Significance
The microbial community of the Amazon River Plume determines the fate of the world’s largest input of terrestrial carbon and nutrients to the ocean. By benchmarking with internal standards during sample collection, we determined that each liter of plume seawater contains 1 trillion genes and 50 billion transcripts from thousands of bacterial, archaeal, and eukaryotic taxa. Gene regulation by taxa inhabiting distinct microenvironments provides insights into micron-scale patterns of transformations in the marine carbon, nitrogen, phosphorus, and sulfur cycles in this globally important ecosystem.
Keywords: metatranscriptomics, metagenomics, biogeochemistry, gene expression ratios, marine bacteria
Abstract
We investigated expression of genes mediating elemental cycling at the microspatial scale in the ocean’s largest river plume using, to our knowledge, the first fully quantitative inventory of genes and transcripts. The bacterial and archaeal communities associated with a phytoplankton bloom in Amazon River Plume waters at the outer continental shelf in June 2010 harbored ∼1.0 × 1013 genes and 4.7 × 1011 transcripts per liter that mapped to several thousand microbial genomes. Genomes from free-living cells were more abundant than those from particle-associated cells, and they generated more transcripts per liter for carbon fixation, heterotrophy, nitrogen and phosphorus uptake, and iron acquisition, although they had lower expression ratios (transcripts⋅gene−1) overall. Genomes from particle-associated cells contributed more transcripts for sulfur cycling, aromatic compound degradation, and the synthesis of biologically essential vitamins, with an overall twofold up-regulation of expression compared with free-living cells. Quantitatively, gene regulation differences were more important than genome abundance differences in explaining why microenvironment transcriptomes differed. Taxa contributing genomes to both free-living and particle-associated communities had up to 65% of their expressed genes regulated differently between the two, quantifying the extent of transcriptional plasticity in marine microbes in situ. In response to patchiness in carbon, nutrients, and light at the micrometer scale, Amazon Plume microbes regulated the expression of genes relevant to biogeochemical processes at the ecosystem scale.
Microbially mediated biogeochemical transformations in the ocean take place within a poorly characterized matrix of particles, colloids, and dissolved phase materials (1) (Fig. 1). This microspatial structure affects the taxonomy and functional gene inventories of microbial communities (2, 3), driven by differences in nutrient concentrations, light exposure, oxygen availability, and predation (4, 5), and undoubtedly affects the types and rates of biogeochemical processes occurring as well. Bacteria associated with particulate material have higher rates of extracellular enzyme activity (6, 7) and metabolism (8, 9) compared with free-living (FL) cells. However, these differences remain difficult to measure and challenging to link to biogeochemistry.
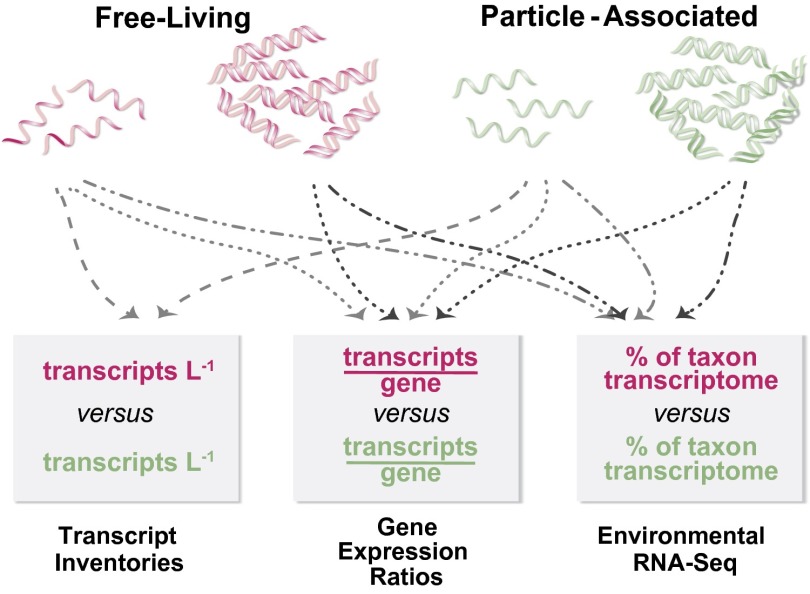
Fig. 1.
Sampling and calculation strategies for gene expression analyses. Maroon text indicates calculations for FL cells and green text for PA. Approaches to expression analysis are indicated in boxes, with the first two making use of internal gene and transcript standards.
The microspatial structure of the Amazon River Plume is strongly influenced by the mixing of riverine dissolved organic carbon (DOC) and particulate organic carbon (POC) into the tropical Atlantic. Fluvial export from the Amazon River amounts to 22.3 Tg⋅y−1 of DOC and 13.7 Tg⋅y−1 of POC (10) and equals that of the next eight largest rivers of the world combined (11). Although relatively dilute compared with other rivers (12), this mixture of dissolved and particulate nitrogen, phosphate, silica, and iron that is delivered to the ocean stimulates marine microbial activity and affects both primary productivity and carbon sequestration at a global scale (13).
Here, a metaomics methodology benchmarked with internal standards (14) was used to assemble, to our knowledge, the first fully quantitative inventories of microbial genes and transcripts in a natural ecosystem, producing a highly resolved view of gene expression driving carbon and nutrient flux through FL and particle-associated (PA) microbes of the Amazon Plume. We compare the level of transcription for the same gene in each microenvironment, enumerate transcripts mediating key carbon and nutrient transformations, and predict the biogeochemical roles of transcriptionally active FL and PA cells.
Results and Discussion
A Quantitative Multiomics Dataset.
Inventories of microbial genes and transcripts were obtained from PA and FL cells (defined operationally as PA, >2.0 μm; FL, 0.2–2.0 μm) in Amazon Plume waters at the outer continental shelf in July 2010 (station 10). Coverage of eukaryotic transcripts was improved by additionally targeting poly(A)-tailed mRNAs in the PA fraction (Fig. 1). Each sequence library obtained in duplicate for the three metaomics data types contained 1.2–11 × 106 possible protein-encoding reads averaging 190 nt. The eukaryotic community was dominated by sequences that binned to the genomes of diatoms Thalassiosira pseudonana, Phaeodactylum tricornutum, and Odontella sinensis and the green picoeukaryote Micromonas sp. RC299; direct microscopic observation confirmed a dense multispecies diatom bloom. We focus here on analysis of the genes and transcripts from Bacteria and Archaea [hereafter referred to as prokaryotes (15)], accounting for ∼70% of the annotated reads.
