being tested in a clinical trial (NCT01264081). Our results identifying microdeletions within the PTEN gene are of particular interest, since such events were not detected by CGH and could play important roles in targeted therapy. Recent improvements in bioinformatic techniques enabling comparison of patient germline to neoplastic tissue for CNV detection will aid future analysis of targeted tumor suppressor sequencing in a clinical setting. The ability to affordably and reliably assay TSG status could aid prognosis, subtyping and therapy in melanoma.
METHODS
Clinical Material
Snap-frozen primary tumor tissues were obtained as part of institutional review board (IRB)-approved protocols at the Lineberger Comprehensive Cancer Center and the University of North Carolina. Samples from four prospective patients were collected under an ongoing clinical trial LCCC1108 (NCT01457196), whereas samples from 14 retrospective patients were collected under IRB#09-0989. For the four prospective patients, a matched normal tissue (whole blood), was also acquired. DNA was isolated with the Puregene DNA Purification Kit (Gentra Systems) or DNAeasy (QIAGEN).
Sequencing Library Preparation and Capture
Custom adapters were designed to match Illumina PE adapters with an additional 4 nucleotide ‘barcode’ sequence located internal to the first and second read sequencing (Supplementary Fig. 2). Barcode sequences were designed to require errors in at least 2 locations to be misidentified, allowing for error correction with the use of paired reads tolerant to up to 3 errors.
Sample workflow is detailed in Supplementary Figure 3A. Libraries were constructed per the SureSelect protocol (Agilent) with custom adapters replacing the Illumina PE adapters. Samples were quantified by bioanalyzer prior to capture, and pooled in equimolar quantity with 10 libraries to a pool in the case of cell lines and 8 libraries to a pool in the case of melanoma tumor samples to a final quantity of 500ng. Four samples for which a matched normal was available were prepared with the SureSelect XT library preparation kit (Agilent) by manufacturer protocols. Capture and post capture amplification for all samples proceeded by SureSelect protocol. Samples were sequenced on the GAII or HiSeq machine in a variety of formats (see Supplementary Table 3).
Bioinformatics Analysis of Unmatched Samples
Bioinformatics workflow is detailed in Supplementary Figure 3B. Custom Java applications were used to assign multiplex libraries to their source sample, after which samples were mapped using the Burrows-Wheeler Aligner (Keniry & Parsons 2008) to the hg18 reference genome. The Genome Analysis Toolkit GATK (version 1.0.5506)(H. Li & Durbin 2009) was used for quality score recalibration, indel realignment, and SNV and indel calling. SNV and indels were then annotated for appearance in prior 1,000 genomes (DePristo et al. 2011; McKenna et al. 2010) and dbSNP version 130 (Consortium 2012) to filter likely germline mutations and identify affected transcripts using the ANNOVAR tool (Sherry et al. 2001). Mutations with any prior detection rate in 1,000 genomes samples as annotated by ANNOVAR were ignored in further analysis, with exception of the NRAS Q61R mutation, which appears in dbSNP 130, as it is a known oncogenic event. Analysis of duplicate reads counts, base quality calls, and read mapping percentages were performed at appropriate steps as quality control. One line sequenced, 1205LU, was proved to be murine in origin, and was excluded from downstream analysis. Two cell lines believed to be distinct were genetically identical (SKMEL78 and SKMEL88), and were treated as a single line in analysis.
Bioinformatics Analysis of Matched Samples
Bioinformatics analysis of tumor samples for which DNA extracted from a same-patient normal tissue was available followed a similar IRB-approved protocol, but somatic variants were called using Varscan (Koboldt et al. 2009) and annotated using the ANNOVAR utility(Wang et al. 2010). Coding somatic variants with “Moderate” or “High” predicted effects were reported. For copy number analysis, each gene targeted to capture is tested for significantly different coverage compared to a normal reference using the DESeq algorithm (Anders & Huber 2010).
Gene Expression Microarray
Previously published gene expression microarray data sets (Carson et al. 2012) were filtered for the top 10% most variable genes (standard deviation of log2 ratio of expression vs. reference) and median centered by gene. Probe sets were clustered by unsupervised hierarchical clustering using uncentered correlation as the similarity metric via the Gene Cluster 3.0 (Carson et al. 2012) tool and visualized with Java TreeView (de Hoon et al. 2004).
