Abstract
The MinC division inhibitor is required for accurate placement of the septal ring at the middle of the Escherichia coli cell. The N-terminal domain of MinC (ZMinC) interferes with FtsZ assembly, while the C-terminal domain (DMinC) mediates both dimerization and complex formation with either MinD or DicB. Binding to either of these activators greatly enhances the division-inhibitory activity of MinC in the cell. The MinD ATPase plays a crucial role in the rapid pole-to-pole oscillation of MinC that is proposed to force FtsZ ring formation to midcell. DicB is encoded by one of the cryptic prophages on the E. coli chromosome (Qin) and is normally not synthesized. Binding of MinD or DicB to DMinC produces complexes that have high affinities for one or more septal ring-associated targets. Here we show that the FtsZ-binding protein ZipA is required for both recruitment of the DMinC/DicB complex to FtsZ rings and the DicB-inducible division block normally seen in MinC+ cells. In contrast, none of the known FtsZ-associated factors, including ZipA, FtsA, and ZapA, appear to be specifically required for targeting of the DMinC/MinD complex to rings, implying that the two MinC/activator complexes must recognize distinct features of FtsZ assemblies. MinD-dependent targeting of MinC may occur in two steps of increasing topological specificity: (i) recruitment of MinC from the cytoplasm to the membrane, and (ii) specific targeting of the MinC/MinD complex to nascent septal ring assemblies on the membrane. Using membrane-tethered derivatives of MinC, we obtained evidence that both of these steps contribute to the efficiency of MinC/MinD-mediated division inhibition.
In Escherichia coli, cell constriction is mediated by the septal ring organelle and normally occurs at the midpoint of the long axis of the mother cell. Formation of the septal ring involves the ordered assembly of at least 10 essential division proteins. Assembly is thought to initiate with polymerization of FtsZ on the cytoplasmic face of the inner membrane. Polymeric FtsZ recruits FtsA and ZipA through direct protein-protein interactions, giving rise to a higher-order membrane-associated structure (Z ring) to which the remaining components are recruited until a mature septal ring is generated (1, 6, 16, 51, 60).
Equal partitioning of cell components to daughter cells requires that septal ring placement be accurately restricted to the middle of the mother cell. This specific placement appears to be accomplished primarily by two negative regulatory mechanisms that act independently, albeit in a partially redundant manner, to prevent FtsZ assembly at off-center sites (50). The first mechanism couples Z ring placement to positioning of the nucleoid(s) via nucleoid occlusion, which causes Z rings to preferentially form on membrane sites that do not directly surround the nucleoid(s) (57, 73). The molecular basis for this phenomenon is still unclear. The second mechanism is controlled by the protein products of the minB operon (MinC, -D, and -E), referred to collectively as the Min system. minB mutants display either a minicell phenotype (Min−), where cells frequently divide close to either one of the cell poles (MinC− and/or MinD−), or a MinCD-dependent filamentation phenotype (Sep−), where cells fail to divide altogether (MinE−) (13).
The MinC protein is a division inhibitor that negatively regulates FtsZ ring formation (5, 12, 37, 39, 41, 46, 59). The 231-residue polypeptide folds into two domains of approximately equal size. The amino-terminal domain (or Z-domain, henceforth ZMinC) is necessary and sufficient to inhibit FtsZ polymerization in vitro and Z ring assembly in vivo. The carboxy-terminal domain (D domain, DMinC) is required both for homodimerization of MinC and for binding to the so-called activators MinD and DicB (8, 34, 41, 69).
In the absence of either activator, MinC blocks cell division only when present at a concentration at least 25-fold greater than normal. At the physiological concentration of the division inhibitor, either MinD or DicB is required for efficient MinC-mediated division inhibition (12, 15).
Whereas DicB is not an integral part of the Min system, MinD plays several important roles in the proper functioning of the system. The protein is well conserved and belongs to a large family of ATPases of bacterial origin which also includes the ParA/SopA-type proteins involved in chromosome and plasmid segregation systems (9, 11, 19, 30, 48, 65). When bound to ATP, MinD interacts with itself, MinC, and MinE, as well as with phospholipid membrane (33, 35, 38, 45, 67). Binding of MinD-ATP to the membrane occurs in a cooperative fashion (45, 53) and likely involves the formation of polymeric filaments on the membrane surface (33, 66, 67). MinC and MinE have little affinity for membrane themselves. However, both MinC and MinE are recruited to membrane-associated MinD-ATP (33, 36, 38, 45, 61, 62). MinE interferes with the interaction between MinC and MinD-ATP, causing release of MinC from the MinD-ATP membrane complex (38, 45). In addition, MinE stimulates hydrolysis of the nucleotide by MinD (35), resulting in release of MinD-ADP, as well as MinE itself, from the phospholipid surface (33, 45).
The interactions between MinC, MinD, MinE, nucleotide, and membrane culminate in highly dynamic behaviors of all three Min proteins in normally dividing cells, where they accumulate alternately on the membrane at either end of the cell, oscillating back and forth in a rapid cycle that averages about 45 s (17, 26, 36, 61, 63, 66). These dynamics cause the time-averaged concentration of MinC at the membrane to be highest at the cell poles and lowest at the cell center. In turn, this concentration differential is proposed to define the center as the site where membrane-associated FtsZ polymers are most stable, as they are least subject to ZMinC-induced disassembly (26, 32, 42, 52). Although the proper dynamic positioning of ZMinC is the functionally relevant output of the Min system, MinC's role in generating the dynamics is passive (26, 63). Rather, Min protein oscillation requires both MinD and MinE. In the absence of MinD, both MinC and MinE are found throughout the cytoplasm, and cells are Min− (13, 36, 61, 62). In the absence of MinE, both MinD and MinC accumulate along the entire membrane without showing obvious bulk movement, and cells are Sep− (13, 36, 61, 63, 64).
The present study concerns the interaction(s) of MinC with the division apparatus and, specifically, the roles of the activators MinD and DicB in stimulating MinC-mediated division inhibition. The DicB protein is encoded on the cryptic prophage Kim (Qin), and its production is actively repressed under normal conditions (2, 7). When produced, however, DicB causes a rapid MinC-mediated division block, which is independent of MinD and insensitive to MinE (3, 12, 43, 44).
We showed previously that DicB competes with MinD for binding to the C-terminal domain of MinC (DMinC) and that DicB, like MinD, stimulates the disassembly of Z rings by MinC in vivo (41).
The finding that MinD recruits MinC to the membrane suggested a simple explanation for how MinD might stimulate MinC-mediated division inhibition, namely, that the accumulation of MinC on the membrane increases its local concentration to an extent sufficiently high to efficiently counteract FtsZ polymerization (36, 61). To assess if DicB acted in a similar fashion, we previously studied the subcellular localization of functional green fluorescent protein (GFP)-tagged versions of DicB and MinC (41). In contrast to the situation in MinC/MinD-induced filaments, in which both MinC and MinD are found distributed along the membrane, MinC and DicB are primarily found throughout the cytoplasm in MinC/DicB-induced filaments (41). This indicated that, unlike MinD, DicB does not direct MinC to the membrane per se. Interestingly, the distribution of proteins was strikingly different when either MinD or DicB was coexpressed with mutants of MinC (DMinC) that lack a functional Z-domain. In these cases, DMinC together with MinD or DicB was found to decorate septal rings, implying that the DMinC/MinD and DMinC/DicB complexes bind to one or more septal ring components and that these interactions are independent of that between ZMinC and its FtsZ substrate (41). These observations led us to propose that specific targeting of DMinC/activator complexes to septal ring assemblies is likely to contribute to the efficiency of activator-dependent, ZMinC-mediated division inhibition (41).
Here, we describe experiments aimed at elucidating what septal ring molecule(s) is recognized by the DMinC/activator complexes, as well as to what extent the specific targeting of the DMinC/activator complexes to septal ring components contributes to the stimulation of MinC-mediated division inhibition by the activators.
We present several lines of evidence that DMinC/DicB is targeted specifically to ZipA-decorated FtsZ polymers, likely involving a direct interaction between the DMinC/DicB heteromer and ZipA, and that this interaction is crucial to DicB-induced division inhibition. In contrast, DMinC/MinD targeting to FtsZ rings does not seem to specifically require any one of the other known septal ring components, including ZipA, FtsA, and ZapA. DMinC/MinD may interact with FtsZ polymers directly or with some associated molecule that still needs to be identified.
We further find that, even though a membrane-tethered derivative of MinC localizes to the membrane in a MinD-independent fashion, MinD still causes it to target to rings and enhances its division-inhibitory activity. This result suggests that, as in the case of DicB-dependent targeting, specific MinD-dependent targeting of MinC to FtsZ polymeric structures on the membrane is likely to contribute to the efficacy of ZMinC action.
Although it is clear from this work that DMinC/MinD and DMinC/DicB recognize distinct targets on FtsZ assemblies, both MinD and DicB appear capable of activating MinC by serving as obligate coassembly factors, allowing the Z-domain of MinC to be brought in close proximity to its substrate.
MATERIALS AND METHODS
E. coli strains.
The most relevant E. coli strains, plasmids, and phages used in this study are listed in Table 1.
TABLE 1.
