Abstract
An atypical, Stx2-producing, pathogenic Escherichia coli O157:H− strain has been isolated with increasing frequency from hemolytic uremic syndrome patients in Germany. The lack of the H7 antigen coupled with the strain's ability to ferment sorbitol and express β-glucuronidase have complicated its detection and identification. In this study, we have determined that the loss of motility in these German sorbitol-fermenting (SF) O157 strains is due to a 12-bp in-frame deletion in flhC that is required for transcriptional activation of genes involved in flagellum biosynthesis. Either complementation with a functional flhC or repair of this mutation restored H7 antigen expression and motility. PCR analysis of several nonmotile E. coli O157 strains from various geographical sources confirmed that the 12-bp flhC deletion is found only in the cluster of German SF O157 strains, providing a potentially useful marker by which these atypical strains can be identified. The loss of motility via mutations in the flhDC operon that we observed in the German SF O157 strains is consistent with a similar phenomenon currently observed in a significant subset of other important gram-negative pathogens.
Enterohemorrhagic Escherichia coli (EHEC) of serotype O157:H7 is routinely identified by its inability to ferment sorbitol, its lack of β-glucuronidase activity, and the presence of somatic (O)157 and flagellar (H)7 antigens. There are many O157:H7 phenotypic variants, but the Shiga toxin 2 (Stx2)-producing, sorbitol-fermenting (SF) O157:H− strains originally isolated from hemolytic uremic syndrome (HUS) patients in Germany (17) have emerged as important human pathogens in Europe (4, 7, 8, 20). The German SF O157 strains, which have β-glucuronidase activity and are nonmotile (NM) (21), have been found in up to 40% of HUS cases (20). These strains are not detected by assays routinely used for O157:H7 because of their atypical phenotype, namely, the ability to ferment sorbitol, express β-glucuronidase activity, and are NM. As a consequence, German SF O157 strains are only identified by testing first for the stx gene or Stx, with those Stx-positive isolates being further characterized for the presence of the other O157:H7-associated phenotypes and virulence markers. Despite the many phenotypic differences, however, the German SF O157 strains are a unique clone that is closely related genetically to O157:H7 (13, 21) and is postulated to have diverged from O157:H7, in part by the loss of motility (26).
Motility in E. coli is under complex genetic control and can be induced or repressed physiologically by environmental factors, including glucose (catabolite repression), temperature, and high-salt conditions (24). Most O157:NM strains that produce Stx also possess other O157:H7 trait markers and, in some cases, can be induced to express the H7 antigen that restores motility, suggesting that these strains are NM due to environmental factors (12). The German SF O157 strains, however, seem to be an exception, as motility cannot be restored by passages in motility medium (12).
Flagellum biosynthesis in E. coli requires ca. 40 genes (3), 30 of which encode structural components of the basal body, motor, and filament. These flagellar genes are under a three-class regulatory system and are sequentially expressed in the order of assembly. Class I genes consist of flhDC, the master control operon whose products are required to activate class II genes that encode the structural proteins for the basal body and hook, as well as FliA and FlgM, two regulators of class III genes. Class III genes encode the motor (Mot), chemotaxis (Che), and filament (FliC) proteins (3).
The German SF O157 strains, although NM, carry the entire fliC gene that exhibits the restriction fragment length polymorphism profile characteristic of strains in the O157:H7 group (16). Previously, the fliC gene of 493-89, a German SF O157 strain, was reported to contain two single-site nucleotide insertions that produced a frameshift mutation that could, presumably, account for the loss of motility (30). In this study, we examined the motility-related regulatory and structural genes to identify the genetic defect(s) that caused the NM phenotype in the German variant.
MATERIALS AND METHODS
Bacterial strains and characterization.
The bacterial strains used in this study are listed in Table 1. All strains were tested by five-product PCR (14), which confirmed the presence of genes encoding Shiga toxin (stx1, stx2), γ-intimin (eae), enterohemolysin (ehxA), and the +92 uidA mutation that is unique to O157:H7 and its Stx-producing NM variants (Fig. 1 and Table 1). All strains, before and after genetic manipulations, were serologically typed for the O157 and H7 antigens using the RIM O157:H7 test (REMEL, Lenexa, Kans.). Motility was determined by microscopic examination of wet mounts of cell suspensions for the presence or absence of motility. NM strains were passed in motility agar (BD Diagnostic Systems, Franklin Lakes, N.J.) several times and reexamined for motility.
TABLE 1.
E. coli strains used in the study
| Strain | Serotypea | Motb | stx1c | stx2d | uidAe | eaef | ehxAg | ΔflhCh | Sourcej |
|---|---|---|---|---|---|---|---|---|---|
| 35150 | O157:H7 | + | + | + | + | + | + | − | ATCC |
| 43890 | O157:H7 | + | + | − | + | + | + | − | ATCC |
| 493-89 | O157:H− | − | − | + | + | + | + | + | Karch, Germany |
| CB569 | O157:H− | − | − | + | + | + | + | + | Karch, Germany |
| CB1009 | O157:H− | − | − | − | + | + | + | + | Karch, Germany |
| 5412 | O157:H− | − | − | + | + | + | + | + | Karch, Germany |
| 514-91 | O157:H− | − | − | + | + | + | + | + | Karch, Germany |
| 210-89 | O157:H− | − | − | − | + | + | + | + | Karch, Germany |
| TT7 | O157:H− | − | + | − | + | + | + | − | Takeda, Japan |
| 3204-92 | O157:NM | − | − | + | + | + | + | − | CDC |
| H0482 | O157:NM | − | − | + | + | + | + | − | CDC |
| 7123 | O157:NM | − | − | − | − | − | − | − | USDA |
| DEC5A | O55:H7 | + | − | − | − | + | − | − | Whittam, MSU |
| DEC5D | O55:H7 | + | − | − | − | + | − | − | Whittam, MSU |
| DH5α | − | − | − | − | − | − | NDi |
Designated H− or NM by original source.