Based on internal standard recoveries, the sequencing depth for each sample (percentage of sampled genes or transcripts sequenced) ranged from 1.5 × 10−6 to 8.7 × 10−4 (SI Appendix, Table S1), leading to estimates of prokaryotic gene and transcript numbers per liter of seawater of ∼1 × 1013 and 5 × 1011 (Table 1). Two independent measures of prokaryotic cell abundance provided a check on quantitation from internal standard recoveries. First, flow cytometric counting indicated 5.8 × 109 cells⋅L−1 at this station, whereas standard-normalized metagenomic read counts estimated 3.6 × 109 cells⋅L−1 (Table 2). Second, direct microscopic counts of phycoerythrin- and chlorophyll-a fluorescing cells indicated 6.9 × 108 cells⋅L−1 for Synechococcus (16), whereas calculated genome equivalents based on standard-normalized metagenomic data were 6.8 × 108 cells⋅L−1 (Table 2).
Table 1.
Metagenome and metatranscriptome datasets from Amazon River Plume station 10 in June 2010
| Parameter | 0.2–2.0 μm size fraction* | ≥2.0 μm size fraction | ||||||||
| Metatranscriptomes | Metagenomes | Metatranscriptomes | Metagenomes | Poly(A)-selected metatranscriptomes | ||||||
| ACM11 | ACM29 | ACM4 | ACM36 | ACM12 | ACM30 | ACM3 | ACM37 | ACM8 | ACM27 | |
| Total reads | 3.22 × 107 | 2.04 × 107 | 1.32 × 107 | 1.44 × 107 | 4.22 × 107 | 3.08 × 107 | 1.22 × 107 | 4.66 × 106 | 3.66 × 107 | 4.46 × 107 |
| Paired reads† | 5.39 × 106 | 3.70 × 106 | 4.66 × 106 | 1.33 × 106 | 5.08 × 106 | 5.36 × 106 | 4.21 × 106 | 1.31 × 106 | 1.16 × 107 | 8.05 × 106 |
| Genes or transcripts⋅L−1 | ||||||||||
| Bacterial | 1.60 × 1011 | 2.89 × 1011 | 6.43 × 1012 | 6.12 × 1012 | 9.51 × 1010 | 3.82 × 1011 | 3.45 × 1012 | 2.59 × 1012 | — | — |
| Archaeal | 1.75 × 109 | 2.92 × 109 | 7.33 × 1010 | 6.18 × 1010 | 1.31 × 109 | 1.56 × 109 | 3.28 × 1010 | 2.61 × 1010 | — | — |
| Eukaryotic | 1.25 × 1010 | 2.49 × 1010 | 1.46 × 1011 | 5.96 × 1011 | 5.42 × 1010 | 1.03 × 1012 | 3.55 × 1011 | 1.01 × 1012 | 5.40 × 1010 | 2.57 × 109 |
| Viral | 7.88 × 109 | 3.92 × 1010 | 8.01 × 1011 | 6.71 × 1011 | 6.70 × 109 | 5.90 × 1010 | 5.99 × 1011 | 2.73 × 1011 | — | — |
| Expression Ratio | ||||||||||
| Bacterial | 0.034 | 0.075 | — | |||||||
| Archaeal | 0.033 | 0.046 | — | |||||||
| Eukaryotic | 0.048 | 0.755 | — | |||||||
| Viral | 0.030 | 0.072 | — | |||||||
Per liter calculations are based on recovery of internal standards (SI Appendix, Table S1).
Replicate samples analyzed for each size fraction and data type are shown.
Post-quality control sequences.
Table 2.
Contribution of the most abundant prokaryotic taxa (out of 2,999 total) to Amazon Plume metagenomes and metatranscriptomes
| Genome bin | Genes per L | PA genes, % | % of total genes | Genes in reference genome | Genome equivalents per L | % of cell count* | Protein % identity | Transcripts per L | PA transcripts, % | % of total transcripts |
| Synechococcus sp. CB0205 | 1.60 × 1012 | 39.09 | 15.96 | 2,719 | 5.87 × 108 | 10.13 | 89.5 | 1.90 × 1010 | 16.7 | 7.36 |
| Candidatus Pelagibacter sp. HTCC7211 | 3.62 × 1011 | 30.99 | 3.63 | 1,447 | 2.50 × 108 | 4.31 | 75.9 | 2.07 × 109 | 52.8 | 0.80 |
| Gammaproteobacterium HTCC2080 | 2.51 × 1011 | 27.72 | 2.53 | 3,185 | 7.89 × 107 | 1.36 | 71.2 | 8.25 × 109 | 30.9 | 3.20 |
| Synechococcus sp. CB0101 | 1.50 × 1011 | 40.69 | 1.50 | 3,010 | 4.98 × 107 | 0.86 | 87.8 | 2.83 × 109 | 19.5 | 1.10 |
| Gammaproteobacterium NOR51-B | 1.48 × 1011 | 27.90 | 1.49 | 2,930 | 5.05 × 107 | 0.87 | 70.7 | 7.18 × 109 | 34.7 | 2.78 |
| Synechococcus sp. RS9916 | 1.24 × 1011 | 46.20 | 1.24 | 2,961 | 4.20 × 107 | 0.72 | 83.9 | 2.91 × 109 | 26.6 | 1.13 |
| Muricauda ruestringensis DSM13258 | 9.81 × 1010 | 29.32 | 0.99 | 3,432 | 2.86 × 107 | 0.49 | 69.3 | 3.46 × 109 | 48.5 | 1.34 |
| Gammaproteobacterium HTCC2148 | 9.05 × 1010 | 29.29 | 0.91 | 3,827 | 2.37 × 107 | 0.41 | 66.2 | 2.63 × 109 | 35.6 | 1.02 |
| Fluviicola taffensis DSM 16823 | 7.55 × 1010 | 50.56 | 0.75 | 4,033 | 1.87 × 107 | 0.32 | 64.4 | 7.23 × 109 | 41.9 | 2.80 |
