Abstract
Cfa1 was overproduced in Escherichia coli and Pseudomonas syringae, and the degree of 4′-phosphopantetheinylation was determined. The malonyl-coenzyme A:acyl carrier protein transacylase (FabD) of P. syringae was overproduced and shown to catalyze malonylation of Cfa1, suggesting that FabD plays a role in coronatine biosynthesis. Highly purified Cfa1 did not exhibit self-malonylation activity.
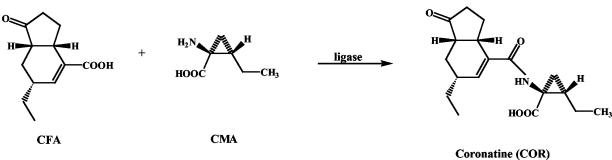
Coronatine (COR) is a non-host-specific chlorosis-inducing phytotoxin produced by several Pseudomonas syringae pathovars (1). COR consists of two moieties joined by an amide linkage: the polyketide coronafacic acid (CFA) and coronamic acid (Fig. 1) (1). The CFA polyketide synthase (PKS) genes include two genes (cfa6 and cfa7) that encode modular type I PKS proteins as well as three genes (cfa1, cfa2, and cfa3) that encode monofunctional proteins similar to the acyl carrier protein (ACP), dehydratase, and β-ketoacyl:acyl carrier protein synthase, respectively, of type II fatty acid synthases (FASs) and PKSs (25, 27). The genes coding for the CFA portion of COR are located on a single long transcript driven by a promoter upstream of the cfl gene that is regulated by a three-component regulatory system present in the COR gene cluster (17, 24).
FIG. 1.
Structures of COR, CFA, and coronamic acid (CMA) and presumed formation of COR by action of a ligase.
A biosynthetic hypothesis for CFA formation was derived from analysis of the CFA PKS (12, 27) and the isolation of a potential biosynthetic intermediate, 2-[1-oxo-2-cyclopenten-2-ylmethyl]-butanoic acid (19, 23, 35). This hypothesis postulates that Cfa1 functions as the carrier in the assembly of a cyclic carboxylic acid that is subsequently transferred from Cfa1 to the ACP domain of Cfa6 via the loading didomain. The important role assigned to Cfa1 and the lack of information concerning PKS ACPs of Pseudomonas prompted us to investigate the properties of Cfa1.
Cloning of cfa1, fabD, and acp from P. syringae.
To clone cfa1 into the FLAG vectors pFLAG-CTC (Sigma) and pSFFLAG-CTC (4), cfa1 was amplified from pSAY10 (38) using the following primers: 5′ GCCAAGCTTGTGAACGAGATC (HindIII site in bold) and 5′ GCCGAATTCCTTGGCCTGCAG (EcoRI site in bold). The PCR product was ligated into the HindIII-EcoRI-digested vectors, and the ligation mixtures were used to transform Escherichia coli DH5α or DH10B, resulting in pFLAG-Cfa1 and pSFFLAG-Cfa1, respectively. For overproduction of Cfa1 in P. syringae pv. glycinea PG4180, pFLAG-Cfa1 was linearized with BamHI and ligated into BamHI-digested pRK415 (13), resulting in pRKFLAG-Cfa1.
Primers were designed to amplify the fabD gene from P. syringae FF5 (9) based upon the sequence of fabD in Pseudomonas aeruginosa (http://Pseudomonas.bit.uq.edu.au/) and P. syringae pv. tomato DC3000 (http://www.tigr.org). The forward primer for fabD amplification was 5′ CCCAAGCTTATGTCTGCATCCCTCGCATTC (HindIII site in bold), and the reverse primer containing a BglII site (bold) was 5′ GTTAGATCTGGCCAGTGCCGCGCGTG. P. syringae FF5 genomic DNA was used as a template for PCR. The PCR product was digested with HindIII and BglII and ligated into HindIII-BglII-digested pSFFLAG-CTC.