Motile phenotype.
Shiga toxin 1 gene.
Shiga toxin 2 gene.
+92 mutation in glucuronidase gene.
γ-Intimin gene.
Enterohemolysin gene.
Presence of 12-bp flhC deletion.
ND, not done.
ATCC, American Type Culture Collection; CDC, Centers for Disease Control and Prevention; USDA, U.S. Department of Agriculture; MSU, Michigan State University.
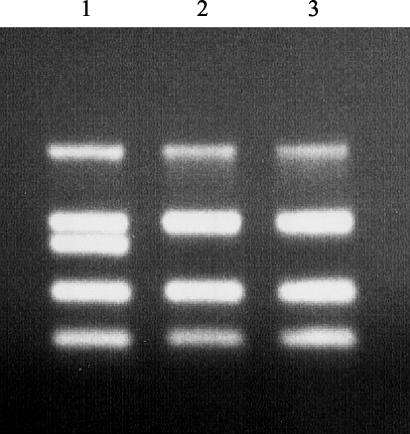
FIG. 1.
Agarose gel electrophoresis of five-product PCR amplification products derived from representative strains used in this study. Lanes: 1, 35150; 2, 493-89; 3, 5412. The products (top to bottom) are stx2 (584 bp), γ-eae (397 bp), stx1 (348 bp), uidA (252 bp), and ehxA (166 bp).
Plasmid constructs used in complementation studies.
The plasmid constructs used in this study are shown in Table 2, and the primer sequences used to amplify the respective genes are shown in Table 3. Constructs were made as described below.
TABLE 2.
Plasmid constructs engineered for this study
| Plasmid | Description | Primers used |
|---|---|---|
| pSM1 | pBluescript SK(−) containing the 35150 fliC at the vector EcoRV site | F-FLIC1/R-FLIC2 |
| pSM2 | pBluescript SK(−) containing the 493-89 fliC at the vector EcoRV site | F-FLIC1/R-FLIC2 |
| pSM3 | pBluescript SK(−) containing the CB569 fliC at the vector EcoRV site | F-FLIC1/R-FLIC2 |
| pSM4 | pTrc99A containing the 35150 fliC ORF for expression from the vector Trc promoter | NAa |
| pSM5 | pBluescript SK(−) containing the 35150 flhDC operon at the vector EcoRV site | SRM11A/SRM16 |
| pSM6 | pBluescript SK(−) containing the 493-89 flhDC operon at the vector EcoRV site | SRM11A/SRM16 |
| pSM7 | pACYC184 containing the 35150 flhDC operon at the vector EcoRV site; insert is oriented with the native flhDC promoter distal to the vector tetracycline (Ter) promoter | SRM11A/SRM20 |
| pSM8 | pACYC184 containing the DEC5A flhDC operon and promoter at the vector EcoRV site; insert is oriented with the native promoter oriented adjacent to the Tcr promoter | SRM11A/SRM20 |
| pSM9 | pACYC184 containing the 493-89 flhDC operon and promoter at the vector EcoRV site; insert is oriented with the native promoter adjacent to the vector Tcr promoter | SRM11A/SRM20 |
| pSM10 | pACYC184 containing the 493-89 flhDC operon and promoter at the vector EcoRV site; insert is oriented with the native promoter distal to the vector Tcr promoter | SRM11A/SRM20 |
| pSM11 | pBluescript SK(−) containing the 493-89 flhDC operon with the 12-bp flhC deletion repaired by two-round PCR; operon is inserted into the vector at the EcoRV site | SRM11A/SRM20 and SRM51/SRM52 |
| pSM12 | pBluescript SK(−) containing the 35150 flhDC operon with the flhC F126A mutation; operon is inserted into the cloning vector at the EcoRV site. | SRM11A/SRM20 and SRM51/SRM59 |
| pSM13 | pBluescript SK(−) containing the 35150 flhDC operon with the flhC V127A mutation; operon is inserted into the cloning vector at the EcoRV site | SRM11A/SRM20 and SRM51/SRM60 |
| pSM14 | pBluescript SK(−) containing the 35150 flhDC operon with the flhC E128A mutation; operon is inserted into the cloning vector at the EcoRV site. | SRM11A/SRM20 and SRM51/SRM61 |
| pSM15 | pBluescript SK(−) containing the 35150 flhDC operon with the flhC S129A mutation; operon is inserted into the cloning vector at the EcoRV site. | SRM11A/SRM20 and SRM51/SRM62 |
NA, not applicable.
TABLE 3.
Primer sequences
| Primer | Sequence (5′ → 3′) | Gene | Nucleotides |
|---|---|---|---|
| F-FLIC1 | CCA TGG CAC AAG TCA TTA ATA CCA AC | fliC | +1-24 |
| R-FLIC2 | CTA ACC CTG CAG CAG AGA CA | fliC | +1758-1739 |
| FliC2 | CGT CAT TCG CAC CAA CCT G | fliC | +457-439 |
| SRM11A | ACT GTA CCG AGA ACA ACC AGG | flhDC | 167-147 downstream of flhC stop |
| SRM16 | GTT GTA TGT GCG TGT AGT GAC G | flhDC | −175-154 |
| SRM17 | TCA CCC TGG ATG CTG TAG | pACYC184 | 1621-1638 (X06403) |
| SRM18 | CGG TCG GAC AGT GCT C | pACYC184 | 1786-1771 (X06403) |
| SRM19 | TGG TGC GGT TTG TTG AAA G | flhC | +368-386 |
| SRM20 | GAT CTG CAT CAC GCA TTA TTG | flhDC | −270-250 |
| SRM51 | CAG CAA CCA AGA CTC TGA CCA TGA CAG GAT GTT CAG | flhC | 126-106 downstream of flhC stop |
| SRM52 | CCT GGA CAT TGG TGC GGT TTG TTG AAA GTG GAT TAC TGC AAC TTT CC | flhC | +359-394 |
| SRM59 | TGG TGC GGG CTG TTG AAA G | flhC | +368-386 |
| SRM60 | GTG CGG TTT GCT GAA AGT GG | flhC | +370-389 |
| SRM61 | CGG TTT GTT GCA AGT GGA TTA C | flhC | +373-394 |
| SRM62 | GGT TTG TTG AAG CTG GAT TAC TGC | flhC | +374-397 |
| SRM86 | GCT AGT TGC TAA CCT AAC GGC T | 16S rDNA | +232-253 (Z83205) |
| SRM87 | GTG GAC TAC CAG GGT ATC TAA TC | 16S rDNA | +793-771 (Z83205) |
| SRM145 | CTG CTG GCA TTA ACC CTG G | flhC | +340-358 |
| SRM146 | CTG CCA ACA GGC TGG TGA G | flhC | +461-443 |
fliC constructs.