Array Based Identification of Copy Number Variation
For array-based determination of copy number in cell lines we used the Agilent 180K SurePrint G3 Custom CGH+SNP arrays (catalog # G4884a). Calls were determined by circular binary segmentation with the DNAcopy package through Bioconductor. Calls were filtered for those with a log2 ratio < −0.5 and p value < 0.01. These were further analyzed for deletions in APC, CDKN2A, MSH6, TP53, RB1, and PTEN tumor suppressors.
Sequencing Based Identification of Copy Number Variation in Cell Lines
Coordinated analysis of these same genes by sequencing utilized the clonality of cultured cell lines to identify additional deletion events. Deletion regions of the genome were those showing zero coverage by high-throughput sequencing reads. As coverage is expected to be low except in regions of capture, we restricted our analysis to exons of APC, CDKN2A, MSH6, TP53, RB1, and PTEN with average coverage > 30x in control cell lines. Genes with zero coverage in a sample with no otherwise notable mutations were marked as deleted by sequencing determined exon loss.
Chemosensitivity Assays
Cells were plated onto 96-well assay plates (Costar #3903) at different cell densities, depending on each cell line’s growth rate, in 100 μl of media per well. Cells were then treated with dilutions of drugs or vehicle (DMSO) in triplicate18 hours after plating. Drugs used included Lapatinib (GlaxoSmithKline), PLX 4032 (Roche), and trametinib (GSK1120212, GalaxoSmithKline). Cell viability was assessed at 72 hours after plating using the CellTiter-Glo Luminenscent Cell Viability Assay (Promega) and a Synergy2 microplate reader (BioTek). Raw data was imported into GraphPad Prism 5.0 and IC50 calculated using non-linear regression analysis of the log inhibitor concentration versus luminescence.
Supplementary Material
Supplementary Figure 1 – Mel537 Expression Coclusters with BRAF/NRAS Mutant Tumors: Unsupervised clustering of the top 10% most dynamic genes quantified by gene expression microarray in melanoma cell lines. Cell line clustering dendrogram appears at top right, with oncogene status and tumor suppressor status indicated. Inset are selected genes demonstrating coclustering of microphthlamia (MITF) high (top), CD200 low (middle), and keratin high (bottom) expression phenotypes.
Supplementary Figure 2 – Custom Barcoding Design: Diagram of custom designed reads, with Illumina PE adapter sequence followed by barcode sequencing, along with resulting reads, with barcode located at the beginning of paired-end sequencing reads.
Supplementary Figure 3 – Schema for Multiplexed Sequencing: (A) Sequencing of melanoma cell lines and tumors utilized a pre-capture pooling of libraries followed by in solution capture and sequencing. (B) Deconvoluted cell line data were mapped and analyzed independently using the Genome Analysis Toolkit followed by filtering, annotation with the SIFT tool, and exclusion of SNPs based on 1000 genomes data.
Genes targeted for capture for the three sample types analyzed.
Nonsynonymous variants not in 1,000 genomes annotations or dbSNP tabulated by cell line. Mutations in NRAS appearing in dbSNP are included due to known relevance in melanoma.
Relevant sequencing data statistics for each sample, including number of reads, number of reads mapping, percent duplicates and percentage of reads within the target region.
SIGNIFICANCE.
