Abstract
We report here that wild-type Escherichia coli grows on N-acetylmuramic acid (MurNAc) as the sole source of carbon and energy. Analysis of mutants defective in N-acetylglucosamine (GlcNAc) catabolism revealed that the catabolic pathway for MurNAc merges into the GlcNAc pathway on the level of GlcNAc 6-phosphate. Furthermore, analysis of mutants defective in components of the phosphotransferase system (PTS) revealed that a PTS is essential for growth on MurNAc. However, neither the glucose-, mannose/glucosamine-, nor GlcNAc-specific PTS (PtsG, ManXYZ, and NagE, respectively) was found to be necessary. Instead, we identified a gene at 55 min on the E. coli chromosome that is responsible for MurNAc uptake and growth. It encodes a single polypeptide consisting of the EIIB and C domains of a so-far-uncharacterized PTS that was named murP. MurP lacks an EIIA domain and was found to require the activity of the crr-encoded enzyme IIA-glucose (EIIAGlc), a component of the major glucose transport system for growth on MurNAc. murP deletion mutants were unable to grow on MurNAc as the sole source of carbon; however, growth was rescued by providing murP in trans expressed from an isopropylthiogalactopyranoside-inducible plasmid. A functional His6 fusion of MurP was constructed, isolated from membranes, and identified as a polypeptide with an apparent molecular mass of 37 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis. Close homologs of MurP were identified in the genome of several bacteria, and we believe that these organisms might also be able to utilize MurNAc.
N-Acetylmuramic acid (MurNAc; 2-acetamido-2-deoxy-3-O-[(R)-1-carboxyethyl]-d-glucopyranose) is a monosaccharide compound present in the cell wall of almost all bacteria and missing in only few bacterial groups, such as mycoplasmas (32). Together with N-acetylglucosamine (GlcNAc), it forms the backbone of the cell wall peptidoglycan of both gram-positive and gram-negative bacteria (34). Cleavage of peptidoglycan by lysozyme and other muramidases has been studied intensively since the 1960s (15, 35). Interestingly, the uptake and further catabolism of the products of lysozyme degradation have not been investigated. In contrast, degradation of cellulose and chitin, β-glycans constituting the cell walls of plants, fungi, and the exoskeleton of arthropods, has attracted tremendous attention in recent years, and the catabolism of these polysaccharides has been elucidated in many microorganisms (10, 16, 21, 44, 45, 46). The polysaccharides are cleaved by a set of exo- and endo-acting hydrolases or transglycosylases; degradation products are then taken up by specific transport systems and are further metabolized. One important family of transport systems for carbohydrates in bacteria is the phosphotransferase system (PTS), which mediates uptake and phosphorylation of carbohydrates and controls the metabolism. The system consists of general PTS components (enzyme I [EI] and histidine protein [HPr]) and sugar-specific PTS components (EIIA and EIIB) as well as transmembrane transporter proteins for different sugars (EIIC).
Recently, Keyhani and Roseman reported that Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose (17). The genes responsible for uptake and degradation of the disaccharide, previously assigned to the cryptic cellobiose operon, were identified as components EIIB, -C, and -A of a phosphoenolpyruvate (PEP)-dependent PTS specific for N,N′-diacetylchitobiose (chbBCA), an N,N′-diacetylchitobiose-inducible repressor (chbR), a phospho-N,N′-diacetylchitobiose phosphorylase (chbF), and a gene of unknown function (chbG).
Furthermore, uptake and degradation of GlcNAc have been described extensively in E. coli. The nag operon (nagEBADC) is involved in GlcNAc transport and metabolism (29). nagE encodes a permease responsible for the transport and phosphorylation of GlcNAc (19). The NagE molecule contains the complete GlcNAc-specific PTS components EIIA, -B, and -C on a single polypeptide. GlcNAc is transported via EIIC, and phosphate is subsequently transferred from EIIA via the EIIB component onto the C6-hydroxyl group of the imported GlcNAc molecule. EIIA itself is phosphorylated by PEP, involving the general enzymes of the PTS cascade (EI and HPr). The product, GlcNAc 6-phosphate, is then deacetylated and further deaminated by the enzymes NagA and NagB, yielding fructose 6-phosphate, which enters glycolysis. NagC acts as a repressor of GlcNAc catabolism and as an activator of the UDP-GlcNAc anabolic pathway (26, 27).
To date, there is no information available concerning MurNAc as a carbon source; however, it has been known for a long time that E. coli breaks down almost half of its murein sacculus in each generation (8, 9). During enlargement of the cell wall, new material has to be synthesized and integrated into the existing sacculus. This is accomplished by the concerted action of synthetic and lytic enzymes in a sophisticated process presumably involving a multienzyme complex that enables the safe insertion of new material in the stress-bearing cell wall without the risk of lysing the cell (42). It is believed that continuous murein turnover is intrinsically connected to the process of cell elongation and division. The cell wall peptides are efficiently reutilized, without further degradation (i.e., recycled), to form new murein (24, 25).
Recently, evidence was provided to suggest that recycling of cell wall sugars occurs in E. coli (23). However, turnover of the murein sacculus in E. coli involves lytic transglycosidases, i.e., autolysins that do not cleave the glycan chain hydrolytically, as lysozyme does, but introduce an intramolecular glycosidic bond to yield 1,6-anhydro-N-acetylmuramic acid rather than the free sugar (12). The peptidyldisaccharides GlcNAc β-(1,4)-anhydro-MurNAc, with l-alanyl-d-glutamyl-meso-diaminopimelic acid attached to the carboxyl group of MurNAc and with zero, one, or two d-alanine residues linked to the l-anomer of diaminopimelic acid, are taken up by the secondary transporter AmpG (5) and further degraded within the cytoplasm by an N-acetylglucosaminidase, NagZ (4, 43); by an amidase, AmpD (14); and by an l,d-carboxypeptidase, LdcpA (39). The further processing of the turnover product, anhydro-MurNAc, is unknown, as is the degradation of free MurNAc.