The fliC gene from strains 35150, 493-89, and CB569 was amplified using the F-FLIC1 and R-FLIC2 primers (16), except the forward primer was modified to match the GenBank fliC sequences (accession numbers L07388 and AE005415) and also to introduce a NcoI restriction site at the 5′ terminus. The PCR mix contained 1× Thermopol buffer (New England Biolabs [NEB], Beverly, Mass.), 6 mM MgSO4, 200 μM deoxynucleoside triphosphate (dNTP), 300 nM phosphorylated primers, and ∼1.0 × 105 bacterial cells as template. The reaction mix was heated at 95°C for 10 min, during which 0.5 U of Vent DNA polymerase (NEB) was added, followed by 32 successive cycles, each consisting of 95°C for 1 min, 55°C for 45 s, and 75°C for 2 min. The reaction was terminated with a 75°C, 7-min incubation. The ∼1,750-bp product was isolated from a 0.8% Tris-borate-EDTA (TBE) agarose gel, ligated into an EcoRV-digested and phosphatase-treated pBluescript SK(−) vector (Stratagene, San Diego, Calif.), and transformed by electroporation into E. coli DH5α cells (31). Transformants were selected on tryptic soy agar (TSA) containing 100 μg of ampicillin/ml, and plasmids from those ampicillin-resistant (Ampr) clones carrying the fliC gene from strains 35150, 493-89, and CB569 were designated pSM1, pSM2, and pSM3, respectively. Construct pSM4 was made by digesting pSM1 with NcoI and HindIII and ligating the resulting 1,750-bp fragment that contained fliC into a similarly digested pTrc99A plasmid expression vector (6).
flhDC constructs.
The flhDC operons from 35150 and 493-89 were amplified using Vent polymerase and the SRM16 and SRM11A primers. Amplification reactions were set up and performed as described above, except annealing was done at 56°C for 30 s and extension was at 75°C for 1.5 min. Properly sized amplicons were isolated, cloned into pBluescript SK(−), and transformed into DH5α. Several constructs were screened by SmaI restriction digestion to determine insert orientation. The pBluescript SK(−) constructs with the 35150 flhDC and 493-89 flhDC operons were designated pSM5 and pSM6, respectively (Table 2).
To study gene functionality, the flhDC operons from strains 35150, 493-89, and DEC5A (O55:H7) were also cloned into the pACYC184 vector (NEB). The operon from the respective strains was amplified with Vent polymerase and the SRM11A and SRM20 primer pair using conditions described above, except with a 58°C annealing temperature and for 35 cycles. Amplicons were isolated from agarose gels, ligated into EcoRV-digested, phosphatase-treated pACYC184, and transformed into DH5α. Transformants were selected on TSA containing 20 μg of chloramphenicol/ml. Plasmids from several chloramphenicol-resistant (Cmr) clones were isolated and characterized by SphI restriction digestion to determine insert orientation. The construct pSM7 contained the 35150:flhDC operon with the native promoter oriented distal to the vector tetracycline resistance (Tcr) promoter. Construct pSM8 had the DEC5A:flhDC operon with the native promoter adjacent to and transcribed in the same direction as the Tcr promoter, and pSM9 and pSM10 had the 493-89:flhDC operon inserted in opposite orientations, with pSM9 having the flhDC native promoter adjacent to and transcribed in the same direction as the Tcr promoter while pSM10 contained the insert in the opposite orientation.
Constructs for site-directed mutagenesis repair.
The plasmid construct used to repair the 12-bp deletion in 493-89 flhC was made using a two-round PCR technique (25) that introduced site-specific mutations in target sequences. In the first-round PCR, ∼150 ng of the pSM9 construct, carrying the 493-89 flhDC, was amplified by Vent polymerase in two separate reactions, one using the primer set SRM52 and SRM11A and the other using SRM20 and SRM51. Both 32-cycle PCRs were performed as described above using 54°C for 45 s for annealing, but the primer extensions at 75°C were for 25 s and 1.5 min for the SRM52 plus SRM11A and SRM20 plus SRM51 reactions, respectively. Products from each reaction mixture were purified from a 1% TBE agarose gel, mixed, and used as templates in the second-round PCR, which was performed using the SRM20 plus SRM11A primer pair and a 75°C for 1.5 min primer extension. The properly sized amplicon was isolated from a 1% agarose gel, ligated into an EcoRV-digested, phosphatase-treated pBluescript SK(−) vector, and transformed into DH5α. Plasmids from Ampr clones were screened by PCR for the presence of the insert using the vector-specific T3 and T7 promoter primers and for the absence of the 12-bp flhC deletion using deletion-specific PCR primers (see below). The construct in which the 12-bp deletion was repaired, as verified by sequencing, was designated pSM11.
