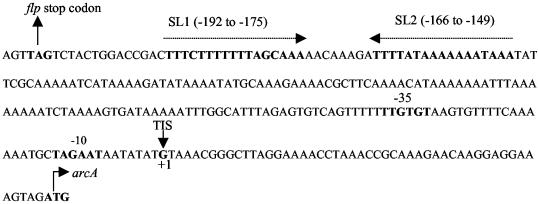Abstract
In Streptococcus gordonii DL1, inactivation of the ccpA gene and a gene encoding an Fnr-like protein (Flp) demonstrated that CcpA was essential for carbohydrate catabolite repression and that Flp was required for optimal expression and anaerobic induction of the arginine deiminase system.
The arginine deiminase system (ADS) (encoded by the arc operon) is a multienzyme pathway that catalyzes the conversion of arginine to ornithine, ammonia, and CO2, with the concomitant production of ATP. The ADS is widely distributed among prokaryotes, and the primary structures of the enzymes involved in the AD pathway have been reasonably conserved throughout evolution. However, the genetic regulation characteristics of ADS differ among organisms. For instance, both Pseudomonas aeruginosa (17) and Bacillus licheniformis (26) utilize the ADS exclusively under anaerobic conditions through a transcriptional regulator belonging to the Anr (for “anaerobic regulation of arginine catabolism and nitrate reduction”)/Fnr (for “fumarate nitrate reductase”) family. Induction by Anr/Fnr can be further enhanced in the presence of arginine by ArgR, the transcriptional regulator. In some lactic acid bacteria, such as Streptococcus sanguis (8) and Lactobacillus sakei (31), the expression of the operon is under the control of carbon catabolite repression (CCR) and is inducible by arginine; however, the mechanisms for CCR and arginine induction are not fully understood. Furthermore, in some oral bacteria (such as S. sanguis NCTC10904 and Streptococcus rattus FA-1) the ADS can be repressed by aeration through an as-yet-undefined pathway (2). In the oral cavity, the ADS is one of two primary pathways for ammonia generation. Ammonia produced by the ADS can neutralize acids generated by bacterial glycolysis, thereby increasing dental biofilm pH. The elevated pH induced by arginine catabolism is thought to be important in inhibiting the development of tooth decay and in modulating the composition of plaque (1, 3). Among the comparatively few ADS-positive species that colonize the mouth, Streptococcus gordonii is one of the more abundant organisms in tooth biofilms and makes up a significant portion of healthy human supragingival dental plaque (3, 19). Thus, the ADS of S. gordonii may play a critical role in the prevention of dental caries.
Dong et al. previously reported that the arc operon in S. gordonii is arranged as arcABCDT. In addition to the genes encoding enzymes involved in the AD pathway, arcR, a gene encoding an activator for the arc operon, is located 3′ to arcT and transcribed in the opposite orientation. We also showed that the expression of the S. gordonii arc operon is inducible by arginine and subject to CCR. Arginine induction is mediated in large part by the activity of ArcR, but the molecular basis for CCR of the arc operon has not been defined (7). In most gram-positive bacteria, binding of CcpA (for “catabolite control protein A”) (13) to a palindromic sequence, the carbon catabolite response element, results in repression of CCR-sensitive operons (28). However, other pathways such as CcpB (4), CcpC (15), and phosphotransferase system-dependent CCR (exerted through phosphorylation of specific transcriptional regulators) are known to be involved in CCR of some gram-positive bacteria (10, 20, 27). To determine whether CcpA is the primary protein exerting CCR of arc operon expression in S. gordonii, an apparent CcpA homolog, which was identified from the partial S. gordonii genome database (The Institute for Genomic Research website [http://www.tigr.org]), was amplified by PCR using primers ccpA-S and ccpA-AS (Table 1), with S. gordonii DL1 chromosomal DNA as the template. An erythromycin resistance cassette was then cloned into the unique EcoRV site within the ccpA gene, and the resulting plasmid was used to transform S. gordonii to generate a CcpA-deficient derivative via double-crossover recombination. The arcA promoter (parcA) (obtained as a 337-bp fragment upstream of the arcA start codon by PCR with primers parcA5′ and parcA3′) (Table 1) was directly fused to a chloramphenicol acetyltransferase (CAT) gene (cat) in plasmid pMC286, which was constructed by insertion of the promoterless cat gene from pC194 (14) into pGEM-Zf3(+) at BamHI and SphI sites. The fusion was constructed such that translation was driven from the arcA ribosome binding site. The transcriptional fusion was then released and cloned into pMJB8 (5), an S. gordonii integration vector that allows insertion of foreign DNA at the gtfG (glucosyltransferase gene) locus with the selection of a kanamycin-resistant phenotype. The construct was then used to transform wild-type and CcpA-deficient strains of S. gordonii. CAT activity (optical density at 600 nm ≅ 0.6 to 0.7) was measured in mid-exponential-phase cells grown in TY medium (29) with 10 mM glucose (a repressing sugar) or 10 mM galactose (a nonrepressing sugar) by the method of Shaw (24). In the wild-type background, the level of CAT activity in cells grown in TY medium with galactose was 17-fold higher than that seen with cells grown in TY medium with glucose (Table 2). In the CcpA-deficient strain, CCR was relieved and there was no significant difference in CAT activity between cells grown in galactose or glucose. These results indicated that (excluding other possible pathways such as those of CcpB, CcpC, and phosphotransferase system-dependent CCR) CcpA is the primary regulator for CCR of the arc operon in S. gordonii.