In this work we investigated the growth of E. coli on MurNAc and identified a previously uncharacterized phosphotransferase system (named MurP) that is involved in the uptake and catabolism of MurNAc.
MATERIALS AND METHODS
Materials, media, and growth conditions.
Reagents for DNA purification were obtained from Qiagen (Hilden, Germany); restriction endonucleases, ligase, and polymerase were obtained from New England Biolabs (Beverly, Mass.) and Roche/Boehringer (Mannheim, Germany). E. coli strain BL21 and His-binding metal chelation resin were from Novagen (Madison, Wis.). Oligonucleotide primers were from MWG Biotech (Ebersberg, Germany). DNA sequencing was performed by GATC (Konstanz, Germany). Growth medium components were obtained from Difco. GlcNAc, MurNAc, antibiotics, vitamins, and amino acids were from Sigma (Taufkirchen, Germany). The expression vector pCS19 was previously constructed by introducing a lacIq gene into pQE60 (Qiagen) to allow constitutive expression of the lac repressor (36). The GenBank database and Swiss-Prot database were used for nucleotide and amino acid sequence searches, and the basic local alignment search tool (BLAST) was used for multiple sequence alignment.
E. coli strains were grown at 37°C on minimal medium A (MMA) (22) containing 0.2% carbon source (i.e., 11.1 mM glucose; 9.0 mM GlcNAc; 6.8 mM MurNAc; 21.7 mM glycerol; 5.3 mM trehalose) and supplemented with 5 μg of thiamine and 40 μg of amino acids per ml if required. A stock solution of 2% MurNAc (68.2 mM) in water was adjusted with KOH to pH 7 and filter sterilized before use. Mini-agar plates (2.6-cm radius) supplemented with 0.1 or 0.2% (wt/vol) MurNAc were used to test cell growth on agar plates. Growth kinetic experiments were performed as follows: 5 to 10 ml of MMA plus 0.2% carbon source and additives was inoculated with an exponential-phase culture (previously washed twice in minimal medium) to reach an optical density at 578 nm of about 0.04. Growth was performed in 100-ml flasks at 37°C under continuous shaking. Changes in turbidity of the cultures were monitored by assaying 60-μl aliquots of medium in disposable microcuvettes (UVette; Eppendorf, Hamburg, Germany) at 578 nm. Expression of MurP from plasmids pCS19YfeV and pCS19YfeV-His6 was achieved by growing E. coli BL21 cells in Luria-Bertani broth containing ampicillin (100 μg/ml). The E. coli strains and plasmids used in this work are listed in Table 1.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Known genotypea | Reference or source |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 flbB5301 deoC1 relA1 rbsR rpsL150 ptsF25 | 3 |
| BL21 | F−ompT hsd SB(rB− mB−) gal dcm | 37 |
| IBPC5321 | thi-1 argG6 argE3 his-4 mlt-1 xyl-5 rpsL ΔlacX74 mlc-1 | 28 |
| IBPC590 | IBPC5321 ΔnagEBADC::Tetr | 28 |
| IBPC531 | IBPC5321 nagA::Cmr | 28 |
| IBPC546 | IBPC5321 nagB::Kmr | 28 |
| IBPC542 | IBPC5321 nagE::Kmr | 28 |
| KM540 | MC4100 treA::Spcrpgi | 13 |
| LR-2-167 | argG6 galT his-1 man1 metB phoA rpsL thi-1 nagE ptsM | 41 |
| LR-2-168 | LR-2-167 ptsG | 40 |
| LM1 | LR-2-167 crr | Josef Lengeler, Osnabrück |
| PPA69 | HfrK16 galR thi glk::Cmr ΔptsHIcrr | Lab stock |
| ET185 | MC4100 φP[(malT-lacZ)(λplacMu50)] glpK::Cmr Δcrr::Kmr | Tanja Eppler, Konstanz |
| ET25 | MC4100 Δcya crp* ilv+ | Tanja Eppler, Konstanz |
| JJ3 | ET25 Δcrr::Kmr | This study |
| CM1 | JJ3 ΔtreA | This study |
| DY330 | W3110 ΔlacU169 gal-490 (λ cI857 Δcro/broA) | 47 |
| CM99 | DY330 yfeV::Kmr | This study |
| CM100 | MC4100 yfeV::Kmr | This study |
| CM101 | KM553 yfeV::Kmr (ptsHIcrr)+ | This study |
| CM102 | KM553 yfeV::Kmr ΔptsHIcrr | This study |
| CM103 | MC4100 ΔyfeV | This study |
| Plasmids | ||
| pKD4 | Apr Kmr plasmid, PCR template for homologous gene displacement | 6 |
| pCP20 | Apr Cmr plasmid that shows temperature-sensitive replication and thermal induction of FLP recombinase synthesis | 6 |
| pCS19 | Expression vector based on pQE31 (Qiagen) Apr, additionally carrying the constitutively expressed lac repressor lacIq | 36 |
| pCS19YfeV | yfeV cloned into pCS19, expression vector for IPTG-inducible MurP expression | This study |
| pCS19YfeV His6 | yfeV cloned into pCS19, expression vector for IPTG-inducible MurP His6 expression | This study |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Spcr, spectinomycin resistant; Tetr, tetracycline resistant.
Construction of transporter deletion mutants.
We used a combination of the protocols for homologous gene replacement (6, 47) to introduce a site-specific insertion of a kanamycin resistance cassette (Kmr) into yfeV, now named murP. Primers that contained 3′-end sequences complementary to the first or last 20 bp of the kanamycin resistance cassette of plasmid pKD4 (6) and 5′-end sequences with flanking regions upstream and downstream murP, based on the published gene sequence (1), were constructed: P1/KD4 murP, TGG ATC AAC ACG GCG GCT TTA TTC GTC AGG TTT TAG ACA AGG AAT AAC CCG TGT AGG CTG GAG CTG CTT CG-3′; and P2/KD4 murP, CTG ACG CTA CAA CTA ACA GCC AGC AGA GAA AGA TAG AGC ATT GTC CGT TTG ATA TGA ATA TCC TCC TTA G-3′.