The primers designed to amplify the FAS ACP gene from P. syringae FF5 were based on the sequence of the FAS ACP gene in P. syringae pv. tomato DC3000. The forward primer contained an XhoI site (bold): GTCCTCGAGATGAGCACCATCGAAGAG. The reverse primer contained a BglII site (bold) to allow insertion into the pSFFLAG-CTC vector with removal of the stop codon: GTAGATCTATAAGCCTGATGGCTG. The PCR product was digested with BglII and XhoI and ligated into pSFFLAG-CTC digested with the same enzymes.
The deduced translation products of the cloned P. syringae FF5 fabD and acp genes exhibited high similarities to the corresponding genes from the COR producer P. syringae pv. tomato DC3000 and from P. aeruginosa as well as lower similarities to homologues found in E. coli and Streptomyces coelicolor (Table 1). P. syringae FF5 was selected as the source of the fabD and FAS acp genes, since unpublished studies have shown that introduction of the entire COR gene cluster into this strain leads to COR production (Y. Zhao, V. Rangaswamy, and C. L. Bender, unpublished data). P. syringae FF5 therefore possesses all the ancillary enzymes required for COR production. In addition, P. syringae FF5 provides a convenient host strain to overproduce proteins from the COR pathway (4).
TABLE 1.
Amino acid similarity of P. syringae FF5 FabD and FAS ACP to some related proteins
| Organism | % Identity to P. syringae FabD | Protein identification no. | % Identity to P. syringae FAS ACP | Protein identification no. |
|---|---|---|---|---|
| P. syringae pv. tomato | 97 | NP_793606 | 97 | P80923 |
| P. aeruginosa | 80 | NP_251658.1 | 88 | NP_251656.1 |
| E. coli | 58 | NP_287226.1 | 87 | NP_287228.1 |
| S. coelicolor A3(2) | 37 | CAB62719.1 | 41 | CAB62721.1 |
Overproduction of Cfa1-FLAG in E. coli.
A starter culture of E. coli DH5α containing pFLAG-Cfa1 in terrific broth (TB) plus ampicillin (100 μg/ml) was used to inoculate 100 ml of the same medium, and cells were grown at 37°C to an optical density at 600 nm (OD600) of 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.5 mM, and the cells were incubated an additional 3 h before harvesting. The cells were lysed with 8 ml of CelLytic B-cell lysis agent (Sigma) containing lysozyme (10 mg/ml) and DNase I (0.02 mg/ml). The suspension was shaken gently for 15 min and then centrifuged at 10,000 × g for 15 min. The supernatant was applied to a column of M2 immunoaffinity resin (Sigma) (1 ml). The eluant was reapplied to the column three times, and the column was then washed with 36 ml of Tris-buffered saline (TBS) buffer (50 mM Tris-HCl [pH 7.4], 150 mM MgCl2). The C-terminal FLAG-tagged Cfa1 protein (Cfa1-FLAG) was eluted from the resin using FLAG peptide (100 μg/ml; TBS buffer) as described in the FLAG manual (Sigma).
Overproduction and purification of Cfa1, FabD, and ACP in P. syringae.