TABLE 1.
Primers used in this study
| Primer | Sequencea | Application |
|---|---|---|
| flp5′ | 5′-CCAGTTTTATATGCCGTA-3′ | flp amplification |
| flp3′ | 5′-GTCCAGTAGACTAACTTTCT -3′ | flp amplification |
| flp SmaI-S | 5′-TCTTTTTTTCTGGAGACCCG GGTGATCGCCTTTTTCTTC-3′ | Adding SmaI site to flp |
| flp SmaI- AS | 5′-GAAGAAAAAGGCGATCACC CGGGTCTCCAGAAAAAAAGA -3′ | Adding SmaI site to flp |
| ccpA-S | 5′-ACAGACGATACAGTAACCAT -3′ | ccpA amplification |
| ccpA-AS | 5′-TAGTCAACATACGCATAC-3′ | ccpA amplification |
| parcA5′ | 5′-AAAAGGTTGAGAGAAGAGCT CCGTATCAGCTATGAG-3′ | parcA amplification |
| parcA3′ | 5′-GGATGTGTAGACATGGATCCT CCTCCTTGTTCTTTG-3′ | parcA amplification |
The introduced restriction recognition site within flp is indicated in boldface characters.
TABLE 2.
CAT-specific activities in wild-type and CcpA-deficient strains of S. gordonii/parcA-cat and wild-type and Flp-deficient strains of S. gordonii/parcA-cat under aerobic and anaerobic growth conditions
| S. gordonii strain | CAT-specific activity under the indicated conditionsa
|
|||
|---|---|---|---|---|
| With galactoseb | With glucoseb | Anaerobicc | Aerobicc | |
| WTd | 1,024 ± 17 | 60 ± 6 | 794 ± 28 | 159 ± 21 |
| CcpA deficient | 907 ± 15 | 882 ± 35 | ||
| Flp deficient | 70 ± 14 | 37 ± 6 | ||
The values presented are the means and standard deviations of three independent experiments and are expressed as nanomoles of chloramphenicol acetylated per minute per milligram of total protein. All CAT assays were done in triplicate.
Cells were grown in 5% CO2 and 95% air and TY medium with the carbohydrate as indicated.
All cultures were grown in TY medium-10 mM galactose-50 mM arginine under anaerobic conditions in a GasPak jar or under aeration conditions with shaking at 300 rpm (see text for details).
WT, wild type.
After the incomplete S. gordonii genome was searched, one 684-bp open reading frame (ORF) (located 263 bp 5′ to arcA) was predicted to code for a homologue of known proteins of the Crp/Fnr family, which has been proven to be responsible for anaerobic versus aerobic gene regulation in many gram-negative bacteria (16, 17, 30) and in some gram-positive bacteria, including Bacillus subtilis and B. licheniformis (6, 18). The proteins with the most similarity to this ORF (Streptococcus pyogenes SPY1548 [43% identical residues], B. licheniformis ArcR [37% identical residues], P. aeruginosa Anr [23% identical residues], L. sakei ArcR [34% identical residues], and Enterococcus faecalis ArcE [35% identical residues]) were putative members of the Crp/Fnr family that were either linked to, or already shown to be involved in, arc regulation. Thus, we designated this ORF flp (for “Fnr-like protein”) in S. gordonii. A multiple-sequence alignment of Flp and selected representatives of the Crp/Fnr family showed that there are conserved residues throughout the proteins (Fig. 1). Although the overall level of similarity between Flp and each of the aligned proteins is not high, the predicted helix-turn-helix motif located in the C-terminal part of the proteins is more conserved. Of note, conserved residues (such as arginine and glutamic acid) (32) shown to be involved in recognition of the DNA binding site by Crp were found in S. gordonii Flp at positions 212 and 213 (Fig. 1). However, Flp in S. gordonii has only two cysteine residues instead of the four conserved cysteine residues that are usually found in other Fnr homologues (12, 21, 25). To date, only two other known Fnr-like proteins containing two cysteine residues that regulate the redox responses in Lactococcus lactis and Lactobacillus casei have been identified (11, 23).