The two-round PCR technique was also used to individually alter to the neutral amino acid alanine each of the four amino acids (phenylalanine 126, valine 127, glutamate 128, and serine 129) in the 35150 flhC open reading frame (ORF) region that are affected by the 12-bp deletion in 493-89. In the first-round PCR, pSM7 (∼100 ng) was amplified with the primer pair SRM20 and SRM51 and also with the primer SRM11A in combination with each of the mutation-specific primers, SRM59 (F126A), SRM60 (V127A), SRM61 (E128A), and SRM62 (S129A). Reactions were set up as described above, except that a 51°C annealing temperature was used. All amplicons were gel purified, mixed, and used in the second-round PCR with primer pair SRM11A and SRM20. Amplicons were cloned into pBluescript SK(−), screened by PCR, and sequenced to confirm that the expected site-specific alteration had been introduced. These 35150 flhC constructs were designated pSM12 (FlhC F126A), pSM13 (FlhC V127A), pSM14 (FlhC E128A), and pSM15 (FlhC S129A).
DNA sequencing.
All DNA sequencing was done double-stranded by Amplicon Express (Pullman, Wash.), using target-specific primers and the Big Dye chemistry with an Applied Biosystems 377 automated sequencer.
PCR analysis for the 12-bp deletion in flhC.
The presence or absence of the 12-bp flhC deletion in various strains was examined by PCR. The SRM19 primer, whose 3′ terminus is complementary to the 12 nucleotides deleted in 493-89, with SRM11A generates a 379-bp amplicon only from templates that do not contain the flhC deletion. The 50-μl reaction mixture contained 1× Taq polymerase buffer, 2.5 mM MgCl2, 200 μM dNTP, and a 300 nM concentration of the SRM11A and SRM19 primers, as well as a 100 nM concentration of each of the 16S ribosomal DNA gene-specific primers (SRM86 and SRM87) and ∼500 ng of DNA template. The reaction mixture was heated at 95°C for 10 min, during which 2.5 U of Taq DNA polymerase was added. Amplification was done with 32 cycles, each consisting of 95°C for 1 min, 60°C for 45 s, and 72°C for 1 min, and was terminated with a 72°C 7-min incubation. Those strains that generated the 562-bp amplicon from the 16S ribosomal DNA but did not yield the 379-bp flhC amplicon presumably carried the 12-bp flhC deletion.
The strains that tentatively carried the deletion were further tested by PCR with primers SRM145 and SRM146, which flank the 12-bp flhC deletion, to ascertain the size of the deletion. The reaction mix contained 1× Taq PCR buffer (Qiagen), 2 mM MgCl2, 200 μM dNTP, a 300 nM concentration of each primer, and 0.5 U of HotStar Taq DNA polymerase (Qiagen). After 15 min of enzyme activation at 95°C, amplification was done for 30 cycles, each consisting of 95°C for 30 s, 59°C for 30 s, and 72°C for 15 s, and terminated with a 72°C, 7-min incubation. PCR products were examined on a 5% TBE NuSieve (FMC, Rockland, Maine) agarose gel. The expected sizes of amplicons from templates containing and not containing the deletion were 122 and 110 bp, respectively.
Western blotting.
To verify flagellin protein expression, isolates were examined by Western blotting with the 15D8 monoclonal antibody that is specific for enteric flagella (15) and with anti-H7 polyclonal sera (BD Diagnostic Systems) to confirm H7 antigen production. Cell suspensions from overnight cultures grown in TSB were extracted by boiling, fractionated by discontinuous (3% stack-10% separating) sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose paper, and probed with 15D8 (1:2,000) or anti-H7 antibody (1:1,500), as described previously (12).
Electron microscopy.
The surfaces of NM and motile cells were examined for the presence of flagella by negative staining. Cells from TSA plates were suspended to a density of about 109 cells/ml in 2% phosphotungstic acid (pH 6.8), and 15-μl aliquots were applied to the surface of a carbon-coated, 300-mesh copper grid and allowed to settle for 1 min. After removing the excess liquid, the specimen grids were examined at either ×37,000 or ×79,000 magnification using a Philips 400 transmission electron microscope at an accelerating voltage of 80 kV.
Nucleotide sequence accession number.
The nucleotide sequence of the flhDC operon of 493-89 was determined from pSM6 and deposited in GenBank (accession no. AY225162).
RESULTS
Characterization of bacterial strains.
All of the O157:H7 strains and their NM variants used in this study were genetically characterized and confirmed to carry EHEC trait markers and virulence genes (Table 1). An exception was strain 7123, which had none of the markers. This strain had previously been found to cluster with an O157:H16 strain (13) and was suspected to be an O157 NM strain that has other than the H7 antigen (12). Two German SF O157 strains (CB1009 and 210-89) had lost the stx2 gene but carried all the other EHEC markers (Table 1). Each strain was serologically typed, and the NM strains were confirmed to be NM after repeated passages on motility soft agar stabs.
Sequencing of fliC.
The 5′ region of the fliC gene from strains 35150 (pSM1), 493-89 (pSM2), and CB569 (pSM3) were double-strand sequenced with primers F-FLIC1 and Flic2. The sequences for all three strains were consistent with that of the O157:H7 fliC sequence previously deposited in GenBank (accession no. AE005415), indicating that the frameshift mutation reported (30) to exist in this region of 493-89 fliC was not present.
Complementation studies.
Analysis of strain 493-89 with a flagellin-specific monoclonal antibody showed that it did not produce flagellin (data not shown). In contrast, 493-89 transformed with pSM4, which carries the 35150 fliC gene expressed from a strong σ70 vector promoter, produced flagellin as determined by Western blotting of cell lysates (data not shown), although it did not react with anti-H7 latex reagent and remained NM.
