Significance
The diagnosis of tumors during surgery still relies principally on an approach developed over 150 y ago: frozen section microscopy. We show that a validated molecular marker—2-hydroxyglutarate generated from isocitrate dehydrogenase 1 mutant gliomas—can be rapidly detected from tumors using a form of ambient MS that does not require sample preparation. We use the Advanced Multimodality Image Guided Operating Suite at Brigham and Women’s Hospital to demonstrate that desorption electrospray ionization MS could be used to detect residual tumor that would have been left behind in the patient. The approach paves the way for the clinical testing of MS-based intraoperative monitoring of tumor metabolites, an advance that could revolutionize the care of surgical oncology patients.
Keywords: mass spectrometry imaging, oncometabolite, intrasurgical diagnosis, brain cancer, IDH1
Abstract
For many intraoperative decisions surgeons depend on frozen section pathology, a technique developed over 150 y ago. Technical innovations that permit rapid molecular characterization of tissue samples at the time of surgery are needed. Here, using desorption electrospray ionization (DESI) MS, we rapidly detect the tumor metabolite 2-hydroxyglutarate (2-HG) from tissue sections of surgically resected gliomas, under ambient conditions and without complex or time-consuming preparation. With DESI MS, we identify isocitrate dehydrogenase 1-mutant tumors with both high sensitivity and specificity within minutes, immediately providing critical diagnostic, prognostic, and predictive information. Imaging tissue sections with DESI MS shows that the 2-HG signal overlaps with areas of tumor and that 2-HG levels correlate with tumor content, thereby indicating tumor margins. Mapping the 2-HG signal onto 3D MRI reconstructions of tumors allows the integration of molecular and radiologic information for enhanced clinical decision making. We also validate the methodology and its deployment in the operating room: We have installed a mass spectrometer in our Advanced Multimodality Image Guided Operating (AMIGO) suite and demonstrate the molecular analysis of surgical tissue during brain surgery. This work indicates that metabolite-imaging MS could transform many aspects of surgical care.
The microscopic review of tissue biopsies frequently remains the sole source of intraoperative diagnostic information, and many important surgical decisions such as the extent of tumor resection are based on this information. This approach is time-consuming, requiring nearly 30 min between the moment a tissue is biopsied and the time the pathologist’s interpretation is communicated back to the surgeon. Even after the report of the final pathologic diagnosis is issued days later, a lot of diagnostic, prognostic, and predictive information is left undiscovered and unexamined within the tissue. Tools that provide more immediate feedback to the surgeon and the pathologist and that also rapidly extract detailed molecular information could transform the management of care for cancer patients.
MS offers the possibility for the in-depth analysis of the proteins and lipids that comprise tissues (1, 2). We have recently shown that desorption electrospray ionization (DESI) MS is a powerful methodology for characterizing lipids within tumor specimens (3–6). The intensity profile of lipids ionized from within tumors can be used for classifying tumors and for providing valuable prognostic information such as tumor subtype and grade. Because DESI MS is performed in ambient conditions with minimal pretreatment of the samples (7, 8), there is the potential to provide diagnostic information rapidly within the operating room (4, 6, 9). The ability to quickly acquire such valuable diagnostic information from lipids prompted us to determine whether we could use DESI MS to detect additional molecules of diagnostic value within tumors, such as their metabolites.
Recently, recurrent mutations have been described in the genes encoding isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) in a number of tumor types including gliomas (10, 11), intrahepatic cholangiocarcinomas (12), acute myelogenous leukemias (13), and chondrosarcomas (14). These mutant enzymes have the novel property of converting isocitrate to 2-hydroxyglutarate (2-HG) (15). This oncometabolite has pleiotropic effects on DNA methylation patterns (16–18), on the activity of prolyl hydroxylases (19), and on cellular differentiation and growth (20–22). Whereas 2-HG is present in vanishingly small amounts in normal tissues, concentrations are extremely high in tumors with mutations in IDH1 and IDH2—several micromoles per gram of tumor have been reported (15). Several groups have reported that 2-HG can be detected by magnetic resonance spectroscopy and imaging, hence providing a noninvasive imaging approach for evaluating patients (23–27). Although such imaging approaches may provide information to plan surgery and to follow the response to chemotherapeutics, applying them to guide decision making during an operation is currently impractical.
The ability to detect 2-HG intraoperatively would be particularly useful because infiltrating gliomas such as IDH1 and IDH2 mutant gliomas are difficult to visualize with conventional means, which contributes to the high prevalence of suboptimal surgical resection. Multiple studies suggest that the more residual tumor remains after surgery, the shorter the patient survival for both low- and high-grade gliomas (28–32). Detecting infiltrating glioma cells by microscopic review is challenging on well-prepared H&E-stained permanent sections, and even more so on H&E-stained frozen sections, which frequently harbor processing artifacts. Thus, 2-HG detection could help to define surgical margins, thereby allowing for more complete resection and for longer survival (31, 32). Moreover, directing patients toward appropriate clinical trials for targeted therapeutics (33) would be facilitated by more rapid molecular categorization of tumors.