| Haliscomenobacter hydrossis DSM1100 | 7.55 × 1010 | 49.87 | 0.75 | 6,858 | 1.10 × 107 | 0.19 | 61.7 | 1.90 × 109 | 48.2 | 0.74 |
| Kordia algicida OT-1 | 7.46 × 1010 | 34.22 | 0.75 | 4,514 | 1.65 × 107 | 0.29 | 67.5 | 1.91 × 109 | 48.0 | 0.74 |
| Gammaproteobacterium IMCC3088 | 6.52 × 1010 | 27.83 | 0.65 | 2,855 | 2.28 × 107 | 0.39 | 67.6 | 1.98 × 109 | 35.3 | 0.77 |
| Gammaproteobacterium HTCC2207 | 6.34 × 1010 | 27.14 | 0.64 | 2,388 | 2.66 × 107 | 0.46 | 62.4 | 2.26 × 109 | 23.7 | 0.88 |
| Zobellia galactanivorans | 6.29 × 1010 | 30.77 | 0.63 | 4,732s | 1.33 × 107 | 0.23 | 68.6 | 1.80 × 109 | 44.6 | 0.70 |
| Erythrobacter litoralis HTCC2594 | 6.27 × 1010 | 62.52 | 0.62 | 3,011 | 2.08 × 107 | 0.36 | 76.9 | 3.36 × 109 | 45.3 | 1.30 |
| Total | 33.04 | 4.87 × 108* | 21.39 | 26.66 |
Data are based on replicates ACM3, ACM4, ACM11, and ACM12, and size fractions are summed.
Genome equivalents for the 250 most abundant taxa (75% of prokaryotic genes) = 2.7 × 109. The direct cell count at station 10 was 5.80 × 109 cells⋅L−1.
Natural History of Amazon Plume Prokaryotes.
Protein sequence comparisons against reference genomes indicated that the prokaryotic assemblage was dominated by Cyanobacteria in the Synechococcaceae family and heterotrophic bacteria in the Gammaproteobacteria, Bacteroidetes, and Alphaproteobacteria. The average percentage identity between the metagenomic reads and the reference proteins they binned to ranged from 62% to 92% for the dominant taxa (Table 2). About 0.1% of metagenomic reads contained 16S rRNA gene fragments, and the taxonomic distribution of these matched the metagenomic protein binning and the patterns of PCR-amplified 16S rRNA genes (SI Appendix, Fig. S1).
Prokaryotic genes in the Amazon Plume were present in higher copy number than the mRNAs produced from them. This is consistent with earlier studies of Escherichia coli (17) and marine bacterioplankton (18, 19) and reflects a small and dynamic mRNA pool in environmental cells. Genes were split 68%/32% between the FL and PA fractions, and transcripts were split 49%/51%. Thus, per-gene expression levels were twofold greater for PA cells than FL cells (Table 1), a finding in line with previous reports of higher rates of growth and enzymatic activity for attached cells (6). Approximately 99% of prokaryotic genes and transcripts were bacterial and ∼1% were archaeal. The compositional difference between FL and PA prokaryotes was small at station 10. Although disruption of the microspatial structure during sampling could homogenize the communities, samples collected identically from other stations had distinct compositional differences between microenvironments (SI Appendix, Fig. S2). There were also substantial differences in transcription patterns between size fractions within a single genome bin at this station (SI Appendix, Fig. S3).
Transcript Inventories of Biogeochemically Relevant Genes.
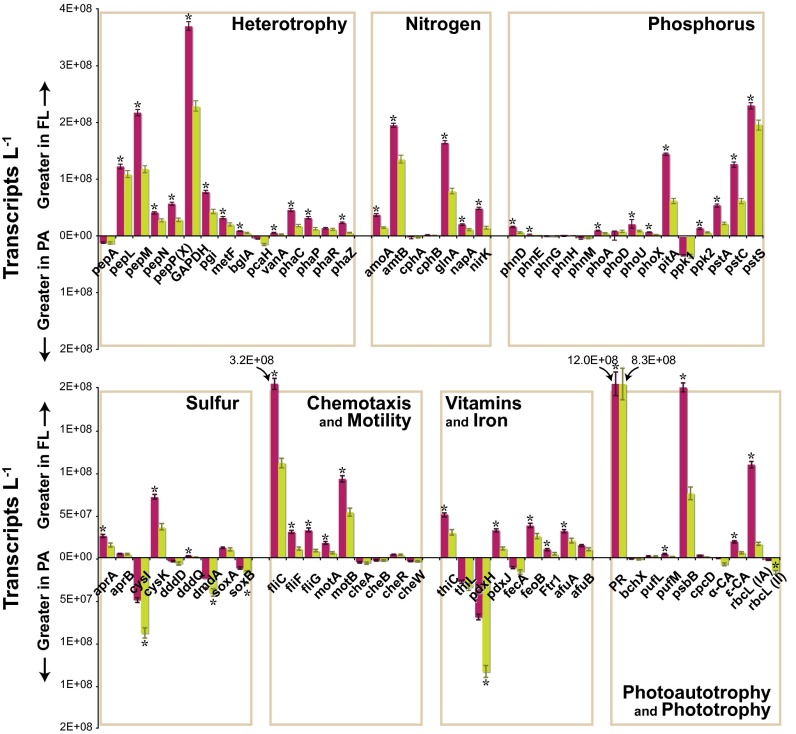
Transcript abundance per liter of seawater (Fig. 1) was determined for prokaryotic genes involved in transport, fixation, and metabolism of C, N, P, and S (SI Appendix, Table S2). Counts ranged from near the detection limit at ∼105 transcripts⋅L−1 for phosphonate utilization gene phnG up to ∼2 × 109 transcripts⋅L−1 for proteorhodopsin (Fig. 2). Most genes had higher transcript counts in the FL microenvironment (Fig. 2) due in part to the greater abundance of FL genomes (Table 2). These included genes involved in heterotrophic C metabolism (12 of 15), phototrophy (5 of 10), N cycling (5 of 7), P cycling (10 of 15), and motility (5 of 5). Only a few genes had transcript inventories biased toward the PA community; these included vitamin B6 biosynthesis gene pdxH; S cycling genes cysI, soxB, and dmdA; and aromatic ring cleavage gene pcaH (Fig. 2). Summing across genes with related functions, a liter of seawater at this plume station contained more transcripts mediating C and N cycling originating from genomes of FL cells but more transcripts for vitamin biosynthesis and S cycling from PA cells.