E. coli strains, plasmids, and phages used in this study
| Strain, plasmid, or phage | Relevant genotypeb | Source |
|---|---|---|
| Strainsa | ||
| PB103 | dadR trpE trpA tna | 14 |
| CH3 | PB103, recA::Tn10 | 23 |
| LL1 | PB103, minCDE::aph lon::Tn10 | 41 |
| DR109 | PB103, minCDE::aph recA::Tn10 | This work |
| CH28 | PB103, minCDE<>frt zapA<>cat | This work |
| JE14 | PB103, ftsA0minCDE::aph | This work |
| JE15 | PB103, ftsA0minCDE::aph recA::Tn10 | This work |
| JE23 | PB103, minCDE<>frt | This work |
| JE24 | PB103, minCDE<>frt zipA::aph | This work |
| JE25 | PB103, minCDE<>frt zipA::aph recA::Tn10 | This work |
| JE32 | PB103, zipA::aph ftsA* | This work |
| JE33 | PB103, minCDE<>frt zipA::aph ftsA* | This work |
| JE34 | PB103, ftsA* | This work |
| JE35 | PB103, minCDE<>frt ftsA* | This work |
| Plasmids | ||
| pBAD33 | cat araC PBAD:: | 22 |
| pJE80 | cat araC PBAD::sfiA | 41 |
| pZAQ | tet ftsQAZ | 71 |
| pDR119 | bla laclq Plac::gfp-t-minD | 63 |
| pDR121 | bla laclq Plac::gfp-t-minC | 61 |
| pJE39 | bla laclq Plac::gfp-t-dicB | 41 |
| pDB326 | aadA repA(Ts) | 23 |
| pCH32 | aadA repA(Ts) zipA ftsZ | 23 |
| pCH204 | aadA repA(Ts) minD | This work |
| pDB280 | cat repA(Ts) ftsA | 24 |
| pJE102 | cat repA(Ts) ftsA* | This work |
| Phages | ||
| λCH178 | imm21bla laclq Plac::zipA(1-183)-gfp | This work |
| λDR121 | imm21bla laclq Plac::gfp-t-minC | 61 |
| λPC100 | imm21bla laclq Plac::zipA(1-183)-gfp-t-minC | This work |
| λLL49 | imm21bla laclq Plac::zipA(1-183)-gfp-t-minC(108-231) | This work |
| λJE64 | imm21bla laclq Plac::t-dicB-gfp-t-minC(108-231) | This work |
| λJE65 | imm21bla laclq Plac::gfp-t-minC(108-231) minD | This work |
Note that strains JE14, 15, 24, and 25 required an appropriate complementing plasmid or phage for survival.
Strains MG1655 (ilvG rfb50 rph1) (21), PB114 (dadR trpE trpA tna minCDE::aph) (13), DX1 (dadR trpE trpA tna minCDE::aph recA::Tn10) (11), CH4/pCH32 [dadR trpE trpA tna zipA::aph/aadA repA(Ts) zipA ftsZ] (23), CH1/pDB280 [dadR trpE trpA tna ftsA0/cat repA(Ts) ftsA] (24), and DY329 [rph1 IN(rrnD-rrnE) Δ(argF-lac)U169 nadA::Tn10 gal490 λcI857 Δ(cro-bioA)] (72) have been described previously.
Strain DR109 was obtained by P1-mediated transduction (transduction, for short) of recA::Tn10 from DX1 to PB114, JE14/pDB280 was obtained by transduction of minCDE::aph from PB114 to CH1/pDB280, and JE15/pDB280 was obtained by transduction of recA::Tn10 from DX1 to JE14/pDB280.
Strain TB10 [MG1655, nadA::Tn10 λcI857 Δ(cro-bioA)] was obtained by cotransduction of nadA::Tn10 and λcI857 Δ(cro-bioA) from DY329 to MG1655, selecting for tetracycline resistance and screening for Ts growth.
Construction of JE23 and CH28 involved λ Red recombineering (10, 72). For JE23, we made use of TB14 (MG1655, lacIZYA<>frt minCDE<>aph), whose construction will be detailed elsewhere. The minCDE<>aph allele from TB14 was transduced to PB103, yielding TB33 (PB103, minCDE<>aph). The aph cassette was next evicted as described elsewhere (10), resulting in JE23. In JE23 and derivatives, the complete minB operon has been replaced with an frt scar sequence (10). For CH28, the cat cassette from pKD3 (10) was amplified with primers 5′-TAAAGACGAAGGCAGCGCAGTCAATCAGCAGGAAGGTGGCGTGTAGGCTGGAGCTGCTTCG-3′ and 5′-GTCTTCACGGTTACTCTACCACAGTAAACCGAAAAGTGCATATGAATATCCTCCTTAG-3′, resulting in a 1,092-bp fragment with end sequences homologous to those flanking the zapA (ygfE) gene on the chromosome (underlined). Recombination of this fragment with the chromosome of TB10 yielded CH21 (TB10, zapA<>cat), in which the complete zapA gene is replaced with cat and transcription of the latter is directed away from the downstream ssrS gene. Transduction of zapA<>cat from CH21 to JE23 resulted in CH28.
Strains JE24/pCH32 and JE24/pZAQ were obtained by transduction of zipA::aph from CH4/pCH32 to JE23/pCH32 and JE23/pZAQ, respectively. Strain JE25/pCH32 was obtained by transduction of recA::Tn10 from DX1 to JE24/pCH32.
Strains JE32, JE33, JE34, and JE35 were obtained by recombination of the ftsA* allele on the pMAK705 derivative pJE102 [cat repA(Ts) ftsA*] (see below) with the chromosomal wild-type ftsA allele of strains CH4/pCH32, JE24/pCH32, PB103, and JE23, respectively, using the method of Hamilton et al. (28).
Lysogenic derivatives of strains were obtained as described before (13).
E. coli plasmids and phages.
Plasmids pZAQ (71), pDB173 (13), pBAD33 (22), pDB326 and pCH32 (23), pDB280 (24), pDR119 (63), pDR121 and pDR155 (61), pDB341 (27), pDB387 (26), pJE39, pJE80, and pPC105 (41), and pCH151 (4) have been described previously. pET plasmids were obtained from Novagen, and pBluescript KS was obtained from Stratagene. In the descriptions below, t and h stand for sequences encoding the T7 tag (Novagen) and oligohistidine, respectively.
Plasmid pCH204 [aadA repA(Ts) minD] was obtained by replacing the 1,335-bp PacI-HindIII fragment of pDB387 [aadA repA(Ts) minDE] with the 596-bp PacI-HindIII fragment of pDR155 (Plac::minD). Transcription of minD from pCH204 is in the direction opposite from that of the aadA gene and is driven from the native P2 promoter embedded in upstream minC sequences (13). The plasmid lacks the ribosome-binding site and translation start codon of minC and does not express MinC.
Plasmid pPC100 [Plac::zipA(1-183)-gfpmut2-t-minC(5-231)] was constructed in two steps. A PCR product, obtained with primers 5′-ACAGAGATCCATATGATGCAGGATTTGCGTCTG-3′ and 5′-ACGCCTCGAGTGGAGCAGGTTCCGCTAC-3′, was digested with AgeI (internal to fragment) and XhoI (underlined), and the resulting 241-bp fragment was used to replace the 676-bp AgeI-XhoI fragment of pDB341 (PT7::zipA-gfpS65T), yielding pCH100 [PT7::zipA(1-183)-gfpS65T]. The 191-bp XbaI-NcoI fragment of pDR121 [Plac::gfpmut2-t-minC(5-231)] was replaced with the 780-bp XbaI-NcoI fragment of pCH100, resulting in pPC100.
For pCH178 [Plac::zipA(1-183)-gfpmut2], the 2,264-bp NcoI-ClaI fragment of pPC100 was replaced with the 1,391-bp NcoI-ClaI fragment of pCH151 (Plac::zipA-gfpmut2).
Plasmid pLL49 [Plac::zipA(1-183)-gfpmut2-t-minC(108-231)] was obtained by replacing the 562-bp NcoI-SphI fragment of pCH178 with the 976-bp NcoI-SphI fragment of pPC105 [PλR::gfpmut2-t-minC(108-231)].
For pJE60 [Plac::gfpmut2-t-minC(108-231)], the 1,527-bp ApaI-NcoI fragment of pLL49 was replaced with the 937-bp ApaI-NcoI fragment of pDR121.
Plasmid pJE64 [Plac::t-dicB-gfpmut2-t-minC(108-231)] was constructed in three steps. A PCR product, obtained with primers 5′-GACGGATCCATGAAAACGTTATTACCAAACG-3′ and 5′-ATGCTGCTCGAGTTGTGCACATCCTTTTGGCATC-3′, was treated with BamHI and XhoI (sites underlined), and the 192-bp fragment was ligated to similarly digested pET21a, giving rise to pJE40 (PT7::t-dicB-h). Next, the 270-bp XbaI-XhoI fragment of pJE40 was used to replace the 1,026-bp XbaI-XhoI fragment of pCH151, generating pJE42 (Plac::t-dicB-gfpmut2). Finally, pJE64 was obtained by replacing the 938-bp ApaI-NcoI fragment of pJE60 with the 1,193-bp ApaI-NcoI fragment of pJE42.
The construction of pJE65 [Plac::gfp-t-minC(108-231) minD] also took three steps. The minD gene was amplified using primers 5′-CAAGGAATTCATATGGCACGCATTATTGTTG-3′ and 5′-GTACCGTCGACTTATCCTCCGAACAAGCG-3′, and the 815-bp NdeI-SalI fragment (sites underlined) was ligated into similarly digested pET21a, yielding pDB313 (PT7::minD). The 596-bp PacI-HindIII fragment of pDB313 was next used to replace the 774-bp PacI-HindIII fragment of pDB173. This yielded pDR5 (Plac::minC minD), which is similar to pDB173 (13) except that it lacks all minE sequences. Finally, pJE65 was obtained upon replacement of the 1,200-bp BstEII fragment of pDR5 with the 1,766-bp BstEII fragment of pJE60.
The construction of plasmid pJE102 [repA(Ts) ftsA*] involved two PCRs using pDB280 [repA(Ts) ftsA] as a template. The ftsA* mutation (R286W) creates an NcoI site (18), which was included in one of the primers of each set. Primers 5′-AGCTTCTAGATCAGGCACAGGCAGAAC-3′ and 5′-GACTCCATGGCGGACACCACCTACGC-3′ were used to amplify the 5′ portion of ftsA*, and the product was digested with XbaI and NcoI (sites underlined). The 879-bp fragment was used to replace a 39-bp XbaI-NcoI fragment of plasmid pDB289 (PT7::h-ftsA ftsZ), a pET21 derivative whose lineage is detailed below, resulting in pJE100. Subsequently, primers 5′-TCCGCCATGGAGTCTGCAACGTCAGAC-3′ and 5′-TTAGGCACCCCAGGCTTTACAC-3′ were used to amplify the 3′ portion of ftsA*. The product was treated with NcoI (underlined) and EcoRI (internal to fragment), and the 582-bp fragment was used to replace the 1,499-bp NcoI-EcoRI fragment of pJE100, yielding pJE101 (PT7::ftsA* ftsZ). Finally, the 819-bp BglII-BlpI fragment of pDB280 was replaced with that of pJE101, resulting in pJE102.