Previously, pBluescript KS:Yersinia enterocolitica fliA and pACYC184-Y. enterocolitica flhDC were shown to complement respective mutations in Salmonella enterica serovar Typhimurium. Given the relatedness of these loci in E. coli and Salmonella (3), we attempted to restore motility to 493-89 by complementation using these same constructs. Strain 493-89 transformed with the fliA construct failed to express flagellin or regain motility. But, when transformed with the flhDC construct, 493-89 produced flagellin, became motile, and reacted with anti-H7. Similarly, complementation with pSM7 and pSM8, which carried the flhDC operon of motile E. coli strains 35150 and DEC5A, respectively, also restored motility and H7 antigenicity to 493-89, and examination by electron microscopy confirmed that the cells produced flagella (Fig. 2C and D). In contrast, transformation of 493-89 with its own flhDC operon, cloned in either orientation (pSM9 or pSM10), did not restore motility.
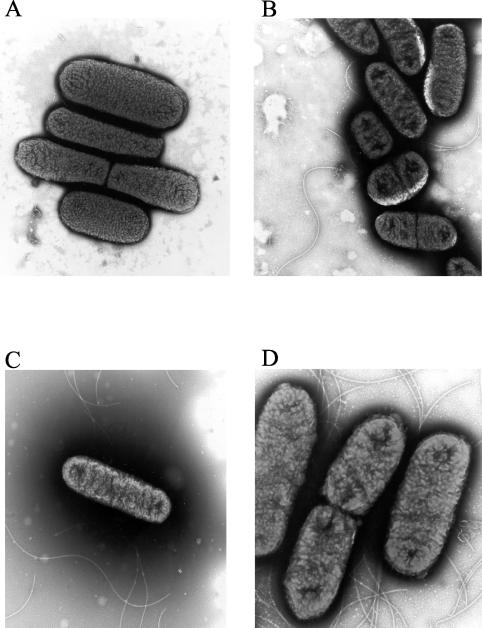
FIG. 2.
Electron micrographs showing the presence or absence of flagellum synthesis. The strains shown are NM 493-89 (A), motile 35150 (B), 493-89 strain transformed with pSM7 (C), and 493-89 strain transformed with pSM8 (D).
Sequencing of flhDC.
The DNA sequence of the 35150 flhDC operon obtained from pSM5 was identical to the annotated sequence for this region previously submitted to GenBank for O157:H7 (accession no. AE005411) and was ∼99% homologous to that of the E. coli K-12 flhDC locus (accession no. D90831) (Fig. 3). Comparing the sequences of the individual genes in the operon showed that the 35150 flhD ORF sequence had several nucleotide substitutions that differed from that of K-12. However, all except one of these mutations were silent, and the single nucleotide difference that resulted in an amino acid change is probably insignificant in terms of motility, since both K-12 and 35150 are motile and the FlhD proteins of both of these strains shared greater than 99% amino acid homology. Similarly, although the 35150 flhC ORF had three nucleotides that differed from that of K-12, none resulted in any alteration of the amino acid sequence, and the two predicted protein sequences showed 100% identity.
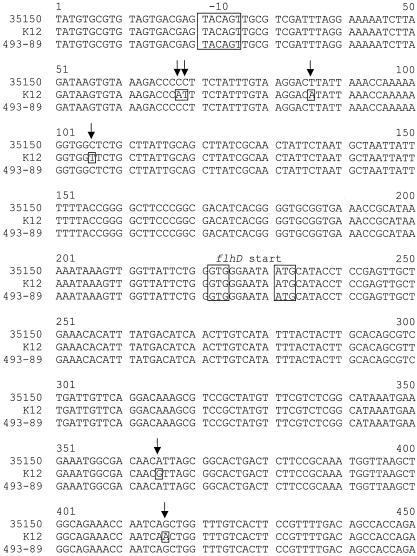
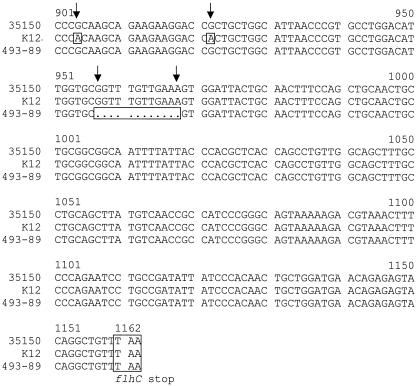
FIG. 3.
Comparison of flhDC sequences from E. coli K-12, 35150 (O157:H7), and German SF O157 strain 493-89 (O157:H−). Dissimilarities are boxed and indicated with arrows. The −10 promoter site and ORF start and stop codons are in boxes.
The nucleotide sequence of the flhDC operon of 493-89 was determined from pSM6 (accession no. AY225162). The sequences of the 5′ untranslated region containing the native promoter for the flhDC operon were found to be identical in both 493-89 and 35150; however, there were two sites within the flhDC coding sequences where differences occurred. The first, located within the flhD ORF, was a single-nucleotide polymorphism at position +239 (assuming the GTG start codon as defined in GenBank accession no. AE005411) that caused an L80Q amino acid substitution in the 35150 FlhD. The second mutation site where the 493-89 sequence differed from that of 35150 was the presence of an in-frame, 12-bp deletion within the 493-89 flhC ORF at +374 to 385 that caused the loss of phenylalanine, valine, glutamate, and serine at positions 126 to 129, respectively.
Site-directed mutagenic repair of flhC complements motility.
Transformation of strain 493-89 with pSM11, which carried the 493-89 flhDC operon with the 12-bp deletion in flhC repaired, restored motility and H7 antigenicity to 493-89.