Linear DNA that carries the kanamycin resistance cassette of plasmid pKD4 and the homologous regions of the E. coli chromosome was generated by PCR. The PCR mixture contained 0.8 μM primers and 0.2 mM concentrations of the four desoxynucleoside triphosphates in 100 μl of DNA polymerase buffer. About 5 ng of plasmid DNA (pKD4) was added to the PCR mixture. The reaction mixture was heated to 95°C, and then 1 μl of Pwo DNA polymerase was added. Thirty PCR cycles (60 s at 94°C, 30 s at 56°C, and 90 s at 72°C) were performed in a thermal cycler and revealed a single 1.6-kb fragment, as analyzed by agarose gel electrophoresis. The linear PCR fragment was transformed by electroporation with a GenePulser (Bio-Rad, Munich, Germany) into strain DY330 (47). This strain carries a defective λ prophage, and expression of the λ red recombinase genes for 15 min at 42°C allows recombination of linear DNA fragments into the chromosome.
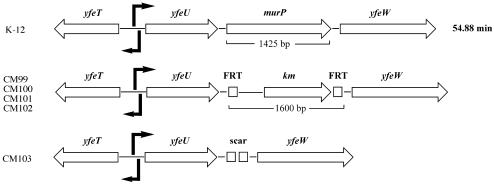
By P1 transduction, the yfeV::Kmr insertion from CM99 was introduced into strains MC4100 and KM553, resulting in strain CM100 (MC4100 yfeV::Kmr) and two KM553 derivatives, CM101 (yfeV::Kmr glk::cm ptsHIcrr+) and CM102 (yfeV::Kmr glk::cm ΔptsHIcrr). The kanamycin resistance cassette was eliminated from strain CM100 with the Flp recombinase provided from plasmid pCP20 (6). The resulting strain, CM103, contained a pKD4 scar in the place of murP (Fig. 5). Colony PCR was performed to confirm the site-specific insertion and deletion of murP in strains CM99 and CM103.
FIG. 5.
Maps of the yfeT-yfeW locus of wild-type E. coli (top) and mutant strains constructed by homologous gene displacement that carry a kanamycin resistance cassette insertion (km) instead of murP (CM100; middle) or a complete deletion of murP (CM103; bottom). In the last mutant strain, the kanamycin resistance cassette insertion flanked by DNA recombinase recognition sites (FRT) was cut out with the yeast Flp recombinase, leaving an 80-bp-long scar sequence in place of murP. A putative divergent promoter was identified in the yfeT-yfeU intergenic region, and an additional promoter was identified within murP.
Construction of plasmids pCS19YfeV and pCS19YfeV-His6 and overexpression of the encoded His6 fusion protein.
DNA preparations, restriction enzyme digests, ligation, and transformations were performed by standard techniques. The gene yfeV was amplified by PCR with genomic DNA from E. coli K-12 MG1655 (genomic sequencing strain) and the following oligonucleotide primers, based on the published gene sequence (1): yfeV BamHI/F, AACCATGGGAGGATCCGCCAAAGAGATCAGCAGTG-3′; and yfeV BglII-HindIII/R, TTCCCAAGCTTAAGATCTGTCCAGATTGACGTTACGG-3′.
The PCR mixture contained 0.8 μM primers and 0.2 mM concentrations of the four desoxynucleotides in 100 μl of DNA polymerase buffer. About 100 ng of genomic DNA was added to the PCR mixture. The reaction was heated to 95°C, and then 1 μl of Pwo DNA polymerase was added. Thirty PCR cycles (45 s at 94°C, 45 s at 56°C, and 90 s at 72°C) were performed in a thermal cycler and revealed a single 1.4-kb fragment, as analyzed by agarose gel electrophoresis. The fragments were subsequently cloned into pCS19 vector in two ways: digestion with BamHI and BglII for cloning of a C-terminal His6 fusion protein and digestion with BamHI and HindIII for cloning the native protein. The opened vector was dephosphorylated with alkaline phosphatase and purified prior to ligation.
Expression of YfeV and separation of the protein extracted from the membrane fraction.
E. coli MC4100 carrying pCS19YfeV-His6 was induced with isopropylthiogalactopyranoside (IPTG) and grown for an additional 3 h. IPTG-induced cells from a 200-ml Luria-Bertani broth culture were collected by centrifugation and resuspended in a minimal volume of buffer (20 mM Tris-HCl). The cells were ruptured by passing them three times though a French pressure cell. Debris and unbroken cells were removed by centrifugation at 30,000 × g for 30 min. The supernatant was then centrifuged at 130,000 × g for 1 h to collect the membrane fraction. The membrane fraction was boiled for 5 min in Laemmli sample buffer to solubilize membrane proteins, which were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to fluoropolymer membranes (Fluorotrans; Pall GmbH, Dreieich, Germany) and analyzed with antihistidine-horseradish peroxidase conjugates with the QIAexpress detection kit according to the manufacturer's protocol (Qiagen, Hilden, Germany).
RESULTS
E. coli grows on MurNAc.
We found that MurNAc is utilized as the sole source of carbon and energy by E. coli K-12 and B derivatives and several other bacteria (Salmonella enterica serovar Typhimurium LT2, Bacillus subtilis, and Enterococcus faecalis; data not shown). However, growth of E. coli on MurNAc was slow compared to growth on GlcNAc and glucose (Table 2). The doubling time for E. coli MC4100 growing on MurNAc in minimal medium at 37°C with continuous shaking was determined to be about four times longer than for growth on glucose or GlcNAc. MurNAc depletion from the growth medium was observed by high-pressure liquid chromatography analysis and indicates uptake of MurNAc by E. coli cells (data not shown).