Fig. 2.
Transcript inventories for biogeochemically relevant genes. Bars above the x axis indicate more transcripts contributed by the FL prokaryotes (maroon), and those below indicate more contributed by PA prokaryotes (green). Asterisks indicate significant differences between fractions. afu, iron transport; amoA, ammonium monooxygenase; amtB, ammonium transporter B; apr, adenosine-5′-phosphosulfate reductase; bchX, chlorophyll iron protein; bglA, beta-glucosidase; CA, carbonic anhydrases; che, chemotaxis-related; cpcD, phycocyanin-associated linker polypeptide; cph, cyanophycin-related; cysI, sulfite reductase; cysK, cysteine synthase; ddd, DMSP lyase; dmdA, DMSP demethylase; fecA, Fe dicitrate transporter; feoB, Fe(II) G protein-like transporter; fli, flagellar proteins; Ftr1, high affinity Fe(II) permease; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; glnA, glutamine synthetase; metF, methylenetetrahydrofolate reductase; mot, flagellar motor; napA, nitrate reductase; nirK, nitrite reductase; pcaH, protocatechuate 3,4-dioxygenase (3,4-PCD); pdx, vitamin B6 synthesis; pep, aminopeptidases; pgi, glucose-6-phosphate isomerase; pha, polyhydroxyalkanoate-related; phn, phosphonate assimilation; pho, alkaline phosphatase; pitA, low affinity PO4 transporter; ppk, polyphosphate kinase; PR, proteorhodopsin; psbB, photosystem II; pst, high affinity PO4 transporter; puf, photosynthetic reaction center; rbcL, ribulose 1,5-bisphosphate carboxylases/oxygenases; sox, sulfur oxidation; thi, thiamine synthesis; vanA, vanillate demethylase.
Expression Ratios of Biogeochemically Relevant Genes.
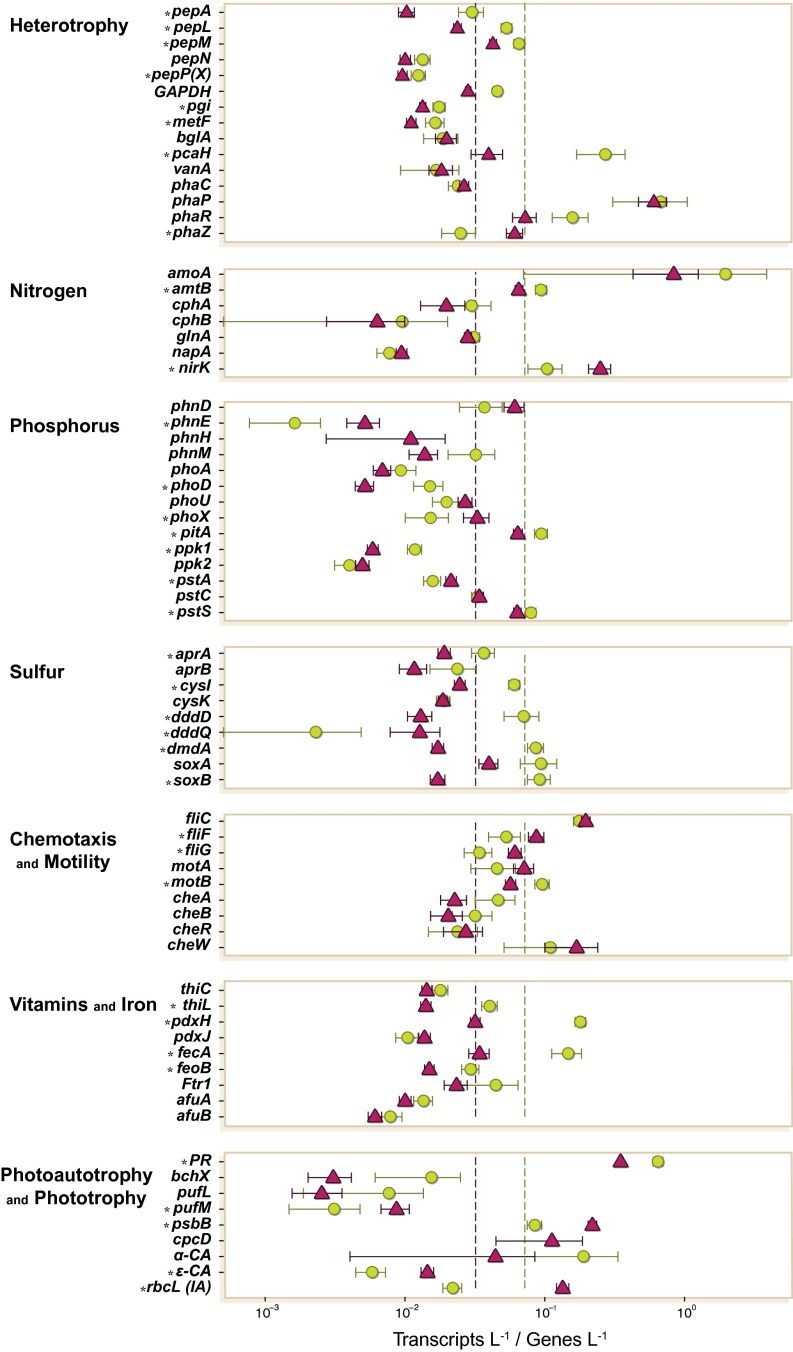
A different calculation approach scaled transcript copies to gene copies (transcripts⋅gene−1) to ask whether the same gene is differentially regulated in the two microenvironments (Fig. 1). Heterotrophic C transformation genes were up-regulated in genomes of PA cells compared with those from FL cells (Fig. 3), including genes for the catabolism of aminopeptidases (pepA,L,M), glycolysis (GAPDH), and aromatic compound metabolism (pcaH but not lignin-related vanA). Only one heterotrophy gene was significantly up-regulated in FL cells, and this mediates the degradation of cellular carbon reserves (phaZ) and does not contradict the higher organic C availability signal for PA cells. Up-regulated genes in PA prokaryotes also included those for synthesis of vitamin B6 (pdxH) and B1 (thiL) required for amino acid and central carbon metabolism (20, 21). Genes for uptake of siderophore-bound Fe and Fe+2 were up-regulated in genomes from PA cells as well, possibly linked to the role of iron–sulfur clusters and heme in respiratory chains and TCA cycle enzymes. Thus, although PA prokaryotes contributed fewer transcripts to the water column, they were more transcriptionally active per gene in C heterotrophy than FL cells.