Plasmid pDB289 (PT7::h-ftsA ftsZ) was used as an intermediate for the construction of pJE102 (see above) and was itself the result of a series of manipulations. Primers 5′-CCGAATTCCCATATGATCAAGGCGACGGAC-3′ and 5′-GTGATGTTTAACCCC-3′ were used to amplify the 5′ part of the ftsA gene. The product was treated with NdeI (underlined) and HindIII (internal to fragment), and the 953-bp fragment was ligated to similarly digested pET21b. The resulting plasmid, pDB273 [PT7::ftsA(1-319)-h], encodes the N-terminal 319 residues of FtsA fused to a His6 tag (h). The 1,433-bp BclI-EcoRI fragment, containing virtually all of ftsA and part of ftsZ, was isolated after partial digestion and inserted in the BamHI and EcoRI sites of pET21c. This yielded pDB267 (PT7::t-ftsA), which encodes a fusion in which the starting methionine of FtsA is replaced with the T7 tag (t). The 2,215-bp KpnI-PstI fragment of pDB273 was replaced with the 2,717-bp KpnI-PstI fragment of pDB267, yielding a plasmid (pDB274 [PT7::ftsA]) encoding native FtsA. Plasmid pDB277 (PT7::h-ftsA) was next obtained by replacing the 1,037-bp ApaI-NdeI fragment of pDB274 with the 1,099-bp ApaI-NdeI fragment of pET16b. To generate pDB289, the 1,900-bp HindIII fragment from pZAQ was inserted into the HindIII site of pBluescript KS. This yielded pDB190, in which ftsZ is transcribed in the direction opposite to that of the lac promoter in the vector. One of the HindIII sites of this plasmid was removed by deletion of a 12-bp ClaI fragment, resulting in pDB260. This plasmid was treated with Bsu36I and HindIII, blunt ended by treatment with Klenow enzyme and deoxynucleoside triphosphates, and recircularized. Finally, the 1,418-bp EcoRI-SalI fragment of the resulting plasmid (pDB261) was used to replace the 13-bp EcoRI-SalI fragment of pDB277, yielding pDB289.
Lysogenic λ phages were obtained by crossing the appropriate plasmid with λNT5 as described before (13). Each phage name corresponds to that of the plasmid used in the cross.
E. coli growth conditions.
Unless indicated otherwise, cells were grown in Luria-Bertani medium at 37°C. M9 minimal salts medium supplemented with tryptophan (50 μg/ml), Casamino Acids (0.2%), and maltose (0.2%) was used in some experiments. Where appropriate, antibiotics were used at 12.5 (tetracycline), 25 (chloramphenicol), and 50 (ampicillin and spectinomycin) μg/ml. Isopropyl β-d-thiogalactopyranoside (IPTG) and arabinose were used as indicated. Cultures to be analyzed by microscopy were typically inoculated by a 200-fold dilution of an overnight culture into fresh medium containing IPTG and/or arabinose at the indicated concentrations. Cultures were then incubated until the optical density at 600 nm (OD600) reached a value between 0.1 and 0.5.
Microscopy and other methods.
To determine localization patterns of GFP fusions, cells were viewed both live and upon chemical fixation. Patterns corresponded under both conditions in all cases. Fixed samples are shown, unless stated otherwise. Fixation, immunostaining with affinity-purified FtsZ antibodies, microscopy, image collection, and cell length measurements were performed as described previously (41).
Western analyses using FtsZ-, FtsA-, ZipA-, MinC-, or GFP-specific primary antibodies (4, 15, 24, 41) and horseradish peroxidase-conjugated secondary antibodies were performed essentially as described elsewhere (4, 41). Antigen bands were visualized using the SuperSignal West Pico Western blotting analysis system (Pierce). Digital images of blots were collected and analyzed with a Bio-Rad Fluor-S-Max MultiImager using the accompanying Quantity One software. For relative quantitation, the amount of antigen in a whole-cell extract sample was normalized to that in a standard sample, as indicated. To improve accuracy, the photon counts of both the minimal area corresponding to an antigen band and an equal area immediately adjacent were measured, and the value of the latter was subtracted from the former. The total amount of protein loaded per lane was kept between 0.5 and 5.0 μg, as signal intensities with all five antigens corresponded almost linearly with amounts of sample within this range. Protein concentrations were determined using the Non Interfering Protein Assay (Genotech) according to the manufacturer's instructions with bovine serum albumin as a standard.
Yeast strains, plasmids, and two-hybrid analyses.
Strains PJ69-4A (MATa trp1-901 leu2-3,112 ura3-52 his3-200 gal4Δ gal80Δ LYS::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) (40), and SL3004 (MATα trp1-901 leu2 ura3 his3 gal4 gal80 lys2-801 ade2-101) (31) have been described previously.
The two-hybrid plasmids listed below in Table 4 are derivatives of the pGBDU-C(X) (URA3+) or pGAD-C(X) (LEU2+) vector series (40) and encode fusions to the C terminus of the Gal4 DNA-binding domain (BD; pGBDU derivatives) or the Gal4 activation domain (AD; pGAD derivatives). In yeast, transcription of the fusions is under control of the ADH1 promoter (PADH1). Plasmid pJE55 (PADH1::gal4BD-dicB) was described before (41).
TABLE 4.
Yeast two-hybrid interactions
| Plasmids (BD/AD) | BDa | ADa | Unitsb (±SD) |
|---|---|---|---|
| pJE55/pGAD-C1 | DicB | —c | 1.9 ± 0.2 |
| pJE55/pJE87 | DicB | ZipA(23-328) | 2.7 ± 0.5 |
| pJE92/pGAD-C1 | GFP-DMinC | — | 6.0 ± 2.2 |
| pJE92/pJE87 | GFP-DMinC | ZipA(23-328) | 6.3 ± 2.1 |
| pJE72/pGAD-C1 | DicB-GFP-DMinC | — | 2.9 ± 0.1 |
| pJE72/pJE87 | DicB-GFP-DMinC | ZipA(23-328) | 17.2 ± 4.0 |
| pJE88/pGAD-C1 | ZipA(23-328) | — | 3.2 ± 0.1 |
| pJE88/pJE93 | ZipA(23-328) | GFP-DMinC | 2.9 ± 0.1 |
| pJE88/pJE86 | ZipA(23-328) | DicB-GFP-DMinC | 7.3 ± 0.8 |
BD and AD indicate fusions to the yeast Gal4 DNA binding domain and transcription activating domain, respectively.
Units given are the means of three measurements.
—, unfused AD.
To construct pJE72 [PADH1::gal4BD-dicB-gfp-t-minC(108-231)], pJE64 (see above) was digested to completion with SalI and partially with BamHI. The 1,341-bp fragment was used to replace the 195-bp BamHI-SalI fragment of pJE55 (41), giving rise to pJE72.
For pJE86 [PADH1::gal4AD-dicB-gfp-t-minC(108-231)], the 1,347-bp XmaI-SalI fragment of pJE72 was used to replace the 18-bp XmaI-SalI fragment of pGAD-C1.
To construct pJE87 [PADH1::gal4AD-zipA(23-328)], we performed a PCR with primers 5′-GCTTTAGAATTCCATGGTTTCTGGACCAGCCGT-3′ and 5′-TTATGTCGACTTAGGCGTTGGCGTCTTTGACTTCGCGG-3′. The product was cut with EcoRI and SalI (sites underlined), and the 927-bp fragment was used to replace the 24-bp EcoRI-SalI fragment of vector pGAD-C1.
For pJE88 [PADH1::gal4BD-zipA(23-328)], the 927-bp EcoRI-SalI fragment of pJE87 was used to replace the 24-bp EcoRI-SalI fragment of vector pGBDU-C1.
Plasmids pJE92 [PADH1::gal4BD-gfp-t-minC(108-231)] and pJE93 [PADH1::gal4AD-gfp-t-minC(108-231)] were obtained by deletion of the 222-bp EcoRI fragment of pJE72 and pJE86, respectively, thereby removing all dicB sequences by an in-frame deletion.
Two-hybrid assays were performed as described before (41). Values given represent the means of at least three measurements.
RESULTS
Targeting of the tripartite fusion DicB-GFP-DMinC to septal rings in vivo.
Previously, we determined the cellular distribution of DMinC/DicB and DMinC/MinD complexes using strains in which either one of the partners was fused to GFP and the other was expressed in trans from a plasmid or lysogenic phage (41). To simplify genetic manipulations required for the present study, we constructed lysogenic phages λJE64 (Plac::dicB-gfp-DminC) and λJE65 (Plac::gfp-DminC minD). λJE64 encodes a chimeric protein in which the N terminus of GFP is fused to DicB, and its C terminus is fused to the D-domain of MinC (residues 108 to 231, DMinC). λJE65 encodes GFP-DMinC together with native MinD, allowing synthesis of these proteins in cis.
Phages λJE64 and λJE65 were introduced into the chromosome of the ΔminCDE strain DR109, and the resulting lysogens were grown in the presence of IPTG prior to microscopy. Both DR109(λJE64) and DR109(λJE65) cells showed pronounced fluorescent rings (Table 2). To ascertain that this accumulation of GFP-DMinC reflected its decoration of septal ring structures, the lysogens were transformed with either pJE80 (PBAD::sfiA), encoding the FtsZ polymerization inhibitor SfiA (56) under control of the araBAD promoter, or its parent vector, pBAD33. Growth of either lysogen carrying pBAD33 in the presence of 0.1% arabinose did not affect the accumulation of fluorescence in rings. However, growth with arabinose led to dispersal of the fluorescent signal throughout the cytoplasm of DR109(λJE64)/pJE80 filaments and to accumulation of fluorescence along the membrane of DR109(λJE65)/pJE80 filaments (Table 2). These results were consistent with our previous experiments, in which DMinC was expressed in trans with either DicB or MinD and where the resulting DMinC/DicB and DMinC/MinD complexes decorated rings in ΔminCDE cells in an FtsZ-dependent manner (41). Thus, the DicB-GFP-DMinC fusion encoded by λJE64 can serve as a convenient marker for DicB-dependent targeting of MinC to septal ring assemblies, and λJE65 provides a convenient means to similarly study the targeting of GFP-DMinC/MinD complexes.
TABLE 2.