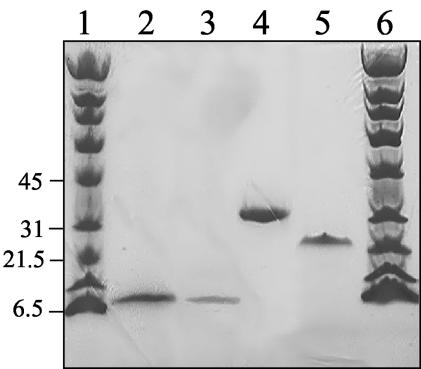
The pSFFLAG-Cfa1 construct was electroporated into P. syringae FF5 cells, and the pRKFLAG-Cfa1 plasmid was mobilized into P. syringae pv. glycinea PG4180 using the helper plasmid pRK2013 (2). An overnight culture (20 ml) of P. syringae FF5 containing pSFFLAG-Cfa1 in King’s medium B (KMB) (14) plus ampicillin (100 μg/ml) was used to inoculate 1 liter of KMB, which was then incubated at 28°C and 250 rpm. After 5 h, the incubation temperature was reduced to 18°C, and cells were grown for 16 h or to an OD600 of 0.5 to 1.0. The cells were then induced with IPTG (1 mM), incubated for 24 h at 18°C, and harvested by centrifugation (8,000 × g), and the pellets were stored at −80°C. P. syringae PG4180 containing pRKFLAG-Cfa1 was grown in 1 liter of TB plus tetracycline (25 μg/ml) at 18 or 28°C to an A600 of 0.5, induced with 1 mM IPTG, and incubated an additional 16 h at the initial incubation temperature (18 or 28°C). Cells were harvested as described for FF5, and pellets were stored at −80°C. Starter cultures of P. syringae transformants containing fabD and acp in pSFFLAG-CTC were grown overnight as described above, added to 1 liter of KMB with ampicillin, and grown at 28°C until the OD600 was 0.5 to 0.8. The cultures were then induced with IPTG (1 mM) and incubated for 16 h at 28°C. The cells were harvested by centrifugation and frozen at −80°C. Frozen pellets were thawed to room temperature and resuspended in a lysis buffer (3 ml per g of cells) containing Tris buffer (100 mM; pH 8), EDTA (150 mM), and lysozyme (0.32 mg/ml). The mixture was incubated for 30 min at 37°C. After incubation, a solution (0.3 ml per g of cells) containing NaCl (150 mM), MgCl2 (100 mM), CaCl2 (100 mM), and DNase I (0.2 mg/ml) was added, and the mixture was incubated for 15 min at room temperature with stirring and then centrifuged at 26,000 × g. Supernatants were applied three times to a column of M2 resin (1.5 ml), which was washed with TBS (three times, 20 ml). FLAG-tagged proteins were eluted with 4 ml of FLAG peptide (100 μg/ml) and concentrated to a volume of approximately 200 μl with an Amicon concentrator. Proteins were further purified on a Mono Q column equilibrated with 100 mM Na2HPO4, 2 mM EDTA, and 2 mM dithiothreitol. An NaCl gradient (0 to 500 mM) in the same buffer was used to elute the proteins. The purity of the eluted proteins was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the total protein concentrations were determined with the Advanced protein assay reagent (Cytoskeleton Inc.) using bovine serum albumin or gamma globulin as the standard. The overproduced proteins were analyzed by SDS-PAGE. The mass of Cfa1-FLAG was approximately 12 kDa, which is close to the predicted size of 11.5 kDa (Fig. 2, lanes 2 and 3). The Cfa1-FLAG purified from P. syringae FF5 was further purified on a Mono Q column, although no contaminating proteins were visible by SDS-PAGE (Fig. 2, lane 2). After purification, the mass of FabD-FLAG was found to be 34.2 kDa by SDS-PAGE (Fig. 2, lane 4) and 58.7 kDa by native PAGE (data not shown), suggesting that the native protein is a dimer, as reported for FabD from Streptomyces glaucescens (10). ACP-FLAG appeared at a mass of approximately 22 kDa by SDS-PAGE (Fig. 2, lane 5), a value which is significantly larger than the expected one (10.76 kDa). Similar behavior has been reported for the FAS ACP of P. aeruginosa (15) and is consistent with the fact that some ACPs run anomalously on SDS-PAGE due to their highly charged nature and low hydrophobic amino acid content (15, 21, 32). The predicted mass was obtained for ACP-FLAG when the protein was analyzed by mass spectrometry.
FIG. 2.
SDS-PAGE (8 to 25% gradient) of recombinant Cfa1 expressed in P. syringae FF5 at 18°C (lane 2), Cfa1 expressed in P. syringae pv. glycinea PG4180 at 18°C (lane 3), FabD expressed in P. syringae FF5 at 28°C (lane 4), and FAS ACP expressed in P. syringae FF5 at 28°C (lane 5). All proteins contain a C-terminal FLAG tag. Proteins were purified by affinity chromatography followed by anion exchange chromatography (Mono Q), except for lanes 2 and 3, which show Cfa1 purified only by affinity chromatography. The molecular masses of the standards (lanes 1 and 6) were 200, 116, 97.4, 66, 45, 31, 21.5, 14.4, and 6.5 kDa.