Fig. 3.
Gene expression ratios of biogeochemically relevant genes. Dotted lines indicate the average expression ratio for all genes in each size fraction. Maroon, FL; green, PA. Asterisks indicate significant differences between size fractions. Gene abbreviations are as in the Fig. 2 legend.
Among nutrient acquisition genes, those encoding P uptake had the greatest differences in regulation between microenvironments. Low-affinity phosphate transporter pitA was up-regulated in the PA transcriptome, and high-affinity pstA was up-regulated in the FL transcriptome (Fig. 3). Together with the up-regulation of phosphonate transporter gene phnE by FL genomes, turnover of labile P in bulk seawater is predicted to be more rapid and competition more intense compared with particle microenvironments. For N cycle genes, differences in expression levels were smaller, and N availability status was therefore predicted to be more similar between microenvironments. An exception was a dissimilatory nitrate reductase homolog binning exclusively to Thaumarchaeota genomes and up-regulated in the FL fraction, although the function of the nirK homolog in Thaumarchaeota is uncertain (22). Expression ratios were higher in PA cells for genes that mediate organic and inorganic S metabolism, including dimethylsulfoniopropionate (DMSP) and sulfate. Thus, although nutrient concentrations at this station (0.2 μM NOx, 0.4 μM PO4) were high compared with more offshore regions of the plume, transcription patterns suggested microscale differences in their availability and turnover. Genes associated with light harvesting were also expressed differently (Fig. 3); FL cells had higher expression of genes for autotrophic light utilization [photosynthesis and C fixation genes pufM, psbB, ε-CA, and rbcL (type IA)], whereas PA cells had higher expression of photoheterotrophic light utilization (proteorhodopsin). FL prokaryotes also had higher expression ratios of flagellar genes (Fig. 3).
Gene Abundance Versus Gene Regulation.
Three reference genomes with high sequence coverage were used for taxon-specific determinations of whether genome abundance (genomes⋅L−1) or gene regulation (transcripts⋅gene−1) contributed more to differences in microenvironment transcriptomes. The potential complication that a genome bin was derived from different populations in each microenvironment was considered. However, analysis of percentage identity patterns for three housekeeping genes (rpoB, gyrB, and recA) indicated that although genomes may recruit more than one population, the relative population abundances did not differ substantively between FL and PA fractions for the Synechococcus sp. CB0205, Pelagibacter sp. HTCC7211, and gammaproteobacterium HTCC2070 bins (SI Appendix, Fig. S4).
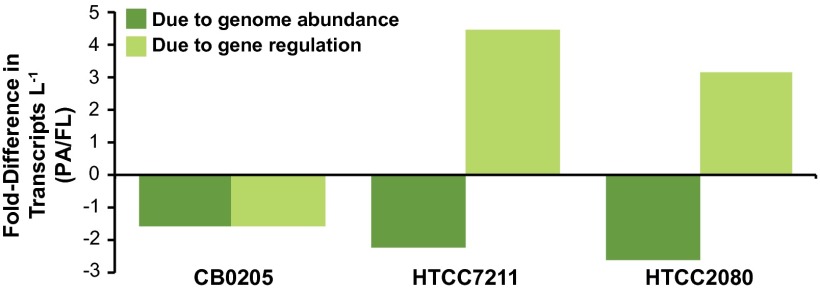
Regarding genome abundance differences, Synechococcus sp. CB0205-like populations had 1.6-fold fewer genomes⋅L−1 in the PA microenvironment. Regarding gene regulation differences, all metabolic pathways in CB0205 that exhibited significant differences in expression levels were down-regulated in PA cells compared with FL cells (62 pathways; Table 3). These included pathways related to autotrophy (photosynthesis, carotenoid biosynthesis, and C fixation), as well as biosynthesis of amino acids, fatty acids, and peptidoglycan (Table 3). At the individual gene level, a total of 516 were down-regulated in PA cells compared with FL (SI Appendix, Table S3). Among them were genes transcribed by picocyanobacteria when experiencing N limitation (ntcA; fourfold down-regulated) and P limitation (pstS; sixfold down-regulated) (23, 24). Only 23 individual genes were up-regulated in PA cells, including stress-related proteins (universal stress protein, peroxiredoxin, and glutaredoxin) and cell-wall–modifying proteins (SI Appendix, Fig. S5 and Table S3). Overall, the transcriptome of CB0205-like cells was affected equally and in the same direction by genome abundance and gene regulation factors; when averaged across the genome, PA genes were less abundant by ∼1.6-fold and less transcriptionally active by ∼1.6-fold (Fig. 4 and SI Appendix, Fig. S5).
Table 3.