FIG. 1.
Alignment of the deduced amino acid sequences of Flp and representative members of the Crp/Fnr family. The identical residues are indicated with boldface characters and gray-shaded boxes; the conserved residues Arg-212 and Glu-213 (potentially involved in protein-DNA interactions) are indicated by asterisks. This alignment was performed with ClustalW alignment and MacVector software.
It was found that the flp gene was preceded by a putative Shine-Dalgarno sequence and began with an ATG codon. In addition, a putative rho-independent terminator (5′-TTTCTTTTTTTAGCAAAAACAAAGATTTTATAAAAAAATAAA-3′ [the bases forming the stem-loop are underlined]) was found between flp and arcA (Fig. 2), in consistency with our finding that arc gene expression is driven from its own promoter (7). We also examined the two ORFs immediately 5′ to the flp gene in S. gordonii; these ORFs were revealed in BLAST searches to be conserved hypothetical proteins.
FIG. 2.
Nucleotide sequence and relevant features of the 5′ region of arcA. The start codon of arcA and the stop codon of flp are indicated with solid arrows. The promoter that directs the transcription of arcA (7) is indicated with boldface characters. The two inverted repeats with the potential to function as terminators, SL1 (positions −192 to −175) and SL2 (positions −166 to −149), are between the flp and arcA genes and are indicated by boldface characters and dotted arrows. TIS, transcription initiation site.
To determine the function of Flp in regulation of arc operon expression, flp was amplified by recombinant PCRs with primers (Table 1) that were designed on the basis of the flp sequence identified from the partial sequence in the S. gordonii genome database. The introduction of a unique SmaI site 141 bp 3′ to the start codon of flp allowed the subsequent cloning of a spectinomycin resistance cassette into the flp gene, and the resulting plasmid was used to transform S. gordonii to generate a Flp-deficient derivative via double-crossover recombination. The parcA-cat fusion was then used to transform the Flp-deficient mutant and wild-type S. gordonii. Early-log-phase cells (optical density at 600 nm ≅ 0.25 to 0.35) grown in TY medium containing 10 mM galactose and 50 mM arginine under anaerobic conditions in a GasPak jar (BBL), or with aeration by shaking at 300 rpm with 50 ml of culture in a 250-ml flask (2), were harvested and used to measure CAT activity. Inactivation of flp resulted in 11- and 4.3-fold decreases in CAT activity compared to the results seen with the wild-type strain grown under anaerobic and aerobic growth conditions (Table 2), respectively, which suggested that Flp was an activator of arc operon expression in S. gordonii. In the Flp-deficient strain, furthermore, CAT activity in cells grown under anaerobic conditions was less than twofold higher than that seen in aerobically grown cells compared to a fivefold difference in the results seen with the wild-type strain, which indicated Flp might be involved in anaerobic induction of the ADS in S. gordonii. So far, there is no evidence to indicate that Flp is a global regulatory protein, as it is in Escherichia coli and P. aeruginosa (9, 22, 30), but the possibility cannot be excluded that Flp regulates genes other than the arc operon.
Acknowledgments
This work was supported by Public Health Service grant DE10362 from the National Institute of Dental and Craniofacial Research.
REFERENCES
- 1.Bowden, G. H., and I. R. Hamilton. 1987. Environmental pH as a factor in the competition between strains of the oral streptococci Streptococcus mutans, S. sanguis, and “S. mitior” growing in continuous culture. Can. J. Microbiol. 33:824-827. [DOI] [PubMed] [Google Scholar]
- 2.Burne, R. A., D. T. Parsons, and R. E. Marquis. 1991. Environmental variables affecting arginine deiminase expression in oral streptococci, p. 276-280. In P. P. Dunny, G. M. Cleary, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci, and enterococci. American Society for Microbiology, Washington, D.C.