Localization of DicB-GFP-DMinC in λJE64 lysogens and of GFP-DMinC in λJE65 lysogens
| Strain | Relevant genotype | Special conditiona | Locationb
|
|
|---|---|---|---|---|
| λJE64 (DicB-GFP-DMinC) | λJE65 (GFP-DMinC/MinD) | |||
| DR109 | ΔminCDE | R | R | |
| DR109/pBAD33 | ΔminCDE/PBAD:: | ARA | R | R |
| DR109/pJE80 | ΔminCDE/PBAD::sfiA | ARA | Cc | Mc |
| JE15/pDB280 | ΔminCDE ftsA0recA/repA(Ts) ftsA | 30°C | R | R |
| 42°C | Rc | Rc | ||
| CH28 | ΔminCDE ΔzapA | R | R | |
| JE25/pCH32 | ΔminCDE ΔzipA recA/repA(Ts) zipA ftsZ | 30°C | R | R |
| 42°C | Cc | Rc | ||
| JE23 | ΔminCDE | R | R | |
| JE35 | ΔminCDE ftsA* | R | R | |
| JE33 | ΔminCDE ΔzipA ftsA* | C | R | |
| JE23/pZAQ | ΔminCDE/ftsQAZ | R | R | |
| JE24/pZAQ | ΔminCDE ΔzipA/ftsQAZ | C | R | |
Unless indicated otherwise, cells were grown at 37°C in LB-AMP medium supplemented with 50 μM IPTG to an OD600 of 0.2 to 0.5. Where indicated, 0.1% arabinose (ARA) was included in the medium.
Over 100 cells were examined in each case. R, concentrated in one or more rings in over 70% of the population; M, concentrated along the cell periphery, less than 1% of cells showed obvious rings; C, cytoplasmic, less than 1% of cells showed obvious rings.
Cells had a Sep− division phenotype, ranging in length from 15 to 60 μm. Cells were Min− in all other cases.
Targeting of DMinC/activator complexes upon depletion of FtsA.
The accumulation of DMinC/activator complexes (DMinC/DicB or DMinC/MinD) in rings depends on the ability of FtsZ to assemble into Z rings (41). In the most straightforward scenario for targeting of DMinC/activator complexes to septal ring assemblies, the complexes simply recognize and bind FtsZ polymers directly. However, as FtsZ assembly triggers recruitment of all other known septal ring proteins, it is also possible that targeting of MinC/activator complexes involves one or more other ring components. If so, proteins that are recruited to FtsZ polymers early in the development of a functional septal ring organelle are the most likely candidates (41). FtsA and ZipA bind FtsZ polymers directly and are recruited in a mutually independent fashion at a very early stage of septal ring assembly (23, 24, 29, 47, 49, 54, 60, 70). Although both proteins are essential for cell division in wild-type cells, a specific mutant variant of FtsA (FtsAR286W, denoted FtsA*) was recently shown to bypass the requirement for functional ZipA, suggesting that the proteins serve partially redundant functions (18).
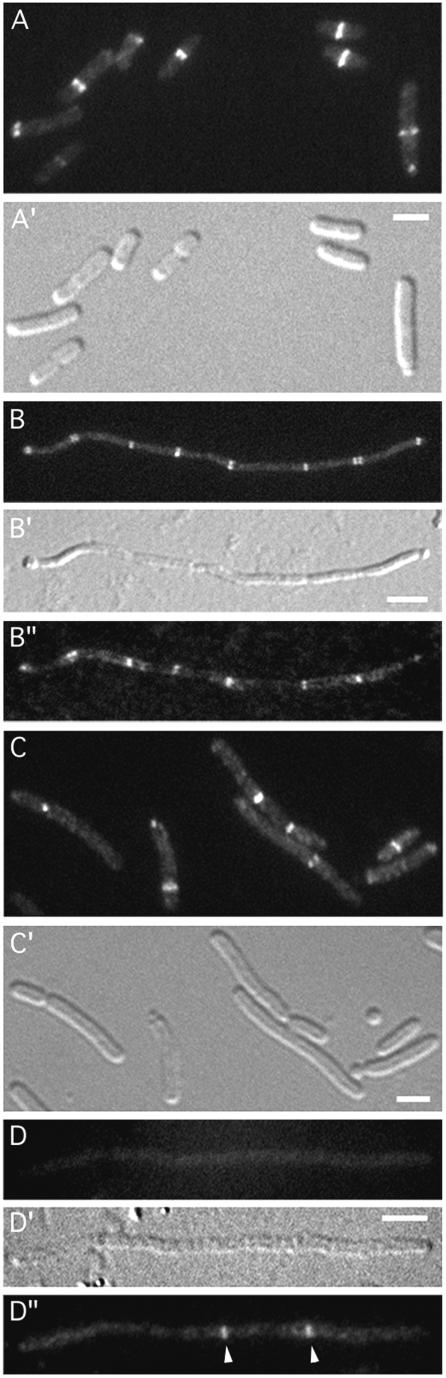
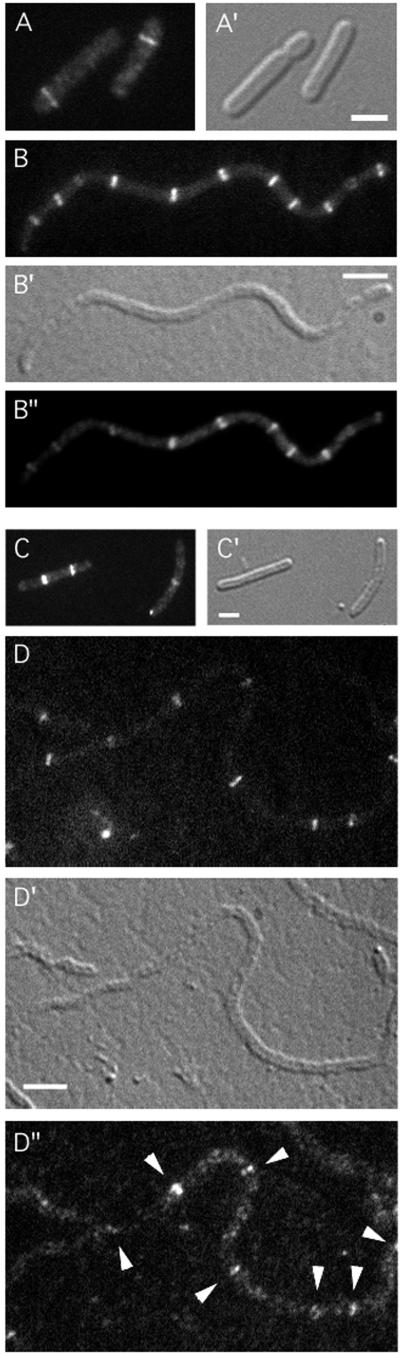
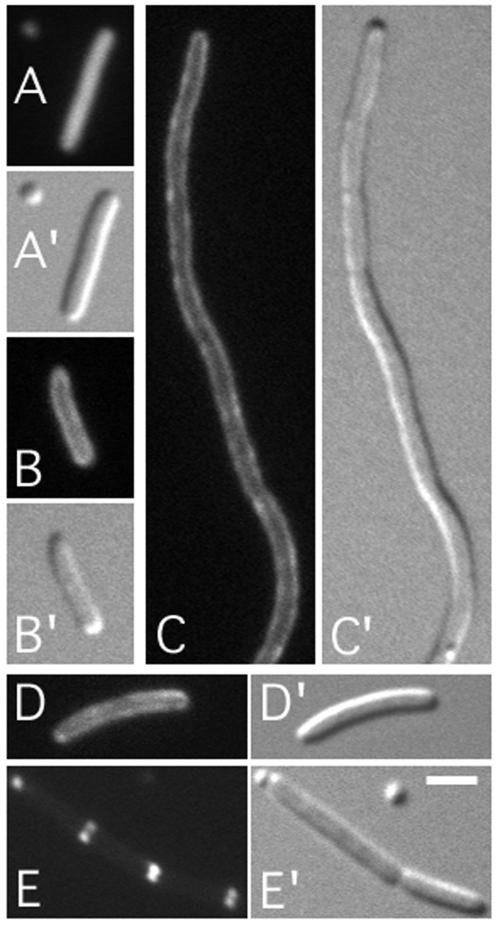
To test for a possible involvement of FtsA in recruitment of DMinC/activator complexes to septal rings, we used a heat-induced depletion (HID) strain, similar to ones described before (24). Strain JE15/pDB280 [ΔminCDE ftsA0/ftsA repA(Ts)] lacks functional copies of the minCDE and ftsA genes on the chromosome but carries a complementing ftsA copy on plasmid pDB280 that is Ts for replication. As expected, JE15/pDB280 cells showed a typical Min− phenotype at 30°C (FtsA+) but failed to divide at 42°C due to depletion of FtsA. The strain was lysogenised with λJE64 (Plac::dicB-gfp-DminC) or λJE65 (Plac::gfp-DminC minD), and lysogens were grown in the presence of IPTG at 30 and 42°C. As expected, the majority of both JE15(λJE64)/pDB280 and JE15(λJE65)/pDB280 cells showed well-defined fluorescent rings at 30°C (Fig. 1A and 2A). Both strains formed long filaments at 42°C, which correlated with a depletion of FtsA to ∼12% of its wild-type level, as determined by quantitative Western analyses (data not shown). Moreover, filaments of either strain showed multiple fluorescent rings distributed along their long axis (Table 2). Filaments were chemically fixed and immunostained for detection of FtsZ, showing that the DicB-GFP-DMinC rings in the JE15(λJE64)/pDB280 filaments (Fig. 1B), and also the GFP-DMinC/MinD rings in the JE15(λJE65)/pDB280 filaments (Fig. 2B), colocalized with FtsZ rings.
FIG. 1.
Localization of DicB-GFP-DMinC in FtsA− and ZipA− filaments. Fluorescence micrographs (A to D, B", and D") and corresponding differential interference contrast micrographs (A′ to D′) showing DicB-GFP-DMinC in dividing (A) and nondividing (B) cells of the FtsA depletion strain JE15(λJE64)/pDB280 [ΔminCDE ftsA0(Plac::dicB-gfp-DminC)/repA(Ts) ftsA] and dividing (C) and nondividing (D) cells of the ZipA depletion strain JE25(λJE64)/pCH32 [ΔminCDE ΔzipA(Plac::dicB-gfp-DminC)/repA(Ts) zipA ftsZ]. Cells were grown to an OD600 of 0.3 to 0.5 in the presence of 50 μM IPTG at either 30°C (A and C) or 42°C (B and D) prior to chemical fixation. Cells in panels B and D were subsequently subjected to immunostaining to localize FtsZ. GFP-specific (A to D) and Cy3-specific (B" and D") filter sets were used to visualize the location of DicB-GFP-DMinC and FtsZ, respectively. Arrowheads in panel D" point to well-defined FtsZ rings that failed to be decorated by DicB-GFP-DMinC. Bars, 2 (A and C) or 4 (B and D) μm.