TABLE 2.
Doubling times and maximal optical densities for MC4100 on different carbon sources
| Mediuma (no. of replicates) | Mean doubling time (h) | Mean maximum optical density at 578 nm |
|---|---|---|
| MMA-glucose (3) | 0.83 ± 0.03 | 2.7 ± 0.3 |
| MMA-GlcNAc (2) | 0.84 ± 0.02 | 2.7 ± 0.3 |
| MMA-MurNAc (6) | 3.4 ± 0.3 | 1.8 ± 0.2 |
E. coli cells were grown at 37°C on MMA supplemented with 0.2% carbon source.
Convergence of the MurNAc and the GlcNAc degradation pathways.
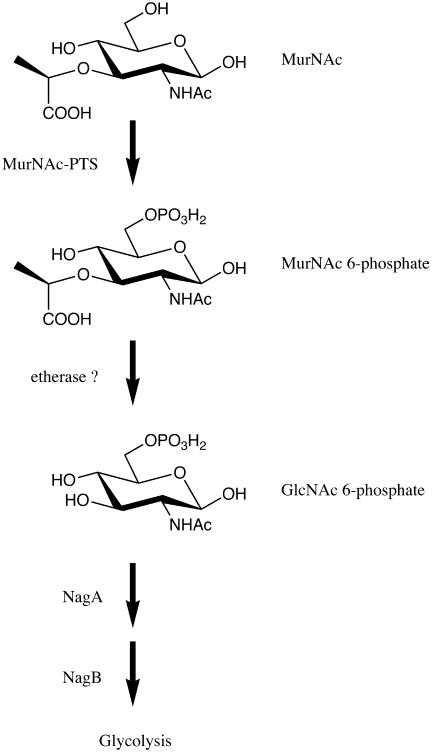
Mutant strains interrupted in the degradative pathway for GlcNAc (deletion of the entire nag operon, ΔnagEBADC; deletion of the GlcNAc 6-phosphate deacetylase, ΔnagA; and deletion of the glucosamine-6-phosphate deaminase, ΔnagB) were found to be unable to utilize MurNAc as the sole source of carbon and energy (Table 3). However, a mutant deficient in glucose 6-phosphate isomerase (pgi) that was not able to utilize glucose efficiently (2) was able to grow on MurNAc and GlcNAc. It can be concluded that the MurNAc and GlcNAc metabolism pathways merge on the level of GlcNAc 6-phosphate, which implies that MurNAc is phosphorylated in the process and that the lactyl ether substituent is cleaved off prior to deacetylation and deamination (Fig. 1).
TABLE 3.
Growth of E. coli strains defective in glucose or GlcNAc metabolisma
| Medium | Growth
|
||||
|---|---|---|---|---|---|
| IBPC 5321 | IBP C590 ΔnagEBACD::Tetr | IBP C531 nagA::Cmr | IBP C546 nagB::Kmr | KM540 pgi | |
| MMA-glucose | ++ | ++ | ++ | ++ | (+) |
| MMA-GlcNAc | ++ | − | − | − | ++ |
| MMA-MurNAc | + | − | − | − | + |
Growth rates on MMA supplemented with 0.2% carbon source (plus thiamine, arginine, and histidine) were classified as follows: ++, fast growth, colonies visible within 1 day at 37°C; +, slow growth, small colonies visible within 1 day but clearly visible within 2 days; (+), residual growth, small colonies visible within 2 days; −, no growth. Only relevant genotypes are indicated.
FIG. 1.
Proposed MurNAc degradation pathway of E. coli. Convergence with the GlcNAc degradation pathway occurs on the level of GlcNAc 6-phosphate. A hypothetical etherase is required for removal of the lactyl ether substituent of MurNAc.
MurNAc is transported by a PTS.
Mutants defective in ptsIHcrr (encoding EI, HPr, and EIIAGlc) were unable to grow on MurNAc. This strongly suggests the involvement of a PTS in MurNAc transport. Since GlcNAc transport and phosphorylation are mediated by the nagE- and the manXYZ-encoded PTS, we tested E. coli nagE manXYZ mutants for growth on MurNAc. Strains LR2-167 (nagE manXYZ) and LR2-168 (nagE manXYZ ptsG) were unable to grow on GlcNAc and were deficient in utilization of glucose, whereas they grew normally on MurNAc (Table 4). Hence, neither nagE, manXYZ, nor ptsG is responsible for MurNAc uptake and phosphorylation.
TABLE 4.
Growth of E. coli strains defective in sugar transporta
| Medium | Growth
|
||||
|---|---|---|---|---|---|
| PPA69 ΔptsHIcrr | IBPC542 nagE::Kmr | LR-2-167 nagE manXYZ | LR-2-168 nagE manXYZ ptsG | LM 1 nagE manXYZ crr | |
| MMA-glucose | − | ++ | ++ | (+) | (+) |
| MMA-GlcNAc | − | ++ | −b | −b | −b |
| MMA-MurNAc | − | ++ | + | + | −b |
Growth rates on MMA supplemented with 0.2% carbon source (plus thiamine, methionine, arginine, and histidine) were classified as defined in Table 3, footnote a.
Revertants appeared after prolonged incubation (see the text for explanations).
It should be noted that after prolonged incubation (>24 h) of strains LR-2-167, LR-2-168, and LM1 on MMA-GlcNAc or MMA-MurNAc, few colonies appeared (Tables 4 and 5). Since the deficiency of nagE, manXYZ, and crr in these strains is the result of point mutations only, a high frequency of back-mutations is expected. When transferred to fresh agar plates, the strains grew as well as if there were no mutations in these genes, and we believe that the colonies represent revertants.