Selected pathways from the Kyoto Encyclopedia of Genes and Genomes database with different expression ratios (transcripts⋅gene−1) between microenvironments for Synechococcus sp. CB0205, Pelagibacter sp. HTCC7211, and Gammaproteobacterium HTCC2080
| Pathway | Description | CB0205 | HTCC7211 | HTCC2080 |
| ko00010 | Glycolysis/gluconeogenesis | FL | PA | PA |
| ko00030 | Pentose phosphate pathway | FL | PA | |
| ko00051 | Fructose, mannose metabolism | FL | PA | |
| ko00071 | Fatty acid metabolism | PA | PA | |
| ko00190 | Oxidative phosphorylation | FL | PA | PA |
| ko00195 | Photosynthesis | FL | ||
| ko00196 | Photosynthesis–antenna | FL | ||
| ko00230 | Purine metabolism | FL | PA | PA |
| ko00240 | Pyrimidine metabolism | FL | PA | PA |
| ko00253 | Tetracycline biosynthesis | FL | PA | |
| ko00260 | Glycine, serine, threonine metabolism | PA | ||
| ko00280 | Val, leu, isoleu degradation | FL | PA | PA |
| ko00310 | Lysine degradation | FL | ||
| ko00330 | Arginine, proline metabolism | FL | PA | PA |
| ko00340 | Histidine metabolism | FL | PA | |
| ko00360 | Phenylalanine metabolism | PA | ||
| ko00380 | Tryptophan metabolism | FL | ||
| ko00430 | Taurine metabolism | PA | ||
| ko00480 | Glutathione metabolism | FL | PA | PA |
| ko00500 | Starch, sucrose metabolism | FL | FL | |
| ko00521 | Streptomycin biosynthesis | FL | ||
| ko00523 | Polyketide biosynthesis | FL | ||
| ko00620 | Pyruvate metabolism | FL | PA | PA |
| ko00630 | Glyoxylate, dicarboxylate metabolism | FL | PA | PA |
| ko00640 | Propanoate metabolism | PA | ||
| ko00650 | Butanoate metabolism | FL | PA | |
| ko00670 | One carbon pool by folate | FL | PA | |
| ko00710 | Carbon fixation | FL | ||
| ko00730 | Thiamine metabolism | FL | ||
| ko00740 | Riboflavin metabolism | FL | ||
| ko00760 | Nicotinate metabolism | FL | PA | PA |
| ko00770 | Pantothenate, CoA biosynthesis | FL | PA | |
| ko00780 | Biotin metabolism | FL | ||
| ko00790 | Folate biosynthesis | FL | PA | |
| ko00860 | Porphyrin, chlorophyll metabolism | FL | PA | PA |
| ko00910 | Nitrogen metabolism | FL | PA | PA |
| ko00920 | Sulfur metabolism | FL | PA | |
| ko01053 | Biosynthesis siderophore NRPs | PA | ||
| ko01055 | Biosyntheis Vancomycin | FL | ||
| ko02020 | Two-component systems | FL | PA | PA |
| ko02040 | Flagellar assembly | FL | ||
| ko03030 | DNA replication | FL | PA | PA |
| ko03060 | Protein export | FL | PA | |
| ko03430 | Mismatch repair | FL | PA | PA |
| ko03440 | Homologous recombination | FL | PA | PA |
FL, greater in FL cells; PA, greater in PA cells.
Fig. 4.
Role of cell abundance versus gene regulation in the differential contribution of PA cells to the Amazon Plume metatranscriptome.
The two heterotrophic taxa likewise had lower abundance of PA genomes (2.2-fold for Pelagibacter sp. HTCC7211 and 2.6-fold for gammaproteobacterium HTCC2080; Fig. 4) but different gene regulation patterns compared with the Synechococcus bin. For HTCC7211, all metabolic pathways with differences in expression levels (39 pathways) were up-regulated in PA compared with FL cells (Table 3), including amino acid, nucleic acid, carboxylic acid, C1 compound, and taurine metabolism pathways (Table 3). For gammaproteobacterium HTCC2080, 31 metabolic pathways were up-regulated in PA cells, including carboxylic and fatty acid metabolism and siderophore production, whereas three were down-regulated, including flagellar assembly (Table 3). At the individual gene level, HTCC7211 had 84 genes up-regulated in PA compared with FL cells, and HTCC2080 had 44 (SI Appendix, Tables S4 and S5). Stress-related functions (heat shock protein, acid tolerance protein, chaperones, and GST) and transporters (ammonium, amino acids, taurine, polyamine, and glycine betaine; SI Appendix, Fig. S5) were represented. Overall, genome abundance and gene regulation differences worked in opposite directions in the HTCC7211 and HTCC2080 bins; when averaged across the genome, cells assigned to these two heterotrophic taxa were less abundant by ∼2.5-fold but more transcriptionally active by ∼three- to fourfold when associated with particles (Fig. 4).
A Prokaryotic Perspective on Amazon Plume Biogeochemistry.
High rates of C fixation and a deficit of dissolved inorganic C in the water column (∼100 μmol⋅L−1) signaled a major system shift to net autotrophy at station 10, driven by release from light limitation and stimulation by river-derived nutrients. This hotspot of primary production was upstream of an area of high POC deposition (25), indicative of sequestration of new production in nearby continental slope sediments. Surface water was dominated by dense populations of coastal diatom species, suggesting substantial contributions by these cells to the particulate material. Indeed, phytoplankton C accounted for ∼80% of POC (676 μg⋅C⋅L−1 estimated from cell count and biovolume measures, compared with 842 μg⋅C⋅L−1 total POC), in agreement with δ13C analysis and terrestrial biomarker concentrations (triterpenoids, C29-sterols, isoprenoids, and selected sugars) that suggested only 10–20% terrestrial POC.