MALDI-TOF analysis of Cfa1 and ACP.
Cfa1-FLAG obtained from E. coli DH5α was analyzed by positive-ion matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometry to determine the degree to which the protein had been converted into its holo form by addition of a 4′-phosphopantetheine moiety. The protein exhibited a peak at mass of 11,516 Da for the apo protein and no peak corresponding to the holo protein (data not shown). The expected sizes for the apo and holo proteins are 11,516.3 and 11,855.3 Da, respectively. Cfa1-FLAG expressed in P. syringae FF5 exhibited a small peak at a mass of 11,511 Da, corresponding to the apo protein, and a larger peak at a mass of 11,854 Da, corresponding to the holo protein (data not shown). The ratio between the areas of the two peaks indicated that the ratio of apo to holo protein was ca. 5:95. Similarly, MALDI-TOF analysis of Cfa1-FLAG overproduced at 18°C in the COR producer P. syringae pv. glycinea PG4180 showed that about 90% of the protein was in the holo form (data not shown). However, when Cfa1-FLAG was overproduced in PG4180 at 28°C, the ratio of apo to holo Cfa1-FLAG was 2:1. The MALDI-TOF spectrum of the ACP-FLAG protein exhibited peaks for both the apo and holo forms, with only about 20% of the protein present in the holo form (data not shown). This might be the consequence of a 4′-phosphopantetheinyl transferase (PPTase) reaction rate that is slow relative to the rate of ACP-FLAG production. Since the primary purpose for ACP-FLAG production was to check the overproduced FabD for malonyltransferase activity, the factors affecting the ratio of apo to holo ACP-FLAG were not further investigated.
FabD malonyl-CoA transferase assay with Cfa1 and FAS ACP.
The malonyl-coenzyme A (CoA) transferase assay was carried out in a total volume of 50 μl, which contained 25 mM Tris (pH 7), 1 mM dithiothreitol, 10 μM FabD, and Cfa1-FLAG and ACP-FLAG concentrations of 10 to 100 μM. To start the assay, 50 μM [2-14C]malonyl-CoA (specific activity, 27.5 mCi/mmol) was added. The reaction mixture was incubated for 5 to 10 min at 25°C, and the reaction was stopped by the addition of 10% trichloroacetic acid (TCA) (400 μl) and bovine serum albumin (10 mg/ml; 10 μl). The pellet obtained after centrifugation was washed three times with 10% TCA. The final pellet was resuspended in Tris buffer (150 μl) and added to 10 ml of aqueous scintillation cocktail for radioactivity determination. In additional experiments, the malonyl-CoA concentration was varied from 10 to 250 μM and the concentration of Cfa1-FLAG was maintained at 20 μM. Inaccurate data were obtained at malonyl-CoA concentrations of less than 10 μM due to the low levels of radioactivity incorporated into protein. All kinetic parameters were measured in the linear range of the reaction for specific enzyme and substrate concentrations, and the experiments were carried out in triplicate. Incubations were also carried out in the absence of Cfa1 or ACP in order to monitor the ability of FabD to load malonyl-CoA. These reactions were treated in a similar manner to those described above.