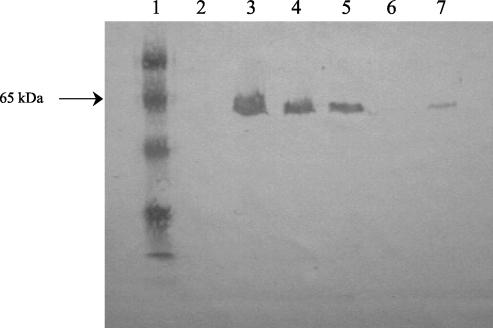
To specifically characterize the effect of the 12-bp flhC deletion on motility, we prepared plasmid constructs in which the flhC sequences for the four amino acids encoded by the 12-bp region were individually replaced by the codon for the neutral amino acid alanine. Western blot analysis of 493-89 strains transformed with each of these constructs revealed that pSM13 (FlhC V127A), pSM14 (FlhC E128A), and pSM15 (FlhC S129A) restored H7 flagellin expression (Fig. 4, lanes 3, 4, and 5, respectively) and motility, as confirmed by both microscopic and motility plate assays. These strains also reacted with anti-H7 latex reagent, confirming that H7 antigen was produced. In contrast, 493-89 transformed with pSM12 (FlhC F126A) remained NM and had no flagellin expression (Fig. 4, lane 2), indicating that phenylalanine 126 in the primary protein sequence is required for functional FlhC activity.
FIG. 4.
Western blot analysis with anti-H7 polyclonal sera to examine H7 flagellin expression in 493-89 strains transformed with 35150 flhDC site-directed mutant constructs. Lanes: 1, low-molecular-mass standard (Bio-Rad); 2 to 6, 493-89 transformed with pSM12, pSM13, pSM14, pSM15, and untransformed, respectively; 7, 35150.
The 12-bp deletion in flhC is clonal for the German SF O157 strains.
The distribution of the 12-bp flhC deletion was examined in other E. coli and O157:NM strains obtained from various geographic areas. The 379-bp PCR product was obtained from the two motile O157:H7 strains and a motile O55:H7 strain, as well as other O157:NM variants, indicating that none of these carried the 12-bp deletion in flhC. However, analysis of the other German SF O157 strains, CB569, CB1009, 5412, 514-91, and 210-89, demonstrated that none of them produced the 379-bp PCR product, indicating that these strains, like 493-89, also carried a defect in the flhC 12-bp deletion region that inhibited binding of the SRM19 primer (data not shown).
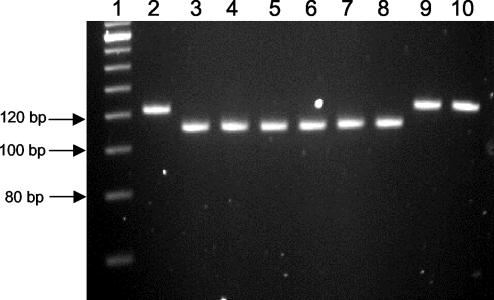
Subsequent PCR analysis with primers flanking the deletion site showed that each of the German SF O157 strains tested had the same size deletion as noted in 493-89 (Fig. 5, lanes 3 to 8), and sequence analysis confirmed the presence of the same 12-bp flhC deletion in each of the German SF O157 strains (data not shown). The results of the PCR deletion analysis are summarized in Table 1.
FIG. 5.
Agarose gel electrophoresis of PCR products from primers SRM145 and SRM146 that flank the 12-bp flhC deletion. Strains generating the 122-bp amplicon do not contain the 493-89 flhC mutation, while those producing a 110-bp product do. Lanes: 1, 20-bp molecular-weight ladder; 2, 35150; 3, 493-89; 4, CB569; 5, CB1009; 6, 5412; 7, 514-91; 8, 210-89; 9, DEC5D; 10, 43890.
DISCUSSION
The pathogenic German SF O157 strains are not easily identified, due to their atypical phenotypes and the lack of motility and H7 antigen expression. Previously, the nonmotility of the German SF O157 strain was postulated to be due to a frameshift mutation caused by two single-site nucleotide insertions at +73 and +78 of the fliC gene (30). We sequenced that region of the fliC ORF encompassing the putative mutations (from +1 to +458) of strain 493-89 and the other German SF O157 strains and found that, in all cases, the sequences in the purported frameshift region were identical to the fliC sequence derived from 35150, a motile O157:H7 strain, confirming that no insertion mutations were present. Consistent with this finding, 493-89 transformed with the functional 35150 fliC gene did not regain motility despite the fact that Western blot analysis of whole-cell lysates showed that the flagellin protein was made. The fact that flagellin production alone, being expressed from the vector Trc promoter, did not lead to flagellin secretion and assembly, as evidenced by the lack of serological H7 antigenicity, suggests that the factor causing nonmotility in 493-89 is epistatic to fliC.
In the three-tiered flagellar regulatory system, the class III fliC gene is regulated by class II genes, which include flgM, fliA, and the genes that encode the structural proteins for the basal body and hook (24). The fliA gene encodes the sigma F (σF) RNA polymerase subunit that is required for promoter recognition of class III genes, while FlgM (anti-σF) directly antagonizes FliA. In the normal sequence of events, when the basal body-hook structure is assembled, FlgM is secreted and its dilution from the cytosol releases FliA, enabling class III gene expression and flagellin production from fliC. In the absence of basal body-hook assembly, FlgM is not secreted and class III gene expression is repressed. We transformed 493-89 with a construct containing the Y. enterocolitica fliA gene, but the cells remained NM and did not express flagellin on Western blot analysis. Since the fliA-dependent expression of fliC did not occur in 493-89 transformed with a functional Y. enterocolitica fliA gene, we suspected that the defect was in another class II gene or, perhaps, in the class I flhDC operon, the transcriptional activator that serves as the master regulator of flagellum biosynthesis and regulates class II gene expression (10, 27, 28, 35).
Transformation of strain 493-89 with constructs carrying the flhDC operon from Y. enterocolitica or other motile E. coli strains (O157:H7 and O55:H7) restored motility to 493-89, confirming our earlier observation that this strain had a functional fliC gene. Furthermore, flagellum biosynthesis could be hyperexpressed when flhDC was cloned in the same transcriptional orientation as the vector Tcr promoter. Western blotting and examination by electron microscopy confirmed that flagella were synthesized by the motility-restored 493-89 strain, and serological typing confirmed that it was the H7 antigen. These results are consistent with the finding that the German SF O157 strains had the same H7 fliC restriction fragment length polymorphism profiles as the O157:H7 strains (16) and that these strains are closely related to the O157:H7 serotype (13, 21).