Within this matrix of marine and terrestrial organic matter, the up-regulation of photosynthesis genes in FL cells of the dominant prokaryotic primary producers in the Synechococcus clade (Fig. 3 and SI Appendix, Fig. S6) accords with their higher abundance in FL microenvironments in this study (Table 2) and others (2). Synechococcus transcription patterns also suggested greater nutrient competition when free living based on N and P stress genes ntcA and pstS, and this may reflect higher nutrient demand in support of photosynthetic activity as well as lower nutrient availability to cells in the FL microenvironment. Expression of nitrate and nitrite transporters in both microenvironments indicated that Synechococcus competed with eukaryotic primary producers for riverine-derived N (Table 3 and SI Appendix, Table S3). Although the ∼1-μm–diameter cells are not typically thought to sink efficiently in the ocean, some studies suggest that picophytoplankton indeed contribute to deep flux in proportion to their biomass (4, 26). At this outer plume station, picocyanobacteria accounted for ∼12% of prokaryotic cells (∼109⋅L−1) (Table 2), a value at the upper range for coastal oceans. In contrast, the photoheterotrophic prokaryotes showed higher expression ratios of light-capturing proteorhodopsin in the particle microenvironment (Fig. 3), with transcripts dominated by Flavobacteria and SAR116. The SAR11 HTCC7211 bin differed from this pattern in having similar proteorhodopsin expression ratios in both microenvironments (1.0 and 0.8 transcripts⋅gene−1), and previous studies agree that SAR11 proteorhodopsin genes are not highly regulated (27). Aerobic anoxygenic phototrophy appeared insignificant here, given gene counts and expression ratios ∼2 orders of magnitude lower for bchX, pufL, and pufM compared with proteorhodopsin and the oxygenic photosynthesis genes (Fig. 3). Overall, transcription patterns suggest that micrometer-scale heterogeneity within a single water depth influences the rates and location of prokaryotic phototrophic activity.
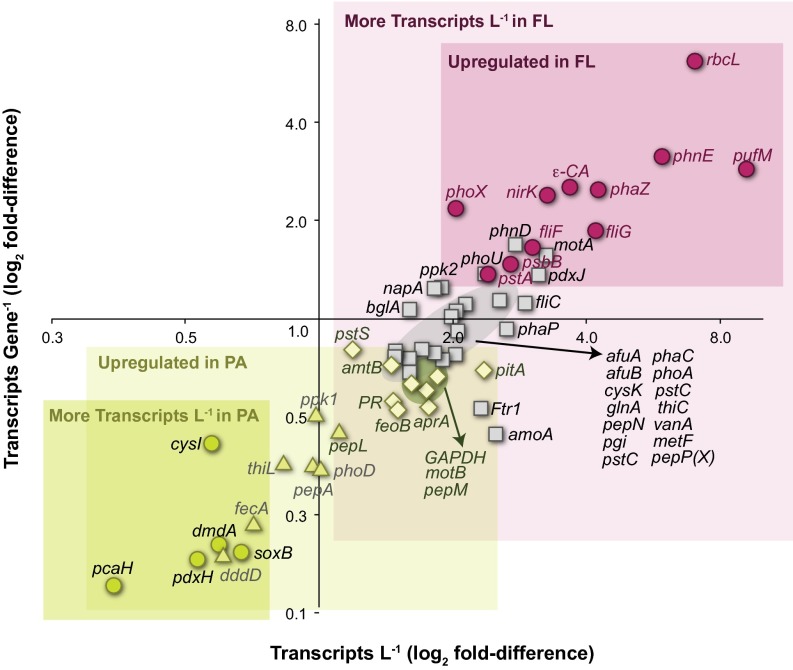
A persistent question in microbial biogeochemistry is whether the typically higher activity of less abundant PA cells outweighs the typically lower activity of more abundant FL cells in mediating biogeochemical transformations (2, 6). For the 74 biogeochemically relevant genes assayed here, there were five answers to the question (Fig. 5). One gene class was up-regulated sufficiently strongly in PA cells (∼fivefold greater than in FL) that they dominated the community transcriptome despite fewer genomes in this microenvironment; this group included functions related to S cycling, vitamin biosynthesis, and aromatic compound degradation. Another gene class was significantly up-regulated in PA cells, but the fold difference (three- to fourfold) was insufficient to overcome genome abundance differences and the community transcriptomes had statistically indistinguishable representation; this group included genes for Fe acquisition, P storage, S transformation, and vitamin biosynthesis. For the three remaining outcomes, significantly more transcripts were contributed by cells in the FL microenvironment. These consisted of a gene group up-regulated in PA cells but at fold differences insufficient to make up for the greater number of FL genomes (∼1.5- to twofold; proteorhodopsin, C heterotrophy, and other Fe and S genes), a group with no difference in expression ratios but for which there were more transcripts produced by the more numerous FL cells (phosphate and phosphonate acquisition genes), and a group up-regulated in FL prokaryotes (∼threefold greater than in PA) and dominating the community transcriptome (up to 10-fold; CO2 fixation, motility, and P acquisition; Fig. 5).
Fig. 5.
Outcome of differential transcript inventories and expression ratios for biogeochemically relevant genes. Inventory ratios are plotted on the x axis and gene expression ratios on the y axis. Green symbols, genes up-regulated in the PA fraction and in greater abundance in the PA community (circles), no difference in abundance between PA and FL communities (triangles), or greater abundance in the FL community (diamonds). Gray symbols, genes not up-regulated in either fraction, but in greater abundance in the FL community. Maroon symbols, genes both up-regulated and more abundant in the FL community. Gene abbreviations are as in the Fig. 2 legend.
The importance of microspatial partitioning of microbial activities in the ocean was first suspected based on differences in taxonomic composition between FL and PA cells (8) and later supported by analysis of gene inventories (2, 3). At the transcriptome level, contributions are determined not just by the abundance of each taxon’s genome but also by the regulation of its genes. By benchmarking with internal standards, differences in gene regulation were determined to be more important than differences in genome counts in explaining transcriptome composition at this Amazon Plume station. Up to 65% of the genes in individual taxa exhibited significantly different expression when FL and PA cells were compared (Fig. 4); on average, PA cells had twice as many transcripts as FL cells (Table 1); and expression of biogeochemically relevant genes varied by up to sixfold between the microenvironments (Fig. 3). With the important caveat that mRNA abundance cannot be interpreted as a proxy for elemental flux (19), we hypothesize that the FL prokaryotic community contributed more to C fixation, heterotrophy, N and P uptake, and iron acquisition than the PA community in this ecosystem, whereas the PA community contributed more to S cycling, aromatic compound degradation, and the synthesis of biologically essential vitamins. The scale of spatial heterogeneity relevant to ecological processes (1) is indeed at the micrometer level in this ecosystem, with patchiness in abundance and regulation of prokaryotic genes within a single water mass signaling the partitioning of functions that drive elemental cycling.