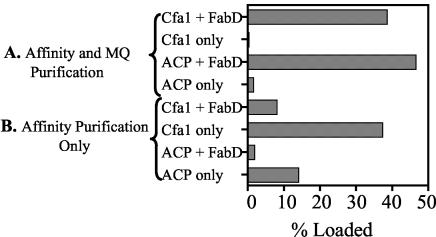
The overproduced Pseudomonas FabD-FLAG was able to malonylate both the ACP-FLAG and Cfa1-FLAG proteins. When the assays were performed with ACP-FLAG and Cfa1-FLAG that were purified only by affinity chromatography, both proteins also exhibited an apparent ability to “self-load” malonate (Fig. 3B). However, when Cfa1 and ACP were further purified by Mono Q chromatography, they no longer exhibited self-malonylation activity (Fig. 3A). Curiously, the addition of FabD-FLAG to the impure Cfa1-FLAG or ACP-FLAG appeared to decrease the rate of self-loading. This behavior can be explained if the Cfa1-FLAG and ACP-FLAG were contaminated with a small amount of native FabD that possesses a specific activity that is significantly greater than that of the overproduced proteins, whose specific activity could be reduced by the presence of the FLAG tag. An analysis of the reaction between FabD-FLAG and excess malonyl-CoA in the absence of Cfa1-FLAG or ACP-FLAG showed that FabD-FLAG was about 25% loaded with malonate after 10 min. Determination of kinetic parameters for the FabD-FLAG-catalyzed malonylation of Cfa1-FLAG revealed Km values of <10 and 22.05 ± 1.10 μM for malonyl-CoA and holo-Cfa1, respectively, and Kcat values of 0.03 ± 0.002 and 0.08 ± 0.004 min−1 for malonyl-CoA and holo-Cfa1, respectively. The only kinetic data available for comparison are those for malonylation of TcmM by FabD of S. glaucescens. The TcmM malonylation reaction was significantly faster than that observed for Cfa1 (8). Kinetic data for the FabD-catalyzed malonylation of other PKS ACPs will be needed before conclusions can be drawn from these differences.
FIG. 3.
Loading of [2-14C]malonate onto Cfa1 and FAS ACP from P. syringae FF5 by FabD. The concentrations of enzymes used were 25 μM Cfa1 or FAS ACP and 10 μM FabD. Reaction mixtures were incubated for 10 min at room temperature, and reactions were stopped by the addition of TCA. The precipitated protein was resuspended in Tris buffer and added to 10 ml of ScintiVerse scintillation cocktail for counting. The results obtained with Cfa1 and ACP further purified by Mono Q (MQ) chromatography (A) or not further purified (B) are shown. The percent loading values for Cfa1 and ACP are corrected to account for the amount of malonyl-CoA on FabD (where added) and for the percentage of holo protein as determined by MALDI-TOF. Assays were done in triplicate.
The type II ACPs are small, acidic proteins that are posttranslationally converted from the enzymatically inactive apo form of the protein to the active holo form by transfer of a 4′-phosphopantetheine moiety from CoA to a conserved serine residue (37). The enzymes that catalyze this transfer, known as PPTases, have been shown to constitute a superfamily (16). Three PPTases are known to be present in E. coli (31), while recent investigations have indicated that P. aeruginosa contains a single PPTase (PcpS) that modifies both fatty acid ACPs and the peptidyl carrier protein domain associated with nonribosomal peptide synthetases (7). In previous investigations where type II ACPs from Streptomyces were produced in E. coli, the apo ACP was the predominant form of the protein observed (5, 6, 32). Interestingly, some FAS ACPs from actinomycetes are produced in E. coli largely in the holo form (29, 30). When Cfa1 was overproduced in E. coli in the present study, the protein was recovered primarily in its apo form. However, when Cfa1 was overproduced in P. syringae FF5 at 18°C, the protein was recovered predominantly in the holo form. Similar results were obtained when Cfa1 was overproduced in P. syringae PG4180 at 18°C, while overproduction at 28°C in the latter strain yielded protein that was about a 2:1 mixture of apo to holo protein. The reasons for the decrease in the amount of holo Cfa1 at the higher temperature are unclear. However, several studies suggest that COR production is thermoregulated at both the transcriptional and posttranslational levels (3, 33). Collectively, these results suggest that a suitable PPTase for Cfa1 exists in P. syringae, whereas Cfa1 is a poor substrate for the E. coli PPTases. Thus, E. coli may not be a suitable host for the heterologous expression of enzymatically active Pseudomonas PKSs, unless a PPTase with broad specificity (7, 20, 31) is simultaneously coexpressed. Our results are consistent with reports that the peptidyl carrier protein from the pyoluteorin pathway of Pseudomonas fluorescens Pf-5 and the thiolation domains of nonribosomal peptide synthetase proteins from the pyochelin pathway of P. aeruginosa are not posttranslationally modified when these proteins are overproduced in E. coli (26, 36).