TABLE 5.
Effect of cAMP and growth of cAMP/CAP-independent E. coli strainsa
| Medium | Growthb
|
||||
|---|---|---|---|---|---|
| LM1 nagE manXYZ crr
|
ET25 Δcya crp* | JJ3 Δcya crp* Δcrr::Kmr | CM1 Δcya crp* Δcrr::Kmr ΔtreA | ||
| 0 mM cAMP | 0.5 mM cAMP | ||||
| MMA-glucose | (+)c | ND | ++ | (+) | (+) |
| MMA-GlcNAc | −c | ND | ++ | ++ | ++ |
| MMA-glycerol | + | ++ | ++ | + | + |
| MMA-trehalose | (+) | (+) | ++ | (+) | − |
| MMA-MurNAc | −c | −c | + | − | − |
Growth rates on MMA supplemented with 0.2% carbon source (plus thiamine, methionine, arginine, and histidine) were classified as defined in Table 3, footnote a.
ND, not determined.
Revertants appeared after prolonged incubation (see the text for explanations).
EIIAGlc dependence of the MurNAc-PTS.
Interestingly, strain LR-2-168 (nagE manXYZ ptsG) was found to grow on MurNAc, whereas strain LM1 (nagE manXYZ crr) did not grow on this carbon source. The difference between LR2-168 and LM1 is that in the first strain, the mutation affects the EIIB and -C components of the glucose-PTS, while in the latter the mutation affects the EIIA component. This indicates that the putative MurNAc-PTS might be dependent on EIIAGlc. However, reduced uptake of MurNAc in the crr strain could possibly be explained by a repression effect involving the catabolite gene activator protein (CAP). EIIAGlc in its phosphorylated form stimulates adenylate cyclase (cya) to produce cyclic AMP (cAMP), required for activation of cAMP- and CAP-dependent transcription (30). We increased the growth of LM1 on the control substrate glycerol by adding cAMP to the medium but found that the strain was still unable to grow on MurNAc (Table 5).
We also constructed strains that contain a mutation in CAP, which renders them independent of cAMP binding (crp*) and additionally carry deletions in the genes encoding adenylate cyclase (cya) and EIIAGlc (crr). These strains were also unable to grow on MurNAc; however, these strains grew on glycerol as the sole source of carbon, confirming the EIIAGlc independence of glycerol metabolism. A dependency on EIIAGlc has been found previously for the trehalose-PTS (treB) (18), and therefore our intention was to use trehalose as a substrate for negative control. Strain JJ3 (Δcya crp* Δcrr::Kmr) showed residual growth in the absence of EIIAGlc caused by hydrolysis of trehalose by the periplasmic trehalase, TreA (18), and a strain (CM1) that additionally carries a deletion in treA was constructed. Indeed, strain CM1 did not grow on trehalose or on MurNAc, proving the EIIAGlc dependence of both PTS. Strain CM1 was able to grow on GlcNAc, and residual growth on glucose independent of a functional glucose-PTS has been described (11).
Genomic database search for a possible PTS-EIIBC.
A genomic database search revealed a candidate PTS-EIIBC (b2429, Swiss-Prot P77272) (Fig. 2, YFEV_ECOLI). This gene was predicted to contain solely an EIIBC domain and was classified as a glucose-glucoside PTS (TC no. 4.A.1) according to the system of Saier and others (38). YfeV showed high similarity (79% overall amino acid sequence similarity according to the BLAST sequence alignment tool) with a putative sucrose PTS from Vibrio cholerae (PPTS_VIBCH) and 50 to 60% similarity with a putative sucrose PTS from Bacillus spp. (Bacillus cereus, YBBF_BACCE; Bacillus subtilis, YBBF_BACSU), Salmonella spp. (Salmonella enterica serovar Typhi, PPTS_SALTY; Salmonella enterica serovar Typhimurium LT2, PPTS_SALTM) as well as Lactococcus lactis (PPTS_LACLA) and Enterococcus faecalis (PPTS_ENTFA). The sucrose PTS of S. enterica serovar Typhimurium LT2 (PTSB_SALTM) and Klebsiella pneumoniae (PTSB_KLEPN) showed 53% overall sequence similarity. Furthermore, some 40% sequence similarity was observed with the trehalose PTS (TREB_ECOLI) and the EIIBC portion of the cryptic β-glycoside PTS (BGLF_ECOLI) of E. coli. Interestingly, genes nearly identical to yfeV could be found within the genome of other E. coli strains (E. coli O157:H7; YFEV_H157:H7 and E. coli O6) and also within the Shigella flexneri chromosome (YFEV_SHIFL). The Shigella protein was 100% identical on the amino acid level and 98% identical on the nucleotide level to the E. coli protein.
FIG. 2.
Partial multiple sequence alignment of the N terminus of the MurNAc-PTS (YfeV; renamed MurP) and selected members of the glucose-glucoside PTS family (4.A.1). Dark shading indicates a conserved sequence motif that includes the Cys residue presumably representing the phosphorylation site of the EIIB domain. SwissProt/TrEMBL identification numbers are given in the first column, and abbreviations are defined in the text.
yfeV knockout mutants fail to grow on MurNAc.
We constructed a genomic yfeV (murP) deletion mutant in strain DY330 by a combination of the methods developed by Donald Court and coworkers (47) and Datsenko and Wanner (6). The yfeV::Kmr deletion was introduced into strains MC4100 and KM553 by P1 transduction, resulting in strain CM100 (MurP−) and two KM553 derivatives, CM101 (MurP−, Glk−, and PTS±) and CM102 (MurP−, Glk−, and PTS−). Recombination of the DNA portion carrying yfeV::Kmr from the transducing P1 phage resulted in both upkeep and correction of the ptsHI crr deletion of strain KM553. A cotransduction frequency of about 50% was observed, consistent with the distance between ptsHI crr and yfeV of 0.3 min on the E. coli chromosome. The kanamycin resistance cassette was eliminated from strain CM100 with the Flp recombinase from plasmid pCP20 (6). The resulting strain, CM103, carries only the characteristic “scar” sequence in place of yfeV (see Fig. 5). All deletion mutants of yfeV constructed were found to be unable to grow on MurNAc. There was no difference between yfeV::Kmr strains CM101 and CM102, although the first strain had deletions in essential PTS genes and the second strain did not.