FIG. 2.
Localization of GFP-DMinC/MinD in FtsA− and ZipA− filaments. Fluorescence micrographs (A to D, B", and D") and corresponding differential interference contrast micrographs (A′ to D′) showing GFP-DMinC in dividing (A) and nondividing (B) cells of the FtsA depletion strain JE15(λJE65)/pDB280 [ΔminCDE ftsA0(Plac::gfp-DminC minD)/repA(Ts) ftsA] and dividing (C) and nondividing (D) cells of the ZipA depletion strain JE25(λJE65)/pCH32 [ΔminCDE ΔzipA(Plac::gfp-DminC minD)/repA(Ts) zipA ftsZ]. Cells were grown as described in the legend for Fig. 1 at either 30°C (A and C) or 42°C (B and D). Cells in panels B and D were subsequently subjected to immunostaining to localize FtsZ. GFP-specific (A to D) and Cy3-specific (B" and D") filter sets were used to visualize the location of GFP-DMinC and FtsZ, respectively. Arrowheads in panel D" point to FtsZ rings decorated by GFP-DMinC. Bars, 2 (A and C) or 4 (B and D) μm.
These results indicate that FtsA is not required for targeting of either MinC/DicB or MinC/MinD to septal rings. In addition, because recruitment of the majority of known septal ring proteins (save FtsZ, ZipA, and possibly ZapA) to FtsZ rings depends on a sufficiently high level of FtsA (1, 6, 16), it can be inferred that FtsK, -Q, -L, -B, -W, -I, and -N are similarly unlikely to be required.
ZapA (YgfE) is not essential for cell division and not required for DMinC/activator targeting to septal rings.
The Bacillus subtilis protein ZapA (YshA) was recently identified as a nonessential septal ring component which binds FtsZ directly and promotes bundling of FtsZ polymers in vitro (20). In addition, the ZapA ortholog of E. coli (YgfE) was shown to similarly localize to the septal ring (20).
To determine if ZapA is required for attracting DMinC/activator complexes to FtsZ assemblies, we constructed CH28 (ΔminCDE ΔzapA), a Min− strain in which the complete zapA gene was removed from the chromosome using λ Red recombineering. Cells of this strain showed a typical Min− phenotype, demonstrating that, as in B. subtilis (20), zapA (ygfE) is not an essential gene in E. coli.
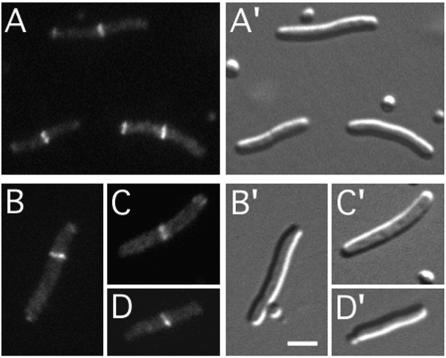
Upon growth in the presence of IPTG, lysogenic derivatives of CH28 carrying either λJE64 or λJE65 showed the same accumulation of fluorescence in rings as observed in ZapA+ cells (Fig. 3; Table 2). We conclude that ZapA is not required for attracting either DMinC/DicB or DMinC/MinD to septal ring assemblies.
FIG. 3.
Localization of DicB-GFP-DMinC and GFP-DMinC/MinD in cells lacking ZapA (YgfE). Fluorescence micrographs (A to D) and corresponding differential interference contrast micrographs (A′ to D′) showing cells of strain CH28 (ΔminCDE ΔzapA) lysogenic for either λJE64 (Plac::dicB-gfp-DminC) (A) or λJE65 (Plac::gfp-DminC minD) (B to D). Cells were grown at 37°C in the presence of 50 μM IPTG. Bar, 2 μm.
DicB-dependent targeting of DMinC requires ZipA.
ZipA is the only known essential septal ring protein that decorates Z rings in FtsA− filaments (24, 60). Therefore, we used λJE64 and λJE65 lysogens of the ZipA(HID) strain JE25/pCH32 [ΔminCDE zipA0/repA(Ts) zipA ftsZ] to assess whether or not ZipA played a role in attracting DMinC/activator complexes to FtsZ assemblies. When grown at 30°C (ZipA+) in the presence of IPTG, cells of both JE25(λJE64)/pCH32 and JE25(λJE65)/pCH32 lysogens showed accumulation of fluorescence in rings (Fig. 1C and 2C). At 42°C, both lysogens formed filaments due to depletion of ZipA. Interestingly, whereas JE25(λJE65)/pCH32 filaments contained multiple well-defined fluorescent ring structures (Fig. 2D), rings were absent in virtually all JE25(λJE64)/pCH32 filaments. Instead, the signal in the latter filaments appeared dispersed throughout the cytoplasm (Fig. 1D). As above, filaments were fixed and subjected to immunostaining to detect FtsZ. Consistent with previous work on ZipA-depleted filaments (24), the majority contained multiple FtsZ rings even though the average number of rings per unit length was relatively low. Figure 1D shows an example of a doubly labeled filament of JE25(λJE64)/pCH32 containing clear FtsZ rings that apparently failed to recruit the DicB-GFP-DMinC fusion. Thus, while MinD-dependent targeting of DMinC to FtsZ rings appeared unaffected in ZipA-depleted filaments, DicB-dependent targeting appeared completely inhibited, indicating an important role for ZipA in attracting MinC/DicB to FtsZ assemblies.
It was recently shown that the normal requirement for ZipA in the division process can be bypassed by mutation of the chromosomal ftsA gene to ftsAR286W (ftsA*), or by supplying cells with plasmid pZAQ, carrying the native ftsQ, -A, and -Z genes (18). These findings provided an opportunity to study targeting of DMinC/activator complexes in cells completely devoid of ZipA. To this end, λJE64 and λJE65 were introduced into a set of five (isogenic) ΔminCDE strains that differ in that they are either zipA+ or zipA0 and in that they either carry the chromosomal ftsA* allele or the pZAQ plasmid to suppress the lethality associated with the absence of ZipA (Table 2). The DicB-GFP-DMinC fusion targeted efficiently to septal rings in JE23 (ΔminCDE), JE23/pZAQ (ΔminCDE/ftsQAZ), and JE35 (ΔminCDE ftsA*) but completely failed to do so in JE24/pZAQ (ΔminCDE zipA0/ftsQAZ) and JE33 (ΔminCDE zipA0 ftsA*) (Fig. 4A and B; Table 2). Rather, the fusion appeared cytoplasmic in the zipA0 strains, as was observed in the ZipA-depleted filaments above. This cytoplasmic distribution of DicB-GFP-DMinC was not caused by a possible enhanced processing of the fusion protein in zipA0 cells, as determined by Western analyses (data not shown). In contrast to DicB-GFP-DMinC in the λJE64 lysogens, the GFP-DMinC fusion accumulated in rings similarly in all five λJE65 lysogens (Fig. 4C and D; Table 2).
FIG. 4.
Localization of DicB-GFP-DMinC and GFP-DMinC/MinD in cells lacking ZipA. Fluorescence micrographs (A to D) and corresponding differential interference contrast micrographs (A′ to D′) showing cells of strains JE33(λJE64) [ΔminCDE ΔzipA ftsA* (Plac::dicB-gfp-DminC)] (A), JE35(λJE64) [ΔminCDE ftsA* (Plac::dicB-gfp-DminC)] (B), JE33(λJE65) [ΔminCDE ΔzipA ftsA* (Plac::gfp-DminC minD)] (C), and JE35(λJE65) [ΔminCDE ftsA* (Plac::gfp-DminC minD)] (D). Cells were grown at 37°C in the presence of 50 μM IPTG. Bar, 4 (A and B) or 2 (C and D) μm.
We conclude that ZipA is required for DicB-dependent targeting, but dispensable for MinD-dependent targeting, of DMinC to FtsZ assemblies.
ZipA0 cells are resistant to MinC/DicB-mediated division inhibition.
Expression of DicB, or overexpression of MinD, in wild-type cells results in a MinC-dependent division block (12, 44). Division inhibition in both cases requires the activities of both ZMinC, which interferes with FtsZ polymerization, and DMinC, which binds MinD or DicB (34, 37, 41). We proposed that DMinC/activator targeting to (nascent) septal ring assemblies is likely to enhance the efficacy of division inhibition by bringing ZMinC in close proximity to the FtsZ polymers in the assembly (41). If so, a defect in DMinC targeting should result in a defect in MinC-mediated division inhibition. The finding that ZipA is required for DMinC/DicB targeting allowed this prediction to be tested.
For these experiments, the minCDE+ strains PB103 (wild type), JE34 (ftsA*), and JE32 (zipA0 ftsA*), carrying either pJE39 (Plac::gfp-dicB) or pDR119 (Plac::gfp-minD) were grown in the absence or presence of IPTG, and the lengths of cells were measured (Table 3). Expression of GFP-DicB in PB103/pJE39 led to the formation of nonseptate filaments with an average length of ∼90 μm, confirming that the fusion retained DicB function. The fusion similarly caused a significant division block in JE34/pJE39, although the filaments were about half the length of those obtained with the ftsA+ strain. This partial resistance to MinC/DicB-induced division inhibition may be due to increased stability of FtsZ rings in FtsA* cells, as suggested previously (18). Interestingly, JE32/pJE39 cells divided readily and had an average length similar to that of cells grown in the absence of IPTG. The different effects of GFP-DicB in the three strains were not due to different expression levels of the fusion, as determined by Western analyses (data not shown). Moreover, JE32/pJE39 cells failed to filament, even when grown with saturating concentrations (>500 μM) of IPTG (data not shown). Thus, the absence of ZipA in this strain rendered it resistant to DicB-induced division inhibition, supporting the notion that DMinC/DicB targeting to FtsZ-ZipA assemblies is crucial for effective stimulation of MinC action by DicB.
TABLE 3.