Type II PKSs generally lack a malonyl CoA:ACP transacylase to catalyze the transfer of a malonyl extender unit onto the PKS ACP (8), although the aromatic polyketide auricin is an exception to this generalization (22). For this reason, the potential role of the FAS malonyl CoA:ACP transacylase (FabD) in the biosynthesis of type II polyketides has been examined by several research groups. For example, the FAS FabDs of S. glaucescens and S. coelicolor can catalyze the transfer of a malonyl group to the ACP components of the tetracenomycin and actinorhodin PKSs, respectively (8, 28, 34, 39). In our experiments, the P. syringae FabD catalyzed the malonylation of Cfa1, which provides evidence that FabD can function in CFA biosynthesis. Some controversy concerning the role of FabD in type II polyketide biosynthesis has arisen because many PKS ACPs appear to self-malonylate (11, 18, 39). This dispute was recently resolved for TcmM, an ACP in S. glaucescens, by a rigorous set of experiments showing that the self-malonylation is an artifact of the expression and purification protocols (8). In the case of Cfa1, the affinity-purified protein isolated from E. coli also exhibited significant self-malonylation activity (Fig. 3B), even though the appearance of the protein on SDS-PAGE suggested a high degree of purity (Fig. 2). Additional purification of the affinity-purified Cfa1 by anion-exchange chromatography eliminated the self-malonylation activity (Fig. 3A), which is consistent with results obtained for TcmM. Our study thus provides additional evidence that the self-malonylation of ACPs is an artifact and further supports a role for FabD in CFA biosynthesis.
Nucleotide sequence accession number.
The nucleotide sequences for fabD and acp of P. syringae FF5 have been deposited in the GenBank database under accession numbers AY391840 and AY391841.
Acknowledgments
We acknowledge support from the National Science Foundation (MCB 9807774 to R.P. and C.L.B. and IBN-0130693 to C.L.B.), the Oklahoma Center for the Advancement of Science and Technology (AR031-005 to C.L.B), and The Robert A. Welch Foundation (C-0729 to R.P.).
Preliminary sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of P. syringae pv. tomato DC3000 at The Institute for Genomic Research was accomplished with support from the National Science Foundation Plant Genome Program.
REFERENCES
- 1.Bender, C. L., F. Alarcon-Chaidez, and D. C. Gross. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, C. L., S. A. Young, and R. E. Mitchell. 1991. Conservation of plasmid DNA sequences in coronatine-producing pathovars of Pseudomonas syringae. Appl. Environ. Microbiol. 57:993-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budde, I. P., B. H. Rohde, C. L. Bender, and M. S. Ullrich. 1998. Growth phase and temperature influence promoter activity, transcript abundance, and protein stability during biosynthesis of the Pseudomonas syringae phytotoxin coronatine. J. Bacteriol. 180:1360-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Couch, R., H. Seidle, and R. J. Parry. 2002. Construction of expression vectors to produce affinity-tagged proteins in Pseudomonas. BioTechniques 32:1230-1236. [DOI] [PubMed] [Google Scholar]
- 5.Cox, R. J., T. S. Hitchman, K. J. Byrom, I. S. C. Findlow, J. A. Tanner, J. Crosby, and T. J. Simpson. 1997. Post-translational modification of heterologously expressed Streptomyces type II polyketide synthase acyl carrier proteins. FEBS Lett. 405:267-272. [DOI] [PubMed] [Google Scholar]
- 6.Crosby, J., D. H. Sherman, M. J. Bibb, W. P. Revill, D. A. Hopwood, and T. J. Simpson. 1995. Polyketide synthase acyl carrier proteins from Streptomyces: expression in Escherichia coli, purification and partial characterisation. Biochim. Biophys. Acta 1251:32-42. [DOI] [PubMed] [Google Scholar]