Materials and Methods
Surface seawater was collected ∼500 km north of the Amazon River mouth in June 2010. Two size fractions were obtained by sequential filtration of 156 μm prefiltered water through 2.0 μm and 0.2 μm pore size membrane filters. Duplicate metagenomes and metatranscriptomes were obtained from each fraction as 2 × 150 bp reads. Internal standards were added immediately before cell lysis in known copy numbers (SI Appendix, Table S1). Assignment of reads to taxonomic bins and functions was based on Blastx searches against RefSeq or a custom database of 74 selected genes. Detailed sample collection, processing, and statistical analysis methods can be found in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We appreciate the help of A. Burd, E. Carpenter, A. Mehring, C. English, J. Mrazek, and R. Nilsen. Resources were provided by the University of Georgia’s Georgia Advanced Computing Resource Center and the Community Cyberinfrastructure for Advanced Microbial Ecology Research and Analysis. This work was funded in part by the Gordon and Betty Moore Foundation through Grants GBMF2293 and GBMF2928 and National Science Foundation Grant OCE-0934095.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive, www.ncbi.nlm.nih.gov/sra [accession nos. SRP039390 (metagenomes), SRP037995 (nonselective metatranscriptomes), and SRP039544 (poly(A)-selected metatranscriptomes)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1402782111/-/DCSupplemental.
References
- 1.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5(10):782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 2.Ganesh S, Parris DJ, DeLong EF, Stewart FJ. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014;8(1):187–211. doi: 10.1038/ismej.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith MW, Zeigler Allen L, Allen AE, Herfort L, Simon HM. Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol. 2013;4:120. doi: 10.3389/fmicb.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiorboe T, Jackson GA. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol Oceanogr. 2001;46(6):1309–1318. [Google Scholar]
- 5.Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc Natl Acad Sci USA. 2008;105(11):4209–4214. doi: 10.1073/pnas.0709765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crump BC, Baross JA, Simenstad CA. Dominance of particle-attached bacteria in the Columbia River estuary, USA. Aquat Microb Ecol. 1998;14(1):7–18. [Google Scholar]
- 7.Karner M, Herndl GJ. Extracellular enzymatic-activity and secondary production in free-living and marine-snow-associated bacteria. Mar Biol. 1992;113(2):341–347. [Google Scholar]
- 8.Delong EF, Franks DG, Alldredge AL. Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38(5):924–934. [Google Scholar]
- 9.Hopkinson CS, Sherr B, Wiebe WJ. Size fractionated metabolism of coastal microbial plankton. Mar Ecol Prog Ser. 1989;51(1-2):155–166. [Google Scholar]
- 10.Richey JE, et al. Biogeochemistry of carbon in the Amazon River. Limnol Oceanogr. 1990;35(2):352–371. [Google Scholar]
- 11.Coles VJ, et al. The pathways and properties of the Amazon River Plume in the tropical North Atlantic Ocean. J Geophys Res-Oceans. 2013;118(12):6894–6913. [Google Scholar]
- 12.Ryther JH, Menzel DW, Corwin N. Influence of the Amazon River outflow on the ecology of the western tropical Atlantic. J Mar Res. 1967;25(1):69–83. [Google Scholar]
- 13.Subramaniam A, et al. Amazon River enhances diazotrophy and carbon sequestration in the tropical North Atlantic Ocean. Proc Natl Acad Sci USA. 2008;105(30):10460–10465. doi: 10.1073/pnas.0710279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Satinsky BM, Gifford SM, Crump BC, Moran MA. 2013. Use of internal standards for quantitative metatranscriptome and metagenome analysis. Meth Enzymol, ed DeLong EF (Academic, Burlington, MA), Vol 531, pp 237–250. [DOI] [PubMed]
- 15.Whitman WB. The modern concept of the procaryote. J Bacteriol. 2009;191(7):2000–2005, discussion 2006–2007. doi: 10.1128/JB.00962-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goes JI, et al. Influence of the Amazon River discharge on the biogeography of phytoplankton communities in the western tropical north Atlantic. Prog Oceanogr. 2014;120:29–40. [Google Scholar]
- 17.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329(5991):533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12(3):679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moran MA, et al. Sizing up metatranscriptomics. ISME J. 2013;7(2):237–243. doi: 10.1038/ismej.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzpatrick TB, et al. Two independent routes of de novo vitamin B6 biosynthesis: Not that different after all. Biochem J. 2007;407(1):1–13. doi: 10.1042/BJ20070765. [DOI] [PubMed] [Google Scholar]
- 21.Koenigsknecht MJ, Downs DM. Thiamine biosynthesis can be used to dissect metabolic integration. Trends Microbiol. 2010;18(6):240–247. doi: 10.1016/j.tim.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollibaugh JT, Gifford S, Sharma S, Bano N, Moran MA. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J. 2011;5(5):866–878. doi: 10.1038/ismej.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindell D, Post AF. Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl Environ Microbiol. 2001;67(8):3340–3349. doi: 10.1128/AEM.67.8.3340-3349.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scanlan DJ, Mann NH, Carr NG. The response of the picoplanktonic marine cyanobacterium Synechococcus species WH7803 to phosphate starvation involves a protein homologous to the periplasmic phosphate-binding protein of Escherichia coli. Mol Microbiol. 1993;10(1):181–191. doi: 10.1111/j.1365-2958.1993.tb00914.x. [DOI] [PubMed] [Google Scholar]
- 25.Chong LS, et al. Carbon and biogenic silica export influenced by the Amazon River Plume: Patterns of remineralization in deep-sea sediments. Deep Sea Res Part I Oceanogr Res Pap. 2014;85:124–137. [Google Scholar]
- 26.Close HG, et al. Export of submicron particulate organic matter to mesopelagic depth in an oligotrophic gyre. Proc Natl Acad Sci USA. 2013;110(31):12565–12570. doi: 10.1073/pnas.1217514110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS ONE. 2011;6(5):e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.