Comparison of the flhDC sequence cloned from strain 493-89 to the flhDC sequence from the motile O157:H7 strain (35150) showed no differences in the operon promoter region and only a single nucleotide change in the flhD ORF, which caused an L80Q substitution. However, a 12-bp deletion was found in the flhC ORF of 493-89 that was not present in the motile 35150 strain. Although the deletion was in frame, this 12-bp flhC deletion was confirmed to be responsible for the loss of motility, as 493-89 transformed with its own flhDC in which the 12-bp deletion had been repaired regained motility.
In the motile O157:H7 strain, the 12 bp covered in the flhC deletion of 493-89 encode phenylalanine, valine, glutamate, and serine. Complementation with constructs in which each of these amino acids was individually replaced with the codon specifying the neutral amino acid, alanine, restored motility and H7 antigen expression to 493-89, except for the one containing the F126A mutation. These results indicate that the phenylalanine at position 126 is essential for biological activity of FlhC. The flhDC operon encodes the FlhD and FlhC proteins, which initially associate to form D2 and C2 dimers that, subsequently, combine to form the C2D2 tetramer protein complex that regulates the class II genes (23). Whether phenylalanine 126 is essential for FlhC dimerization, the subsequent tetramerization with FlhD2 or in DNA binding is currently unclear but should become more evident once the FlhDC crystal structure is resolved.
We looked at the distribution of the 12-bp flhC deletion among other O157:NM strains and found that it occurred only in the German SF O157 strains. The absence of the 12-bp flhC deletion in the other O157:NM strains indicates that nonmotility in these strains may be due to catabolite repression or other physiological mediators (24) or to other uncharacterized genetic defects. The finding that the 12-bp flhC deletion is specific to the German SF O157 strains coincides with the presence of plasmid-encoded fimbriae genes that are unique only to this group (9), supporting previous reports that these strains are in a distinct clonal group (13, 21). The fact that the German SF O157 strains carry the H7 fliC gene (16) and can be manipulated to produce the H7 antigen also confirms their close genetic relation to O157:H7 (13, 21) and is in agreement with our evolution model that they diverged from O157:H7, in part, by the loss of motility (13, 26). As a consequence, the 12-bp flhC deletion provides a potentially useful marker with which strains in this clonal lineage may be identified and traced.
Regulation or loss of motility seems to be a common theme among gram-negative pathogens, as Bordetella pertussis, Shigella spp., and Yersinia pestis are all NM, yet each carries a full complement of flagellar genes (2, 11, 33). Interestingly, like our findings with the German SF O157 strains, these organisms also appear to be NM due to mutations in flhDC (2, 5, 32), which not only serves as the master flagellar control operon but also as a global regulator of numerous other operons (29). Other important pathogens, such as Y. enterocolitica, Yersinia pseudotuberculosis, Bordetella bronchiseptica, and Legionella pneumophila (18, 19, 22), show temperature-sensitive repression of flagellin expression, suggesting that there is a selection for motility repression in the host. The increase in the isolation of NM O157:H7 strains is consistent with this trend. All of these examples of pathogens require type III secretion systems (TTSS) for virulence. The process of flagellar assembly is also a subtype of TTSS (3), and there is evidence to suggest that the components of one TTSS are recognized by other TTSS in the cell (reference 35 and unpublished data). As a consequence, flagellar proteins may out-compete virulence type III proteins for export if both TTSS are simultaneously expressed. Consistent with this assumption, artificial expression of motility in B. bronchiseptica resulted in virulence attenuation (1), confirming the competitive interaction of the flagellar and virulence TTSS. Enteropathogenic E. coli and EHEC carry the locus for the enterocyte effacement pathogenicity island that encodes a TTSS that mediates the injection of virulence factors into the mammalian cell (34). These pathogens, therefore, also have multiple TTSS that can potentially interfere with one another and affect the expression of motility and/or virulence. As to whether the lack of motility in the German SF O157 strains offers any advantages in virulence or is a contributing factor to its increasing frequency in HUS infections remains to be determined.
In conclusion, nonmotility of the German SF O157 strains is due to a 12-bp deletion in the flhC gene of the flhDC master regulator operon. Specifically, phenylalanine 126 in FlhC appears to be the critical amino acid whose deletion resulted in an aberrant protein structure that could no longer function as a transcriptional activator for motility. The highly conserved nature of the 12-bp flhC deletion in the German SF O157 strains may be a useful marker for identifying these strains and also supports not only the clonality of the German SF O157 strains but also its postulated evolutionary divergence from O157:H7. Furthermore, this study has identified yet another pathogen that has selectively sustained a mutation that down regulates flagellar biosynthesis to, perhaps, eliminate competitive interactions between secretory pathways as well as the unnecessary expenditure of cellular energy, thereby resulting in a more virulent phenotype, a trend being more frequently observed among other bacterial pathogens.
Acknowledgments
We thank B. D. Tall and S. K. Curtis for assistance with electron microscopy. Special thanks go to Helge Karch for generously providing the strains used in this study. We also thank David Acheson for critical reading of the manuscript.
S. A. Minnich was supported by grant P20 RR164 54 from the BRIN Program from the National Center for Research Resources (National Institutes of Health).