Growth was rescued by providing yfeV in trans.
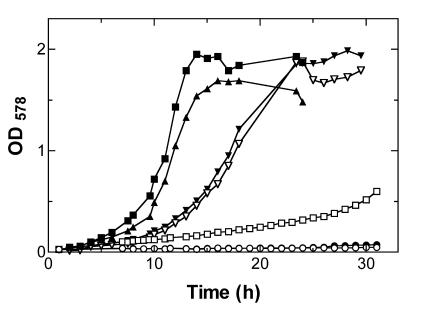
Gene yfeV was amplified by PCR and cloned into the expression vector pCS19 (a derivative of the commercial vector pQE60 from Qiagen, containing a constitutionally expressed lacIq). The cloning was performed in two ways, which allows IPTG-induced overexpression of native YfeV (MurP) enzyme as well as a C-terminal YfeV (MurP)-His6 fusion protein in E. coli. murP deletion strains were rescued for growth on MurNAc by providing the MurNAc-PTS in trans, expressed from these plasmids. The complementation of the MurNAc phenotype was found to be dependent on IPTG (Fig. 3): only slow growth in the absence of IPTG indicates a low level of constitutive expression of MurP, but fast growth was observed in the presence of IPTG. The doubling time for growth of strain CM103 carrying plasmid pCS19YfeV on MurNAc in the presence of IPTG was determined to be 2.2 h.
FIG. 3.
Growth curves of wild-type E. coli and mutant strains growing on MMA supplemented with 0.2% MurNAc (6.8 mM) and induced with 0.5 mM IPTG (solid symbols) or without induction (open symbols): wild-type E. coli (MC4100) carrying control plasmid pCS19 with (▾) and without induction (▿); murP (yfeV) deletion strain CM103 carrying plasmid pCS19 with (•) and without (○) induction; murP deletion strain CM103 carrying plasmid pCS19YfeV with (▪) and without (□) induction; and murP deletion strain CM103 carrying plasmid pCS19YfeV-His6 with induction (▴). Representative data out of three replicates are shown.
Expression of YfeV and Western blot analysis.
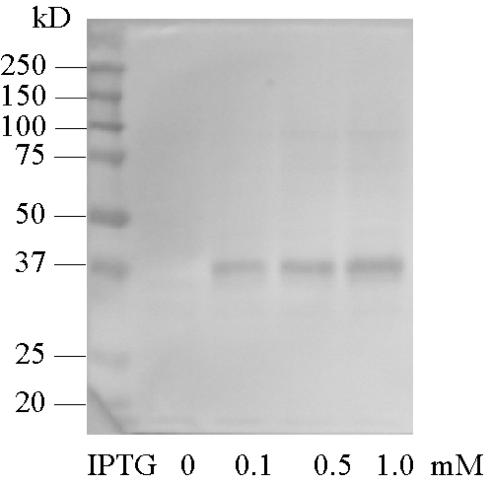
E. coli MC4100 carrying pCS19YfeV-His6 was induced with IPTG and grown for an additional 3 h. Membrane vesicles were isolated as described above, and membrane proteins were separated by SDS-PAGE. Subsequent blotting onto fluoropolymer membranes allowed detection of the His6-tagged fusion protein, YfeV-His6, with antibodies. We found that YfeV-His6, a protein with a theoretical molecular mass of 50 kDa, ran in SDS-PAGE with a mobility similar to that of the 37-kDa marker (Fig. 4). It is known from the literature that membrane proteins show mobilities not consistent with that of soluble globular proteins (7), but we cannot exclude possible proteolytic degradation of the MurP protein. A faint band of approximately 95 kDa was also observed that might represent a multimeric form of MurP or a complex of MurP with soluble PTS components.
FIG. 4.
Western blot of SDS-PAGE with anti-His6 antibodies. Strain CM103 carrying pCS19YfeV-His6 was induced for 3 h at the indicated IPTG concentrations (lanes 2 to 4). Size standards are shown in lane 1.
DISCUSSION
Although MurNAc represents a major natural compound, it was not known until now whether any organism can utilize this ubiquitous sugar or how it is degraded. We found that E. coli and other bacteria are indeed able to utilize MurNAc as a sole source of carbon and energy. However, growth rates were about four times lower than with favorable sugars (glucose and GlcNAc), indicating a less energy-efficient degradation pathway. Although overexpression of the transport system (MurP) from a plasmid increased the growth rate to some extent (see Fig. 3), the amount of MurP did not seem to be the major factor that limited growth. The further degradation of MurNAc 6-phosphate via a hypothetical etherase possibly represents the rate-limiting step in MurNAc degradation. A prolonged lag phase for growth on MurNAc in minimal medium was observed compared to the growth on glucose or GlcNAc (Fig. 3). The lag period was found to be unchanged when cells previously grown to stationary phase on MurNAc were used to inoculate fresh minimal medium supplemented with MurNAc. This shows that a mutation is not required for induction of the MurNAc utilization pathway, and the system therefore cannot be judged as a cryptic or silent system. However, overexpression of the transport system from an IPTG-inducible plasmid abolished the lag period observed for growth of wild-type E. coli on MurNAc. It can be concluded that MurNAc or a product of its processing (e.g., the phosphorylated form) might not be sufficient to fully induce the MurNAc degradation system. Cells growing on MurNAc reached maximal densities of only 1.8 (optical density at 578 nm) compared to 2.7 for growth on glucose and GlcNAc (Table 2). An explanation could be that at higher optical densities, oxygen becomes rate limiting and an oxygen dependency for growth on MurNAc can be postulated either for maximal energy recovery or by an oxygen-dependent degradation step. Growth of batch cultures indicated that oxygen indeed might be necessary for growth on MurNAc. Another explanation for early entry into stationary phase is the accumulation of fermentation products such as acetate or lactate that are produced during removal of the lactyl ether substituent of MurNAc.