- 3.Burne, R. A., and R. E. Marquis. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1-6. [DOI] [PubMed] [Google Scholar]
- 4.Chauvaux, S., I. T. Paulsen, and M. H. Saier, Jr. 1998. CcpB, a novel transcription factor implicated in catabolite repression in Bacillus subtilis. J. Bacteriol. 180:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, Y. Y., M. J. Betzenhauser, and R. A. Burne. 2002. cis-acting elements that regulate the low-pH-inducible urease operon of Streptococcus salivarius. Microbiology 148:3599-3608. [DOI] [PubMed] [Google Scholar]
- 6.Cruz Ramos, H., L. Boursier, I. Moszer, F. Kunst, A. Danchin, and P. Glaser. 1995. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 14:5984-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong, Y., Y.-Y. M. Chen, J. A. Snyder, and R. A. Burne. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68:5549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferro, K. J., G. R. Bender, and R. E. Marquis. 1983. Coordinately repressible arginine deiminase system in Streptococcus sanguis. Curr. Microbiol. 9:145-150. [Google Scholar]
- 9.Galimand, M., M. Gamper, A. Zimmermann, and D. Haas. 1991. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 173:1598-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosalbes, M. J., V. Monedero, and G. Perez-Martinez. 1999. Elements involved in catabolite repression and substrate induction of the lactose operon in Lactobacillus casei. J. Bacteriol. 181:3928-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gostick, D. O., J. Green, A. S. Irvine, M. J. Gasson, and J. R. Guest. 1998. A novel regulatory switch mediated by the FNR-like protein of Lactobacillus casei. Microbiology 144:705-717. [DOI] [PubMed] [Google Scholar]
- 12.Green, J., A. D. Sharrocks, B. Green, M. Geisow, and J. R. Guest. 1993. Properties of FNR proteins substituted at each of the five cysteine residues. Mol. Microbiol. 8:61-68. [DOI] [PubMed] [Google Scholar]
- 13.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 14.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jourlin-Castelli, C., N. Mani, M. M. Nakano, and A. L. Sonenshein. 2000. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J. Mol. Biol. 295:865-878. [DOI] [PubMed] [Google Scholar]
- 16.Lazazzera, B. A., H. Beinert, N. Khoroshilova, M. C. Kennedy, and P. J. Kiley. 1996. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271:2762-2768. [DOI] [PubMed] [Google Scholar]
- 17.Lu, C. D., H. Winteler, A. Abdelal, and D. Haas. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maghnouj, A., A. A. Abu-Bakr, S. Baumberg, V. Stalon, and C. Vander Wauven. 2000. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 191:227-234. [DOI] [PubMed] [Google Scholar]
- 19.Marquis, R. E., G. R. Bender, D. R. Murray, and A. Wong. 1987. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin-Verstraete, I., J. Stulke, A. Klier, and G. Rapoport. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 177:6919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melville, S. B., and R. P. Gunsalus. 1990. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J. Biol. Chem. 265:18733-18736. [PubMed] [Google Scholar]
- 22.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 23.Scott, C., J. R. Guest, and J. Green. 2000. Characterization of the Lactococcus lactis transcription factor FlpA and demonstration of an in vitro switch. Mol. Microbiol. 35:1383-1393. [DOI] [PubMed] [Google Scholar]
- 24.Shaw, W. V. 1979. Chloramphenicol acetyltransferase activity from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 25.Spiro, S., and J. R. Guest. 1988. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol. Microbiol. 2:701-707. [DOI] [PubMed] [Google Scholar]
- 26.Stockley, P. G., A. J. Baron, C. M. Wild, I. D. Parsons, C. M. Miller, C. A. Holtham, and S. Baumberg. 1998. Dissecting the molecular details of prokaryotic transcriptional control by surface plasmon resonance: the methionine and arginine repressor proteins. Biosens. Bioelectron. 13:637-650. [DOI] [PubMed] [Google Scholar]
- 27.Tobisch, S., J. Stulke, and M. Hecker. 1999. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J. Bacteriol. 181:4995-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weickert, M. J., and G. H. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:6238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wexler, D. L., M. C. Hudson, and R. A. Burne. 1993. Streptococcus mutans fructosyltransferase (ftf) and glucosyltransferase (gtfBC) operon fusion strains in continuous culture. Infect. Immun. 61:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye, R. W., D. Haas, J. O. Ka, V. Krishnapillai, A. Zimmermann, C. Baird, and J. M. Tiedje. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 177:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zúñiga, M., M. Champomier-Verges, M. Zagorec, and G. Pérez-Martínez. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zúñiga, M., M. del Carmen Miralles, and G. Pérez-Martínez. 2002. The product of arcR, the sixth gene of the arc operon of Lactobacillus sakei, is essential for expression of the arginine deiminase pathway. Appl. Environ. Microbiol. 68:6051-6058. [DOI] [PMC free article] [PubMed] [Google Scholar]