REFERENCES
- 1.Akerley, B. J., P. A. Cotter, and J. F. Miller. 1995. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell 80:611-620. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., D. M. Monack, S. Falkow, and J. F. Miller. 1992. The bvgAS locus negatively controls motility and the synthesis of flagella in Bordetella brochiseptica. J. Bacteriol. 174:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 4.Aleksic, S., H. Karch, and J. Bockemuhl. 1992. A biotyping scheme for Shiga-like (Vero) toxin-producing Escherichia coli O157 and a list of serological cross-reactions between O157 and other gram-negative bacteria. Zentbl. Bakteriol. 276:221-230. [DOI] [PubMed] [Google Scholar]
- 5.Al Mamun, A. A., A. Tominaga, and M. Enomoto. 1996. Detection and characterization of the flagellar master operon in the four Shigella subgroups. J. Bacteriol. 178:3722-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann, E., B. Ochs, and K.-J. Abel. 1988. Tightly regulated tac promoter vector useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 7.Bielaszewska, M., H. Schmidt, M. Karmali, R. Khakhria, J. Janda, K. Bláhová, and H. Karch. 1998. Isolation and characterization of sorbitol-fermenting Shiga toxin (verocytotoxin)-producing Escherichia coli O157:H− strains in the Czech Republic. J. Clin. Microbiol. 36:2135-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitzan, M., K. Ludwig, M. Klemt, H. Konig, J. Buren, and D. E. Muller-Wiefel. 1993. The role of Escherichia coli O157 infections in the classical (enteropathic) haemolytic uraemic syndrome: results of a central European, multicentre study. Epidemiol. Infect. 110:183-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunder, W., A. Salam Khan, J. Hacker, and H. Karch. 2001. Novel type of fimbriae encoded by the large plasmid of sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H−. Infect. Immun. 69:4447-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, P., P. I. Fields, B. Swaminathan, and T. S. Whittam. 1996. Characterization of nonmotile Escherichia coli O157 and other serotypes by using an anti-flagellin monoclonal antibody. J. Clin. Microbiol. 34:2856-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, P., K. A. Lampel, H. Karch, and T. S. Whittam. 1998. Sequential genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J. Infect. Dis. 177:1750-1753. [DOI] [PubMed] [Google Scholar]
- 14.Feng, P., and S. R. Monday. 2000. Multiplex PCR for detection of trait and virulence factors in enterohemorrhagic E. coli serotypes. Mol. Cell Probes 14:333-337. [DOI] [PubMed]
- 15.Feng, P., R. J. Sugasawara, and A. Schantz. 1990. Identification of a common enterobacterial flagellin epitope with a monoclonal antibody. J. Gen. Microbiol. 136:337-342. [DOI] [PubMed] [Google Scholar]
- 16.Fields, P. I., K. Blom, H. J. Hughes, L. O. Helsel, P. Feng, and B. Swaminathan. 1997. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-RFLP test for the identification of E. coli O157:H7 and O157:NM. J. Clin. Microbiol. 35:1066-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunzer, F., H. Bohm, H. Russmann, M. Bitzan, S. Aleksic, and H. Karch. 1992. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic uremic syndrome. J. Clin. Microbiol. 30:1807-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuner, K., B. C. Brand, and J. Hacker. 1999. The expression of the flagellum of Legionella pneumophila is modulated by different environmental factors. FEMS Microbiol. Lett. 175:69-77. [DOI] [PubMed] [Google Scholar]
- 19.Kapatral, V., J. W. Olson, J. C. Pepe, V. I. Miller, and S. A. Minnich. 1996. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol. Microbiol. 19:1061-1071. [DOI] [PubMed] [Google Scholar]
- 20.Karch, H., and M. Bielaszewska. 2001. Sorbitol-fermenting Shiga toxin-producing Escherichia coli O157:H− strains: epidemiology, phenotypic and molecular characteristics, and microbiological diagnosis. J. Clin. Microbiol. 39:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karch, H., H. Bohm, H. Schmidt, F. Gunzer, S. Aleksic, and J. Heesemann. 1993. Clonal structure and pathogenicity of Shiga-like toxin-producing, sorbitol-fermenting Escherichia coli O157:H−. J. Clin. Microbiol. 31:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leigh, A. F., J. G. Coote, R. Parton, and C. J. Duggleby. 1993. Chromosomal DNA from both flagellate and non-flagellate Bordetella species contains sequences homologous to the Salmonella H1 flagellin gene. FEMS Microbiol. Lett. 111:225-231. [DOI] [PubMed] [Google Scholar]
- 23.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macnab, R. M. 1987. Flagella, p. 70-83. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, D.C.
- 25.Mikaelian, I., and A. Sergeant. 1992. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 20:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monday, S. R., T. S. Whittam, and P. C. H. Feng. 2001. Genetic and evolutionary analysis of mutations in the gusA gene that cause the absence of β-glucuronidase activity in Escherichia coli O157:H7. J. Infect. Dis. 184:918-921. [DOI] [PubMed] [Google Scholar]
- 27.Mytelka, D. S., and M. J. Chamberlin. 1996. Escherichia coli fliAZY operon. J. Bacteriol. 178:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruss, B. M., and P. Matsumura. 1997. Cell cycle regulation of flagellar genes. J. Bacteriol. 179:5602-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pruss, B. M., J. W. Campbell, T. K. van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid, S. D., R. K. Selander, and T. S. Whittam. 1999. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Smith, M. J. 2000. Ph.D. dissertation thesis. University of Idaho, Moscow.
- 33.Tominaga, A., M. A.-H. Mahmoud, T. Mukalhara, and M. Enomoto. 1994. Molecular characterization of intact, but cryptic, flagellin genes in the genus Shigella. Mol. Microbiol. 12:277-285. [DOI] [PubMed] [Google Scholar]
- 34.Yona-Nadler, C., T. Umanski, S.-I. Aizawa, D. Friedberg, and I. Rosenshine. 2003. Integration host factor (IHF) mediates repression of flagella in enteropathogenic and enterohemorrhagic Escherichia coli. Microbiology 149:877-884. [DOI] [PubMed] [Google Scholar]
- 35.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1999. The Yersinia enterocolitica motility master regulatory operon flhDC is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]