Gene sequencing is an essential part of treatment for metastatic melanoma, but current clinical strategies are limited to a few exons of only two genes. Here, we develop a scalable NextGen sequencing assay to identify point mutations and copy number events affecting all actionable genes associated with advanced melanoma. Using this approach, we identify actionable events in 90% of patients, which we disclose to caregivers after validation using a CLIA–approved assay. Importantly, we find that over half of tumors harbor actionable mutations that would not be detected by standard-of-care clinical sequencing, including a novel activating BRAF mutation.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascierto PA, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. The lancet oncology. 2013;14(3):249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- Carnahan J, et al. Selective and potent Raf inhibitors paradoxically stimulate normal cell proliferation and tumor growth. Molecular cancer therapeutics. 2010;9(8):2399–2410. doi: 10.1158/1535-7163.MCT-10-0181. [DOI] [PubMed] [Google Scholar]
- Carson C, et al. A prognostic signature of defective p53-dependent G1 checkpoint function in melanoma cell lines. Pigment cell & melanoma research. 2012;25(4):514–526. doi: 10.1111/j.1755-148X.2012.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan AM, Palanisamy N. Chromosomal aberrations in solid tumors. Progress in molecular biology and translational science. 2010;95:55–94. doi: 10.1016/B978-0-12-385071-3.00004-6. [DOI] [PubMed] [Google Scholar]
- Consortium T.1.G.P. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nature genetics. 2009;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, et al. Open source clustering software. Bioinformatics (Oxford, England) 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Research. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton GM, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nature. 2013 doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R, et al. PLX4032, a selective BRAFV600E kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAFWT melanoma cells. Pigment Cell \$0 Melanoma Research. 2010;23(2):190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanrahan AJ, et al. Genomic complexity and AKT dependence in serous ovarian cancer. Cancer discovery. 2012;2(1):56–67. doi: 10.1158/2159-8290.CD-11-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010 doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman DM, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res. 2006;12(13):3908–3914. doi: 10.1158/1078-0432.CCR-06-0462. [DOI] [PubMed] [Google Scholar]
- Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27(41):5477–5485. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- Kim KB, et al. Phase II trial of imatinib mesylate in patients with metastatic melanoma. Br J Cancer. 2008;99(5):734–740. doi: 10.1038/sj.bjc.6604482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics (Oxford, England) 2009;25(17):2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauthammer M, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nature genetics. 2012;44(9):1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics (Oxford, England) 2012;28(10):1307–1313. doi: 10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, et al. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer cell. 2012;21(6):751–764. doi: 10.1016/j.ccr.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. The $1,000 genome, the $100,000 analysis? Genome Medicine. 2010;2(11):84. doi: 10.1186/gm205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. Available at: http://genome.cshlp.org/content/early/2010/08/04/gr.107524.110.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson K, et al. Tumor genetic analysis of patients with metastatic melanoma treated with the BRAF inhibitor GSK2118436. ASCO Annual Meeting; Chicago, IL. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollau P, Wagener C. Methods for detection of point mutations: performance and quality assessment. IFCC Scientific Division, Committee on Molecular Biology Techniques. Clinical chemistry. 1997;43(7):1114–1128. [PubMed] [Google Scholar]
- Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nature. 2009 doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathirapongsasuti JF, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics (Oxford, England) 2011;27(19):2648–2654. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Research. 2007;67(4):1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- Stahl JM, et al. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Research. 2003 [PubMed] [Google Scholar]
- Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nature genetics. 2012;44(2):165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stones CJ, et al. Comparison of responses of human melanoma cell lines to MEK and BRAF inhibitors. Frontiers in genetics. 2013;4:66. doi: 10.3389/fgene.2013.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermann B, et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS ONE. 2010;5(12):e15661. doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunzer K, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31(14):1767–1774. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemeyer WR, et al. Pattern of retinoblastoma pathway inactivation dictates response to CDK4/6 inhibition in GBM. Proceedings of the National Academy of Sciences. 2010;107(25):11501–11506. doi: 10.1073/pnas.1001613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 – Mel537 Expression Coclusters with BRAF/NRAS Mutant Tumors: Unsupervised clustering of the top 10% most dynamic genes quantified by gene expression microarray in melanoma cell lines. Cell line clustering dendrogram appears at top right, with oncogene status and tumor suppressor status indicated. Inset are selected genes demonstrating coclustering of microphthlamia (MITF) high (top), CD200 low (middle), and keratin high (bottom) expression phenotypes.
Supplementary Figure 2 – Custom Barcoding Design: Diagram of custom designed reads, with Illumina PE adapter sequence followed by barcode sequencing, along with resulting reads, with barcode located at the beginning of paired-end sequencing reads.
Supplementary Figure 3 – Schema for Multiplexed Sequencing: (A) Sequencing of melanoma cell lines and tumors utilized a pre-capture pooling of libraries followed by in solution capture and sequencing. (B) Deconvoluted cell line data were mapped and analyzed independently using the Genome Analysis Toolkit followed by filtering, annotation with the SIFT tool, and exclusion of SNPs based on 1000 genomes data.
Genes targeted for capture for the three sample types analyzed.
Nonsynonymous variants not in 1,000 genomes annotations or dbSNP tabulated by cell line. Mutations in NRAS appearing in dbSNP are included due to known relevance in melanoma.
Relevant sequencing data statistics for each sample, including number of reads, number of reads mapping, percent duplicates and percentage of reads within the target region.