- 7.Finking, R., J. Solsbacher, D. Konz, M. Schobert, A. Schafer, D. Jahn, and M. A. Marahiel. 2002. Characterization of a new type of phosphopantetheinyl transferase for fatty acid and siderophore synthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277:50293-50302. [DOI] [PubMed] [Google Scholar]
- 8.Florova, G., G. Kazanina, and K. A. Reynolds. 2002. Enzymes involved in fatty acid and polyketide biosynthesis in Streptomyces glaucescens: role of FabH and FabD and their acyl carrier protein specificity. Biochemistry 41:10462-10471. [DOI] [PubMed] [Google Scholar]
- 9.Garde, S., and C. L. Bender. 1991. DNA probes for detection of copper resistance genes in Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 57:2435-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han, L., S. Lobo, and K. A. Reynolds. 1998. Characterization of beta-ketoacyl-acyl carrier protein synthase III from Streptomyces glaucescens and its role in initiation of fatty acid biosynthesis. J. Bacteriol. 180:4481-4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hitchman, T. S., J. Crosby, K. J. Byrom, R. J. Cox, and T. J. Simpson. 1998. Catalytic self-acylation of type II polyketide synthase acyl carrier proteins. Chem. Biol. 5:35-47. [DOI] [PubMed] [Google Scholar]
- 12.Jiralerspong, S., V. Rangaswamy, C. L. Bender, and R. J. Parry. 2001. Analysis of the enzymatic domains in the modular portion of the coronafacic acid polyketide synthase. Gene 270:191-200. [DOI] [PubMed] [Google Scholar]
- 13.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 14.King, E. O., M. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescein. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 15.Kutchma, A. J., T. T. Hoang, and H. P. Schweizer. 1999. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD). J. Bacteriol. 181:5498-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambalot, R. H., A. M. Gehring, R. S. Flugel, P. Zuber, M. LaCelle, M. A. Marahiel, R. Reid, C. Khosla, and C. T. Walsh. 1996. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem. Biol. 3:923-936. [DOI] [PubMed] [Google Scholar]
- 17.Liyanage, H., D. A. Palmer, M. Ullrich, and C. L. Bender. 1995. Characterization and transcriptional analysis of the gene cluster for coronafacic acid, the polyketide component of the phytotoxin coronatine. Appl. Environ. Microbiol. 61:3843-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matharu, A. L., R. J. Cox, J. Crosby, K. J. Byrom, and T. J. Simpson. 1998. MCAT is not required for in vitro polyketide synthesis in a minimal actinorhodin polyketide synthase from Streptomyces coelicolor. Chem. Biol. 5:699-711. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell, R. E., H. Young, and M. J. Liddell. 1995. Isolation and structural characterization of 2-[1-oxo-2-cyclopenten-2-ylmethyl]-butanoic acid, a polyketide product of coronatine-producing Pseudomonas sp. Tetrahedron Lett. 36:3237-3240. [Google Scholar]
- 20.Mootz, H. D., R. Finking, and M. A. Marahiel. 2001. 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. J. Biol. Chem. 276:37289-37298. [DOI] [PubMed] [Google Scholar]
- 21.Morbidoni, H. R., D. de Mendoza, and J. E. Cronan, Jr. 1996. Bacillus subtilis acyl carrier protein is encoded in a cluster of lipid biosynthesis genes. J. Bacteriol. 178:4794-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novakova, R., J. Bistakova, D. Homerova, B. Rezuchova, and J. Kormanec. 2002. Cloning and characterization of a polyketide synthase gene cluster involved in biosynthesis of a proposed angucycline-like polyketide auricin in Streptomyces aureofaciens CCM 3239. Gene 297:197-208. [DOI] [PubMed] [Google Scholar]
- 23.Parry, R. J., S. Jiralerspong, S. V. Mhaskar, L. Alemany, and R. Willcott. 1996. Investigations of coronatine biosynthesis. Elucidation of the mode of incorporation of pyruvate into coronafacic acid. J. Am. Chem. Soc. 118:703-704. [Google Scholar]