MurP contains only the PTS domains EIIB and -C on the polypeptide chain. It therefore requires an EIIA molecule for function. Dependence on the glucose-specific EIIA (EIIAGlc) was found for members of the glucose-glucoside family of PTS (4.A.1) (38): e.g., the trehalose PTS TreB (18) of E. coli and the plasmid-encoded sucrose PTS Scr (20). We found that crr, encoding EIIAGlc, is also required for growth on MurNAc. However, it is known that a crr deletion results in continuous catabolite repression because of unstimulated adenylate cyclase and consequently repression of genes dependent on cAMP/CAP activation. We therefore tested a crp* mutation yielding a protein that is independent of cAMP stimulation (33). A crr crp* strain was still not able to grow on MurNAc, and this result unequivocally confirms the EIIAGlc dependence of MurP.
Searching the E. coli GenData base for a possibly EIIAGlc-dependent PTS involved in MurNAc metabolism revealed a possible candidate, yfeV. We show, by testing chromosomal deletion mutants and rescuing growth by providing the PTS on a plasmid, that YfeV (now named MurP) was responsible for growth on MurNAc as the sole source of carbon.
The 1,424-bp open reading frame (ORF) identified as murP starts with an ATG codon and ends with a TGA stop codon. A putative ribosome-binding site (Shine-Dalgarno sequence) was found 7 bases upstream from the start codon. murP is very likely part of an operon flanked by genes of unknown function (Fig. 5, top). An ORF upstream of murP ends three bases before its start codon (yfeU), while another ORF downstream of murP starts four bases after its stop codon (yfeW). This putative operon may be transcribed from two divergently oriented promoters that are positioned face to face (Fig. 5), resulting in transcripts that might overlap in a stretch of 18 bp. The ORF in the upstream direction contains a helix-turn-helix motif near the N terminus, a binding domain found in many sugar phosphate isomerases and sugar phosphate-binding proteins (SIS motif), and displays homology with regulators of the ribose repressor (ripR) family. In the other direction of transcription, as mentioned above, murP lies between two additional ORFs. The first shows similarity with mammalian glucokinase regulatory proteins and also contains a sugar phosphate-binding motif. Interestingly, the last ORF in the row displays similarity with β-lactamases and penicillin-binding proteins. Deletion of the latter gene conferred no detectable phenotype (data not shown).
The putative operon is entirely missing at this position in Salmonella enterica serovar Typhimurium LT2 and Salmonella enterica serovar Typhi; however, it was found at this position on the Shigella flexneri chromosome. We identified an ORF displaying high sequence similarity with murP elsewhere on the chromosome of Salmonella enterica serovar Typhimurium LT2 that might explain growth of this organism on MurNAc (Fig. 2). Since Escherichia, Salmonella, and Shigella are closely related organisms, it might be speculated that the insertion of the putative MurNAc operon into the E. coli chromosome is the result of a horizontal gene transfer event. Genes displaying high sequence similarity with murP and the other genes of the putative MurNAc operon were also found on the Bacillus subtilis chromosome. Interestingly, these genes are linked to a homolog of nagZ, encoding the β-N-acetylglucosaminidase of E. coli that was found to be involved in the hydrolysis of cell wall recycling products (4). It can be presumed that Bacillus possesses a MurNAc degradative pathway similar to that of E. coli. However, NagZ of E. coli was found to be located within the cytoplasm and is believed to cleave the cell wall recycling product GlcNAc-anhydro-MurNAc and peptidyl-substituted derivatives (4, 43), yielding intracellular GlcNAc and anhydro-MurNAc. The NagZ homolog of Bacillus subtilis, however, is predicted to possess a signal sequence and to be lipid anchored and oriented towards the exterior. We believe that this enzyme releases MurNAc that can be taken up by a PTS rather than anhydro-MurNAc. Interestingly, a recent PTS survey within the genome of Bacillus subtilis identified the close homolog of MurP as an uncharacterized PTS of the sucrose-permease family (31); however, MurP itself has not been mentioned in the recent PTS survey within the genome of E. coli (38).
Close MurP homologs have been found within the genomes of several bacteria (Fig. 2). As MurNAc represents half of the repeating backbone of peptidoglycan of nearly all bacteria, the ability to degrade MurNAc is expected to be widely distributed among all bacterial niches. We are currently investigating how MurNAc is degraded further and if there is a connection between MurNAc utilization and cell wall recycling in E. coli.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG project MA2436/1).
We gratefully acknowledge Jacqueline Plumbridge for providing E. coli strains with deletions in genes of the nag operon and Josef Lengeler for strains LM1, LR2-167, and 168. We thank Winfried Boos for critical discussions and Erika Oberer-Bley for helping with the manuscript.