Cells lacking ZipA are resistant to DicB-induced division inhibition
| Strain | Genotype | IPTGa (μM) | Lengthb (μm)
|
nc | |
|---|---|---|---|---|---|
| Avg | Range | ||||
| PB103/pJE39 | wt/Plac::gfp-dicB | 0 | 3.4 | 1.5-12.5 | 63 |
| JE34/pJE39 | ftsA*/Plac::gfp-dicB | 0 | 3.1 | 1.7-5.7 | 61 |
| JE32/pJE39 | ΔzipA ftsA*/Plac::gfp-dicB | 0 | 3.3 | 1.7-11.7 | 60 |
| PB103/pJE39 | wt/Plac::gfp-dicB | 100 | 89.8 | 48.8-158.1 | 56 |
| JE34/pJE39 | ftsA*/Plac::gfp-dicB | 100 | 46.4 | 4.1-123.2 | 54 |
| JE32/pJE39 | ΔzipA ftsA*/Plac::gfp-dicB | 100 | 5.1 | 2.3-12.7 | 60 |
| PB103/pDR119 | wt/Plac::gfp-minD | 0 | 2.6 | 1.1-5.6 | 61 |
| JE34/pDR119 | ftsA*/Plac::gfp-minD | 0 | 2.6 | 1.7-4.0 | 61 |
| JE32/pDR119 | ΔzipA ftsA*/Plac::gfp-minD | 0 | 2.9 | 1.6-6.7 | 61 |
| PB103/pDR119 | wt/Plac::gfp-minD | 500 | 27.5 | 3.2-101.9 | 64 |
| JE34/pDR119 | ftsA*/Plac::gfp-minD | 500 | 4.7 | 2.3-9.1 | 60 |
| JE32/pDR119 | ΔzipA ftsA*/Plac::gfp-minD | 500 | 13.5 | 2.8-78.1 | 60 |
Cells were grown in LB-AMP medium supplemented with IPTG as indicated to an OD600 of 0.3 to 0.5 and analyzed by microscopy.
Each culture contained some nonfluorescent cells. These were presumed to have lost the plasmid and were not included in the analyses.
Number of cells analysed.
As expected, overexpression of GFP-MinD in PB103/pDR119 also led to a division block, yielding filaments with an average length of 28 μm. JE32/pDR119 cells were less sensitive to MinC/MinD action but still formed filaments about half as long as those of PB103/pDR119. The finding that zipA0 ftsA* cells (JE32) were completely resistant to DicB-induced, but not MinD-induced, division inhibition correlated well with the finding that ZipA is needed for DicB-dependent, but not MinD-dependent, targeting of DMinC to septal rings.
The finding that JE32/pDR119 cells were more resistant to MinC/MinD-mediated division inhibition than PB103/pDR119 cells is again consistent with the proposal that the FtsA* protein stabilizes Z-ring assemblies to a significant degree (18). As shown in Table 3, JE34 (zipA+ ftsA*) cells displayed an even greater resistance to GFP-MinD overexpression than JE32 (zipA0 ftsA*) cells. This result suggests that the association of ZipA with FtsA*-decorated Z rings renders these structures even more stable, which is consistent with ZipA and FtsA* being capable of fulfilling similar functions (18).
Yeast two-hybrid assays support a direct interaction between DMinC/DicB and ZipA.
The results above showed that ZipA is required for localization of DicB-GFP-DMinC to septal rings, as well as for rendering cells sensitive to DicB-induced division inhibition. In the simplest scenario, the MinC/DicB complex recognizes ZipA-bound FtsZ polymers via a specific interaction between its DMinC/DicB moiety and ZipA, exposing the FtsZ polymers to the depolymerizing activity of its ZMinC moiety. We used a yeast two-hybrid assay to obtain evidence for an interaction between ZipA and DMinC/DicB.
As summarized in Table 4, we measured a significant interaction (17.2 units) between a fusion protein containing the BD of yeast Gal4 and the DicB-GFP-DMinC chimera used in our localization studies [BD-DicB-GFP-MinC(108-231)] and a fusion containing the transcription activating domain of Gal4 (AD) and the cytoplasmic portion of ZipA [AD-ZipA(23-328)]. In the inverse configuration, the interaction between BD-ZipA(23-328) and AD-DicB-GFP-DMinC was weaker (7.3 units), but still significant. Although activities in both genetic configurations were relatively low, the measured values were both reproducible and significantly higher than those obtained with control combinations where either the ZipA, DMinC, and/or DicB domain was missing (Table 4). These results support the notion that a DMinC/DicB heteromer can bind the cytoplasmic part of ZipA directly.
MinD stimulates division inhibition by a membrane-tethered version of MinC.
As shown above, cells in which DMinC/DicB failed to target to rings were also resistant to DicB-induced division inhibition, indicating that DicB-directed targeting of MinC is crucial for the latter to efficiently interfere with FtsZ ring assembly.
In the case of MinC/MinD-mediated division inhibition, it is unclear whether specific targeting of the complex to FtsZ assemblies contributes to the efficiency of division inhibition. Activation of MinC-mediated division inhibition by MinD is proposed to consist of at least two steps (41). In the first step, bulk recruitment of MinC by MinD from the cytoplasm to the membrane increases the concentration of MinC at the membrane. In the second step, membrane-associated MinC/MinD is more specifically targeted to membrane-associated FtsZ assemblies via an interaction involving the DMinC/MinD moiety of the complex.
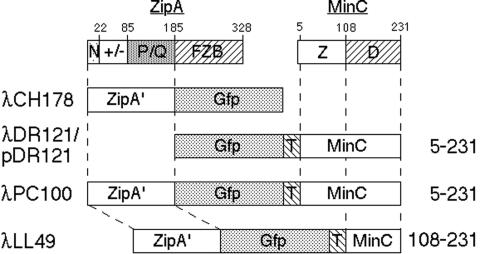
To be able to assess the contribution of the second step in MinC/MinD-mediated division inhibition, we sought to render the first step independent of MinD. To this end, we constructed phage λPC100 (Plac::zipA′-gfp-minC) and derivatives (Fig. 5). λPC100 encodes a fusion protein consisting of GFP fused at its N terminus to residues 1 to 183 of ZipA and at its C terminus to residues 5 to 231 of MinC. The portion of ZipA present in this fusion (ZipA′) includes the transmembrane, charged, and flexible P/Q-rich domains but lacks the FtsZ-binding domain, which is required for ZipA to associate with septal rings (23, 27, 58). Residues 5 to 231 of MinC comprise functional Z and D domains (34, 41). Phage λLL49 [Plac::zipA′-gfp-minC(108-231)] encodes a similar fusion, except that it lacks the ZMinC domain. Phages λDR121 (Plac::gfp-minC) and λCH178 (Plac::zipA′-gfp) are also similar to λPC100, except that λDR121 encodes GFP-MinC without a ZipA′ moiety and λCH178 encodes ZipA′-GFP without a MinC moiety (Fig. 5).
FIG. 5.
Constructs for MinD-independent recruitment of MinC to the membrane. Shown are the domain structures of ZipA and MinC and of GFP fusions containing ZipA and/or MinC domains. Abbreviations for ZipA domains: N, transmembrane; ±, highly charged; P/Q, P/Q rich; FZB, FtsZ binding. Z indicates the division inhibition domain of MinC and D the activator-binding domain. Names of genetic constructs are indicated on the left. Residues of MinC encoded by the constructs are given on the right. T indicates a T7 tag peptide which links the GFP and MinC portions of the fusion proteins.
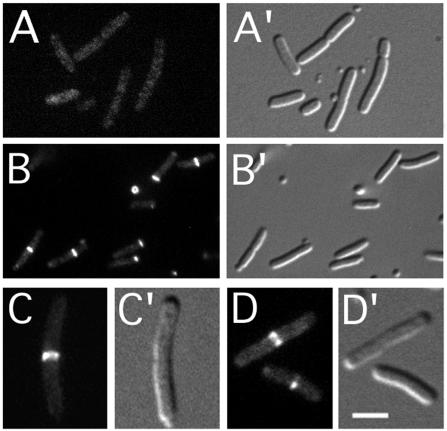
The phages were introduced into strain LL1 (ΔminCDE lon::Tn10), and lysogens were examined after growth in the presence of IPTG. Whereas the GFP-MinC fusion localized throughout the cytoplasm of LL1(λDR121) cells, the other three fusions accumulated along the cell periphery (Fig. 6; Table 5). Interestingly, whereas expression of GFP-MinC, ZipA′-GFP, or ZipA′-GFP-DMinC did not noticeably interfere with cell division, expression of ZipA′-GFP-MinC caused extensive filamentation in LL1(λPC100) cells (Fig. 6; Table 5). The difference in phenotype was not due to large differences in cellular concentrations of the fusions, as determined by Western analyses (Table 5). Like native MinC (15), GFP-MinC is capable of blocking cell division in the absence of MinD and DicB, but it only does so at a cellular concentration that is too high to be reached through transcription from a single copy of λDR121 integrated in the chromosome (Table 5 and data not shown). When GFP-MinC was expressed from plasmid pDR121 instead, cell lengths increased noticeably only at inducer concentrations of and above 75 μM. Table 5 shows that the concentration of GFP-MinC in such cells was at least ninefold higher than the concentration of ZipA′-GFP-DMinC required to cause severe division inhibition in LL1(λPC100) cells.
FIG. 6.
Localization of ZipA′-GFP-MinC and derivatives. (A to C) Cells of LL1 (ΔminCDE lon), lysogenic for λDR121 (Plac::gfp-minC) (A), λCH178 (Plac::zipA′-gfp) (B), and λPC100 (Plac::zipA′-gfp-minC) (C). (D and E) Cells of the lysogen DR109(λLL49) [ΔminCDE recA (Plac::zipA′-gfp-DminC)] carrying either pDB326 (vector) (D) or pCH204 (minD) (E). Cells were grown at 37°C (A to C) or 30°C (D and E) in minimal medium supplemented with 75 (A to C) or 50 (D and E) μM IPTG and were imaged live. Bar, 2 μm.
TABLE 5.