- 24.Penaloza-Vazquez, A., and C. L. Bender. 1998. Characterization of CorR, a transcriptional activator which is required for biosynthesis of the phytotoxin coronatine. J. Bacteriol. 180:6252-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penfold, C. N., C. L. Bender, and J. G. Turner. 1996. Characterization of genes involved in biosynthesis of coronafacic acid, the polyketide component of the phytotoxin coronatine. Gene 183:167-173. [DOI] [PubMed] [Google Scholar]
- 26.Quadri, L. E., T. A. Keating, H. M. Patel, and C. T. Walsh. 1999. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4,2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry 38:14941-14954. [DOI] [PubMed] [Google Scholar]
- 27.Rangaswamy, V., S. Jiralerspong, R. Parry, and C. L. Bender. 1998. Biosynthesis of the Pseudomonas polyketide coronafacic acid requires monofunctional and multifunctional polyketide synthase proteins. Proc. Natl. Acad. Sci. USA 95:15469-15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revill, W. P., M. J. Bibb, and D. A. Hopwood. 1995. Purification of a malonyltransferase from Streptomyces coelicolor A3(2) and analysis of its genetic determinant. J. Bacteriol. 177:3946-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Revill, W. P., M. J. Bibb, and D. A. Hopwood. 1996. Relationships between fatty acid and polyketide synthases from Streptomyces coelicolor A3(2): characterization of the fatty acid synthase acyl carrier protein. J. Bacteriol. 178:5660-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revill, W. P., and P. R. Leadlay. 1991. Cloning, characterization, and high-level expression in Escherichia coli of the Saccharopolyspora erythraea gene encoding an acyl carrier protein potentially involved in fatty acid biosynthesis. J. Bacteriol. 173:4379-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez, C., L. Du, D. J. Edwards, M. D. Toney, and B. Shen. 2001. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC 15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem. Biol. 8:725-738. [DOI] [PubMed] [Google Scholar]
- 32.Shen, B., R. G. Summers, H. Gramajo, M. J. Bibb, and C. R. Hutchinson. 1992. Purification and characterization of the acyl carrier protein of the Streptomyces glaucescens tetracenomycin C polyketide synthase. J. Bacteriol. 174:3818-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smirnova, A. V., L. Wang, B. Rohde, I. Budde, H. Weingart, and M. S. Ullrich. 2002. Control of temperature-responsive synthesis of the phytotoxin coronatine in Pseudomonas syringae by the unconventional two-component system CorRPS. J. Mol. Microbiol. Biotechnol. 4:191-196. [PubMed] [Google Scholar]
- 34.Summers, R. G., A. Ali, B. Shen, W. A. Wessel, and C. R. Hutchinson. 1995. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry 34:9389-9402. [DOI] [PubMed] [Google Scholar]
- 35.Tao, T., and R. J. Parry. 2001. Determination by enantioselective synthesis of the absolute configuration of CPE, a potential intermediate in coronatine biosynthesis. Org. Lett. 3:3045-3047. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, M. G., M. D. Burkart, and C. T. Walsh. 2002. Conversion of L-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem. Biol. 9:171-184. [DOI] [PubMed] [Google Scholar]
- 37.Walsh, C. T., A. M. Gehring, P. H. Weinreb, L. E. Quadri, and R. S. Flugel. 1997. Post-translational modification of polyketide and nonribosomal peptide synthases. Curr. Opin. Chem. Biol. 1:309-315. [DOI] [PubMed] [Google Scholar]
- 38.Young, S. A., S. K. Park, C. Rodgers, R. E. Mitchell, and C. L. Bender. 1992. Physical and functional characterization of the gene cluster encoding the polyketide phytotoxin coronatine in Pseudomonas syringae pv. glycinea. J. Bacteriol. 174:1837-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, P., G. Florova, and K. A. Reynolds. 1999. Polyketide synthase acyl carrier protein (ACP) as a substrate and a catalyst for malonyl ACP biosynthesis. Chem. Biol. 6:577-584. [DOI] [PubMed] [Google Scholar]