REFERENCES
- 1.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 2.Canonaco, F., T. A. Hess, S. Heri, T. Wang, T. Szyperski, and U. Sauer. 2001. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol. Lett. 204:247-252. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, Q., H. Li, K. Merdek, and J. T. Park. 2000. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 182:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Q., and J. T. Park. 2002. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 184:6434-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes, P. B., R. V. Nardi, and S. G. Franklin. 1978. The resolution of membrane proteins based upon size, charge, and hydrophobicity. Anal. Biochem. 91:101-114. [DOI] [PubMed] [Google Scholar]
- 8.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodell, E. W., and U. Schwarz. 1985. Release of cell wall peptides into culture medium by exponentially growing Escherichia coli. J. Bacteriol. 162:391-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazlewood, G. P., and H. J. Gilbert. 1993. Xylan and cellulose utilization by the clostridia. Bio/Technology 25:311-341. [PubMed] [Google Scholar]
- 11.Hernandez-Montalvo, V., F. Valle, F. Bolivar, and G. Gosset. 2001. Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl. Microbiol. Biotechnol. 57:186-191. [DOI] [PubMed] [Google Scholar]
- 12.Höltje, J. V. 1996. Lytic transglycosylases, p. 425-429, In P. Jollès (ed.), Lysosomes: model enzymes in biochemistry and biology. Birkhauser Verlag, Basel, Switzerland.
- 13.Horlacher, R., R. Peist, and W. Boos. 1996. Improved method for the preparative synthesis of labeled trehalose of high specific activity by Escherichia coli. Appl. Environ. Microbiol. 62:3861-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frere. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 15.Jollès, P. 1969. Lysozymes: a chapter of molecular biology. Angew. Chem. Int. Ed. Engl. 8:227-239. [DOI] [PubMed] [Google Scholar]
- 16.Keyhani, N. O., and S. Roseman. 1999. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta 1473:108-122. [DOI] [PubMed] [Google Scholar]
- 17.Keyhani, N. O., and S. Roseman. 1997. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc. Natl. Acad. Sci. USA 94:14367-14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klein, W., R. Horlacher, and W. Boos. 1995. Molecular analysis of treB encoding the Escherichia coli enzyme II specific for trehalose. J. Bacteriol. 177:4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lengeler, J. W., K. Jahreis, and U. F. Wehmeier. 1994. Enzymes II of the phospho enol pyruvate-dependent phosphotransferase systems: their structure and function in carbohydrate transport. Biochim. Biophys. Acta 1188:1-28. [DOI] [PubMed] [Google Scholar]
- 20.Lengeler, J. W., R. J. Mayer, and K. Schmid. 1982. Phosphoenolpyruvate-dependent phosphotransferase system enzyme III and plasmid-encoded sucrose transport in Escherichia coli K-12. J. Bacteriol. 151:468-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leschine, S. B. 1995. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 49:399-426. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, J. T. 1993. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J. Bacteriol. 175:7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17:421-426. [DOI] [PubMed] [Google Scholar]
- 26.Plumbridge, J. 1995. Coordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 14:3958-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plumbridge, J. A. 1990. Induction of the nag regulon of Escherichia coli by N-acetylglucosamine and glucosamine: role of the cyclic AMP-catabolite activator protein complex in expression of the regulon. J. Bacteriol. 172:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plumbridge, J. A. 1991. Repression and induction of the nag regulon of Escherichia coli K-12: the roles of nagC and nagA in maintenance of the uninduced state. Mol. Microbiol. 5:2053-2062. [DOI] [PubMed] [Google Scholar]
- 29.Plumbridge, J. A. 1989. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol. Microbiol. 3:505-515. [DOI] [PubMed] [Google Scholar]
- 30.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reizer, J., S. Bachem, A. Reizer, M. Arnaud, M. H. Saier, Jr., and J. Stulke. 1999. Novel phosphotransferase system genes revealed by genome analysis- the complete complement of PTS proteins encoded within the genome of Bacillus subtilis. Microbiology 145:3419-3429. [DOI] [PubMed] [Google Scholar]
- 32.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholte, B. J., and P. W. Postma. 1980. Mutation in the crp gene of Salmonella typhimurium which interferes with inducer exclusion. J. Bacteriol. 141:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seltmann, G., and O. Holst. 2002. The bacterial cell wall, p. 103-132. Springer-Verlag, Berlin, Germany.
- 35.Sharon, N., J. Jollès, and P. Jollès. 1966. Contribution to the study of the mechanism of action of lysozymes of various origins. Bull. Soc. Chim. Biol. (Paris) 48:731-732. [PubMed] [Google Scholar]
- 36.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 37.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 38.Tchieu, J. H., V. Norris, J. S. Edwards, and M. H. Saier, Jr. 2001. The complete phosphotransferase system in Escherichia coli. J. Mol. Microbiol. Biotechnol. 3:329-346. [PubMed] [Google Scholar]
- 39.Templin, M. F., A. Ursinus, and J. V. Höltje. 1999. A defect in cell wall recycling triggers autolysis during the stationary growth phase of Escherichia coli. EMBO J. 18:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogler, A. P., C. P. Broekhuizen, A. Schuitema, J. W. Lengeler, and P. W. Postma. 1988. Suppression of IIIGlc-defects by enzymes IINag and IIBgl of the PEP:carbohydrate phosphotransferase system. Mol. Microbiol. 2:719-726. [DOI] [PubMed] [Google Scholar]
- 41.Vogler, A. P., and J. W. Lengeler. 1989. Analysis of the nag regulon from Escherichia coli K-12 and Klebsiella pneumoniae and of its regulation. Mol. Gen. Genet. 219:97-105. [DOI] [PubMed] [Google Scholar]
- 42.Vollmer, W., and J. V. Höltje. 2001. Morphogenesis of Escherichia coli. Curr. Opin. Microbiol. 4:625-633. [DOI] [PubMed] [Google Scholar]
- 43.Vötsch, W., and M. F. Templin. 2000. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 275:39032-39038. [DOI] [PubMed] [Google Scholar]
- 44.Warren, R. A. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe, T., K. Kimura, T. Sumiya, N. Nikaidou, K. Suzuki, M. Suzuki, M. Taiyoji, S. Ferrer, and M. Regué. 1997. Genetic analysis of the chitinase system of Serratia marcescens 2170. J. Bacteriol. 179:7111-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe, T., W. Oyanagi, K. Suzuki, and H. Tanaka. 1990. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J. Bacteriol. 172:4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]