MinD-independent membrane localization of, and division inhibition by, a ZipA′-GFP-MinC fusion protein
| Phage or plasmid | Fusion protein | IPTGa (μM) | Relative level of fusion proteinb | Locc | Length (μm)
|
ne | |
|---|---|---|---|---|---|---|---|
| Avg | Ranged | ||||||
| None | 75 | 6.0 | 1.8-31.4 | 131 | |||
| λCH178 | ZipA′-GFP | 75 | 1.5 | M | 5.7 | 1.9-41.9 | 113 |
| λDR121 | GFP-MinC | 75 | 0.7 | C | 5.1 | 1.8-17.7 | 122 |
| λDR121 | GFP-MinC | 250 | 1.1 | C | 5.6 | 1.6-59.0 | 138 |
| pDR121 | GFP-MinC | 75 | 8.6 | C | 9.6 | 1.7-65.0 | 131 |
| λPC100 | ZipA′-GFP-MinC | 75 | 1.0 | M | 20.7 | 3.1-114.4 | 133 |
| λLL49 | ZipA′-GFP-DMinC | 75 | 1.8 | M | 6.2 | 2.1-65.0 | 134 |
Cells of strain LL1 (ΔminCDE lon), harboring the indicated phage or plasmid, were grown at 37°C to an OD600 of 0.2 to 0.3 in minimal medium supplemented with IPTG as indicated.
Protein levels were measured by quantitative Western analyses, using anti-GFP antibodies. Values are normalized to that of the ZipA′-GFP-MinC fusion in LL1 (λPC100).
The localization (Loc) of fusion proteins, and the average cell lengths, were determined by microscopy. M, concentrated along the membrane; C, cytoplasmic.
Note that cultures of LL1 normally contain a small fraction of filaments due to the lon mutation in this strain.
Number of cells measured.
The finding that appending ZipA′ to GFP-MinC rendered it a significantly more potent division inhibitor is consistent with the proposal that bulk recruitment of MinC to the membrane is an important step in the stimulation of MinC activity by MinD (36, 41, 61).
In addition, because the ZipA′-GFP-MinC fusion inserts in the membrane in a MinD-independent fashion, it allowed us to test whether MinD has an effect on MinC-mediated division inhibition beyond this step. For this test, we used strain DR109(λPC100) [ΔminCDE (Plac::zipA′-gfp-minC)] carrying either the minD plasmid pCH204 [repA(Ts) minD] or its parent vector, pDB326 [repA(Ts)]. Cells were grown at 30°C in the presence of IPTG, and the lengths of filaments were compared. Table 6 shows that DR109(λPC100)/pCH204 cells were significantly more filamentous than DR109(λPC100)/pDB326 cells, even though the concentrations of the ZipA′-GFP-MinC fusion in these cells were virtually identical. In addition, virtually all of the fusion detected on Western blotting was intact, indicating that very little (if any) untethered MinC was present in these cells (data not shown). The length difference between MinD+ and MinD− cells was not further exaggerated by lowering the cellular concentration of ZipA′-GFP-MinC, as DR109(λPC100)/pCH204 cells were also about twice as long as DR109(λPC100)/pDB326 cells after growth in the presence of 25 μM IPTG (data not shown).
TABLE 6.
MinD stimulates division inhibition by the membrane-tethered ZipA′-GFP-MinC fusion protein
| Phagea | Fusion protein | Plasmida (genotype) | Relative level of fusion proteinb | Locc | Length (μm)
|
ne | |
|---|---|---|---|---|---|---|---|
| Avg | Ranged | ||||||
| λPC100 | ZipA′-GFP-MinC | pDB326 | 1.0 | M | 18.3 | 1.1-87.0 | 135 |
| pCH204 (minD) | 0.9 | M | 44.7 | 2.7-132.1 | 134 | ||
| λLL49 | ZipA′-GFP-DMinC | pDB326 | 1.1 | M | 4.2 | 1.5-11.5 | 116 |
| pCH204 (minD) | 1.0 | R | 6.0 | 1.7-25.8 | 129 | ||
Cells of strain DR109 (ΔminCDE recA), harboring the indicated phage and plasmid, were grown at 30°C to an OD600 of 0.12 to 0.14 in minimal medium supplemented with 50 μM IPTG. pDB326 was the parent vector.
Protein levels were measured by quantitative Western analyses, using anti-GFP antibodies. Values are normalized to that of the ZipA′-GFP-MinC fusion in DR109(λPC100)/pDB326. The level of ZipA′-GFP-MinC in these cells corresponded to approximately four times the level of native MinC in strain CH3 (recA) as determined by quantitative Western analyses, using anti-MinC antibodies.
The localization (Loc) of fusion proteins, and the average cell lengths, were determined by microscopy. M, concentrated along the membrane; R, accumulated in rings.
Note that cultures of DR109 normally contain a small fraction of filaments due to the ΔminCDE and recA mutations in this strain.
Number of cells measured.
These results indicate that, while the ZipA′-GFP-MinC fusion does not require MinD to localize to the membrane, MinD still stimulates its division-inhibitory activity. An explanation that we favor is that this stimulatory effect reflects the role of MinD in the second step of MinD-mediated activation of MinC, i.e., the specific targeting of the MinC/MinD complex to FtsZ assemblies on the membrane. This explanation can only be valid if the ZipA′-GFP-MinC fusion is indeed still subject to MinD-dependent targeting to septal rings. To explore this point, we compared the localization of the ZipA′-GFP-DMinC fusion in DR109(λLL49) in the presence (pCH204) and absence (pDB326) of minD. Whereas ZipA′-GFP-DMinC accumulated along the membrane in DR109(λLL49)/pDB326 cells (Fig. 6D), it accumulated in rings in DR109(λLL49)/pCH204 cells (Fig. 6E). These results show that even when MinC is tethered to the membrane via the ZipA transmembrane domain, it is indeed still subject to MinD-dependent targeting to its substrate.
DISCUSSION
This study further contributes to our understanding of the mechanisms by which MinD and DicB activate the division-inhibitory activity of MinC. Previous observations revealed that complexes containing DMinC together with either MinD or DicB possess a high affinity for septal ring assemblies in vivo (41).
Here we showed that decoration of septal rings by the DMinC/DicB complex requires the presence of the ring component ZipA. In addition, we showed that the DMinC/DicB complex still recognizes Z rings in the absence of ZapA, as well as in filaments in which FtsA has been severely depleted. Although ZipA still associates with Z rings in FtsA− filaments (24, 47), recruitment of all other known essential septal ring proteins (FtsK, -Q, -L, -B, -W, -I, and -N) is impaired (1, 6, 16, 51). It is unlikely, therefore, that any of these downstream components are required for the recognition of FtsZ/ZipA structures by DMinC/DicB. This argument was further supported by our two-hybrid analyses, which indicated a direct interaction between DMinC/DicB and ZipA.
These observations led to a simple model of DicB-induced division inhibition. DicB binds the D-domain of MinC in the cytoplasm (41), and the complex gains a specific affinity for ZipA, which itself interacts with membrane-associated FtsZ polymers via a specific interaction between its FZB domain and the extreme C terminus of FtsZ (27, 29, 47, 49, 54, 55). This brings the Z domain of MinC in close proximity to the FtsZ polymers, resulting in efficient depolymerization and dissolution of the FtsZ/ZipA polymeric complex.
It is interesting that when FtsZ polymerization is inhibited by SfiA, ZipA disperses along the membrane while the DMinC/DicB complex appears throughout the cytoplasm (41). This suggests that the affinity of DMinC/DicB for ZipA that is associated with polymeric FtsZ is significantly higher than that for ZipA which is not. Binding to polymeric FtsZ may change the conformation of ZipA, or FtsZ itself may contribute a binding surface for DMinC/DicB in the polymeric FtsZ/ZipA complex.
This work also established that the specific Z ring target of the DMinC/MinD complex is distinct from that of DMinC/DicB. Thus, DMinC/MinD still decorated Z rings in the complete absence of ZipA or ZapA, as well as upon depletion of FtsA, implying that none of these three FtsZ-associated proteins are specifically required for DMinC/MinD targeting to FtsZ assemblies. Normally, both FtsA and ZipA are required to recruit all known remaining essential septal ring proteins (1, 6, 16, 25, 51, 60). Again, it is therefore unlikely that any of the known division proteins are specifically required to attract DMinC/MinD to FtsZ polymers.
How then does DMinC/MinD recognize polymeric FtsZ? It is possible that FtsA and ZipA play redundant roles with respect to attracting DMinC/MinD to FtsZ assemblies. Both proteins bind FtsZ in a mutually independent fashion (24) and each may induce some arrangement of the polymeric complex that is attractive to DMinC/MinD. The notion that FtsA and ZipA functions may indeed be partially redundant is supported by the discoveries that the formation of complete FtsZ rings requires the presence of either FtsA or ZipA (60) and that the mutant FtsA* protein renders ZipA dispensable (18). Because at least one of the two proteins is required for Z rings to form in vivo, it will be difficult to test for a potential redundancy of FtsA and ZipA in attracting DMinC/MinD complexes to FtsZ assemblies by the microscopic techniques used here. It is equally possible that neither FtsA nor ZipA are directly involved in attracting DMinC/MinD to FtsZ polymers. Thus, targeting of DMinC/MinD could involve some FtsZ-associated factor whose identity remains to be determined or could simply result from a direct interaction between the DMinC/MinD complex and FtsZ polymers, which would be the simplest mechanism. Despite some effort, we have not yet obtained firm evidence for the latter possibility.
The results of this study also emphasize the importance of DMinC/activator targeting to FtsZ assemblies for stimulation of MinC-mediated division inhibition by the activators. The finding that cells devoid of ZipA (ΔzipA ftsA*) are completely resistant to DicB-induced, but not MinD-induced, filamentation shows the significance of DMinC/DicB targeting to FtsZ/ZipA assemblies in executing the DicB-dependent division block. Taken together, our results indicate that DicB acts as a coassembly factor which allows MinC to connect with ZipA-decorated FtsZ polymers. This property of DicB may well be sufficient to explain its ability to cause a division block.
Stimulation of MinC function by MinD is more complicated. Like DicB, MinD appears to act as a coassembly factor allowing MinC to connect with its substrate (i.e., membrane-associated FtsZ polymers), but targeting is likely to occur in consecutive stages with increased spatial specificity.
In the first stage, MinD-ATP assembles on the membrane and recruits MinC from the cytoplasm (36, 38, 45, 61). In wild-type cells, assembly of the MinC/MinD-ATP complex is typically confined to the membrane of one half of the cell at any one time, as MinE action drives the regular dissolution and reformation of complexes during the oscillation cycle (17, 26, 33, 35, 45, 63, 64, 66, 67). In MinE− cells, MinC/MinD complexes can be found all along the cell membrane (36, 61, 63, 64). In either case, recruitment of MinC to the membrane at this stage occurs independently of FtsZ but will lead to an elevated local concentration of ZMinC at the membrane.
For the second stage, we envision that ZMinC is more precisely directed to its substrate within the two-dimensional plane of the membrane. This search-and-destroy process depends on a specific interaction between the DMinC/MinD-ATP moiety of the complex and membrane-associated FtsZ polymeric complexes.
As a step towards understanding the potential contribution of each stage to the stimulation of MinC-mediated division inhibition by MinD, we studied the properties of membrane-tethered versions of MinC. In the absence of MinD, a ZipA′-GFP-MinC fusion accumulated along the membrane. Furthermore, the fusion was a far more potent inhibitor of division than soluble MinC, supporting the notion that stage 1 (bulk recruitment of MinC to the membrane by MinD) is likely to contribute significantly to activation of MinC function. Very recently, Szeto et al. performed a similar experiment, using a derivative of MinC to which the short C-terminal membrane targeting sequence of B. subtilis MinD had been appended to the C terminus of E. coli MinC (68).
Interestingly, although the ZipA′-GFP-MinC fusion localized to the membrane in a MinD-independent fashion, the presence of MinD enhanced division inhibition by the fusion and also caused a specific accumulation of ZipA′-GFP-DMinC on septal ring structures. Although these results do not constitute conclusive evidence, they are consistent with the proposal that both stages of MinD-directed localization of MinC contribute to the efficiency of ZMinC activity in the cell.
Acknowledgments
We thank Tom Bernhardt, Piyali Chatterjee, and David Raskin for help in strain and plasmid construction.
This work was supported by National Institutes of Health grant GM-57059. In addition, J.E.J. and L.L.L. were supported by NIH NRSA Institutional Training grant T32GM08056.
REFERENCES
- 1.Addinall, S. G., and B. Holland. 2002. The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J. Mol. Biol. 318:219-236. [DOI] [PubMed] [Google Scholar]
- 2.Béjar, S., F. Bouché, and J.-P. Bouché. 1988. Cell division inhibition gene dicB is regulated by a locus similar to lambdoid bacteriophage immunity loci. Mol. Gen. Genet. 212:11-19. [DOI] [PubMed] [Google Scholar]
- 3.Béjar, S., and J.-P. Bouché. 1985. A new dispensable genetic locus of the terminus region involved in control of cell division in Escherichia coli. Mol. Gen. Genet. 201:146-150. [DOI] [PubMed] [Google Scholar]
- 4.Bernhardt, T. G., and P. A. J. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, E., and J. Lutkenhaus. 1993. Cell division inhibitors, SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 175:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 7.Cam, K., S. Béjar, D. Gil, and J.-P. Bouché. 1988. Identification and sequence of division inhibitor gene dicB suggests expression from an internal in-frame translation start. Nucleic Acids Res. 16:6327-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordell, S. C., R. E. Anderson, and J. Lowe. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20:2454-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordell, S. C., and J. Lowe. 2001. Crystal structure of the bacterial cell division regulator MinD. FEBS Lett. 492:160-165. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Boer, P. A. J., R. E. Crossley, A. R. Hand, and L. I. Rothfield. 1991. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J. 10:4371-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1990. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc. Natl. Acad. Sci. USA 87:1129-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1989. A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56:641-649. [DOI] [PubMed] [Google Scholar]
- 14.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1988. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J. Bacteriol. 170:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Boer, P. A. J., R. E. Crossley, and L. I. Rothfield. 1992. Roles of MinC and MinD in the site-specific septation block mediated by the MinCDE system of Escherichia coli. J. Bacteriol. 174:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu, X., Y. L. Shih, Y. Zhang, and L. I. Rothfield. 2001. The MinE ring required for proper placement of the division site is a mobile structure that changes its cellular location during the Escherichia coli division cycle. Proc. Natl. Acad. Sci. USA 98:980-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geissler, B., D. Elraheb, and W. Margolin. 2003. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 100:4197-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes, K., J. Moller-Jensen, and R. Bugge Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 20.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 22.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hale, C. A., and P. A. J. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Hale, C. A., and P. A. J. de Boer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ, and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale, C. A., and P. A. J. de Boer. 2002. ZipA is required for recruitment of FtsK, FtsQ, FtsL, and FtsN to the septal ring in Escherichia coli. J. Bacteriol. 184:2552-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale, C. A., H. Meinhardt, and P. A. J. de Boer. 2001. Dynamic localization cycle of the cell division regulator MinE in E. coli. EMBO J. 20:1563-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hale, C. A., A. C. Rhee, and P. A. J. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haney, S. A., E. Glasfeld, C. Hale, D. Keeney, Z. He, and P. A. de Boer. 2001. Genetic analysis of the E. coli FtsZ-ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J. Biol. Chem. 276:11980-11987. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi, I., T. Oyama, and K. Morikawa. 2001. Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J. 20:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henry, K. R., K. D'Hondt, J. Chang, T. Newpher, K. Huang, R. T. Hudson, H. Riezman, and S. K. Lemmon. 2002. Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of sla2p in yeast. Mol. Biol. Cell 13:2607-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard, M., A. D. Rutenberg, and S. de Vet. 2001. Dynamic compartmentalization of bacteria: accurate division in E. coli. Phys. Rev. Lett. 87:278102. [DOI] [PubMed] [Google Scholar]
- 33.Hu, Z., E. P. Gogol, and J. Lutkenhaus. 2002. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc. Natl. Acad. Sci. USA 99:6761-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu, Z., and J. Lutkenhaus. 2000. Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J. Bacteriol. 182:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu, Z., and J. Lutkenhaus. 2001. Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol. Cell 7:1337-1343. [DOI] [PubMed] [Google Scholar]
- 36.Hu, Z., and J. Lutkenhaus. 1999. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 34:82-90. [DOI] [PubMed] [Google Scholar]
- 37.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu, Z., C. Saez, and J. Lutkenhaus. 2003. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J. Bacteriol. 185:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang, J., C. Cao, and J. Lutkenhaus. 1996. Interaction between FtsZ and inhibitors of cell division. J. Bacteriol. 178:5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson, J. E., L. L. Lackner, and P. A. J. de Boer. 2002. Targeting of DMinC/MinD and DMinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ-rings. J. Bacteriol. 184:2951-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruse, K. 2002. A dynamic model for determining the middle of Escherichia coli. Biophys. J. 82:618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labie, C., F. Bouché, and J.-P. Bouché. 1989. Isolation and mapping of Escherichia coli mutations conferring resistance to division inhibition protein DicB. J. Bacteriol. 171:4315-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Labie, C., F. Bouché, and J.-P. Bouché. 1990. Minicell-forming mutants of Escherichia coli: suppression of both DicB- and MinD-dependent division inhibition by inactivation of the minC gene product. J. Bacteriol. 172:5852-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lackner, L. L., D. M. Raskin, and P. A. de Boer. 2003. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J. Bacteriol. 185:735-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levin, P. A., R. L. Schwartz, and A. D. Grossman. 2001. Polymer stability plays an important role in the positional regulation of FtsZ. J. Bacteriol. 183:5449-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, Z., A. Mukherjee, and J. Lutkenhaus. 1999. Recruitment of ZipA to the division site by interaction with FtsZ. Mol. Microbiol. 31:1853-1861. [DOI] [PubMed] [Google Scholar]
- 48.Lutkenhaus, J., and M. Sundaramoorthy. 2003. MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol. Microbiol. 48:295-303. [DOI] [PubMed] [Google Scholar]
- 49.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 51.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 52.Meinhardt, H., and P. A. J. de Boer. 2001. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. USA 98:14202-14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mileykovskaya, E., I. Fishov, X. Fu, B. D. Corbin, W. Margolin, and W. Dowhan. 2003. Effects of phospholipid composition on MinD-membrane interactions in vitro and in vivo. J. Biol. Chem. 278:22193-22198. [DOI] [PubMed] [Google Scholar]
- 54.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO. J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moy, F. J., E. Glasfeld, L. Mosyak, and R. Powers. 2000. Solution structure of ZipA, a crucial component of Escherichia coli cell division. Biochemistry 39:9146-9156. [DOI] [PubMed] [Google Scholar]
- 56.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulder, E., and C. L. Woldringh. 1989. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J. Bacteriol. 171:4303-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohashi, T., C. A. Hale, P. A. De Boer, and H. P. Erickson. 2002. Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J. Bacteriol. 184:4313-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pichoff, S., and J. Lutkenhaus. 2001. Escherichia coli division inhibitor MinCD blocks septation by preventing Z-ring formation. J. Bacteriol. 183:6630-6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pichoff, S., and J. Lutkenhaus. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raskin, D. M., and P. A. J. de Boer. 1999. MinDE dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 181:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raskin, D. M., and P. A. J. de Boer. 1997. The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91:685-694. [DOI] [PubMed] [Google Scholar]
- 63.Raskin, D. M., and P. A. J. de Boer. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:4971-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rowland, S. L., X. Fu, M. A. Sayed, Y. Zhang, W. R. Cook, and L. I. Rothfield. 2000. Membrane redistribution of the Escherichia coli MinD protein induced by MinE. J. Bacteriol. 182:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakai, N., M. Yao, H. Itou, N. Watanabe, F. Yumoto, M. Tanokura, and I. Tanaka. 2001. The three-dimensional structure of septum site-determining protein MinD from Pyrococcus horikoshii OT3 in complex with Mg-ADP. Structure 9:817-826. [DOI] [PubMed] [Google Scholar]
- 66.Shih, Y. L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suefuji, K., R. Valluzzi, and D. RayChaudhuri. 2002. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc. Natl. Acad. Sci. USA 99:16776-16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szeto, T. H., S. L. Rowland, C. L. Habrukowich, and G. F. King. 2003. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J. Biol. Chem. 278:40050-40056. [DOI] [PubMed] [Google Scholar]
- 69.Szeto, T. H., S. L. Rowland, and G. F. King. 2001. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J. Bacteriol. 183:6684-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward, J. E., and J. Lutkenhaus. 1985. Overproduction of FtsZ induces minicell formation in E. coli. Cell 42:941-949. [DOI] [PubMed] [Google Scholar]
- 72.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu, X.-C., and W. Margolin. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315-326. [DOI] [PubMed] [Google Scholar]