Here, we show that 2-HG can be rapidly detected from glioma samples using DESI MS under ambient conditions, without complex tissue preparation and during surgery, allowing rapid molecular characterization and providing information that is unattainable by standard histopathology techniques. We also present the first implementation, to our knowledge, of MS within an operating room for the molecular characterization of tissue as part of an image-guided therapy program. We cross-validate our findings using standard pathology techniques. Measuring specific metabolites in tumor tissues with precise spatial distribution and under ambient conditions provides a new paradigm for intraoperative surgical decision making, rapid diagnosis, and patient care management.
Results
Identification of 2-HG with DESI MS.
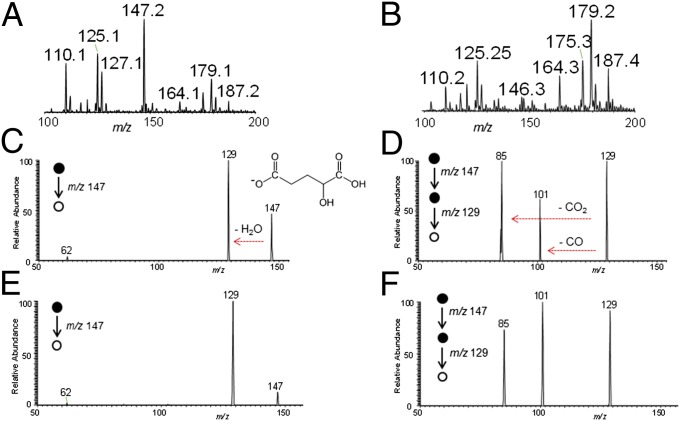
To determine the conditions for detecting 2-HG from glioma frozen tissue sections by DESI MS, we first recorded the negative ion mode mass spectra from two glioma samples: an oligodendroglioma with mutated IDH1 (encoding the amino acid change R132H) and a glioblastoma with wild-type IDH1. The product of mutant IDH1, 2-HG, is a small organic acid containing two carboxylic acid functional groups in its structure. In the negative ion mode, the deprotonated form of 2-HG should be detected at an m/z of 147.03 (C5H7O5−). Together with the rich diagnostic lipid information commonly observed from gliomas by DESI MS in the mass range m/z 100–1,000, we detected a significant peak at m/z 147 in an IDH1 mutated sample (Fig. 1A), but not in an IDH1 wild-type sample (Fig. 1B).
Fig. 1.
Detecting 2-HG in gliomas using DESI MS. Negative ion mode DESI mass spectra obtained using a linear ion trap mass spectrometer from m/z 100–200 for samples G23, an oligodendroglioma with the IDH1 R132H mutant (A), and G31, a glioblastoma with wild-type IDH1 (B). Tandem mass spectra of m/z 147 detected from sample G42, an oligodendroglioma with the IDH1 R132H mutant [MS2, (C); MS3, (D)] and from a 2-HG standard [MS2, (E); MS3, (F)].
We used tandem MS analysis (MS2) with a linear ion trap mass spectrometer to characterize the signal at m/z 147 (Fig. S1 and Fig. 1 C–F). In an oligodendroglioma with the IDH1 R132H mutation, the main fragment ion generated from m/z 147 was m/z 129, which corresponds to loss of a water molecule from 2-HG (Fig. 1C). Further characterization of m/z 129 with an additional stage of MS analysis (MS3) yielded two additional fragment ions at m/z 101 and m/z 85, corresponding to neutral losses of CO and CO2, respectively (Fig. 1D). We obtained identical MS2 and MS3 fragmentation patterns when we subjected purified l-α-hydroxyglutaric acid to tandem MS experiments (Fig. 1 E and F). A purified standard of the wild-type IDH metabolite alpha-ketoglutarate was detected at m/z 145, and MS2 and MS3 fragmentation patterns yielded peaks at m/z 101 and at m/z 73 and m/z 57, respectively (Fig. S2). We further characterized the 2-HG peaks using a high-resolution and high-mass accuracy linear trap quadrupole (LTQ) Orbitrap mass spectrometer (Fig. S3). DESI mass spectra from an IDH1 R132H mutant sample showed a prominent peak at m/z 147.0299 in the negative ion mode, which matched the molecular formula of the deprotonated form of 2-HG (C5H7O5−) with a mass accuracy of 0.3 ppm. In all, these results confirm the ability to reliably and rapidly detect 2-HG from human glioma tissue sections with DESI MS.
Levels of 2-HG Correlate with Mutational Status and Tumor Cell Content.
We next monitored the levels of 2-HG using DESI MS in a panel of 35 human glioma specimens (Table S1) including primary and recurrent oligodendrogliomas, oligoastrocytomas, and astrocytomas of different grades (3). We first characterized the samples using a clinically validated antibody that selectively recognizes the R132H mutant epitope and not the wild-type epitope from IDH1 (34) (Table S1). Twenty-one of the 35 samples had the R132H mutation. We then measured 2-HG levels in these samples directly from frozen tissue sections using a linear ion trap LTQ DESI. In some samples we detected a peak at m/z 147 and assigned it to 2-HG by MS2 analysis, thereby providing strong independent evidence that these samples were mutated for one of the IDH genes. To account for the variability in desorption and ionization efficiency throughout the tissue and between samples, we normalized the 2-HG signal to the combined intensity of the 40 most abundant lipid species that were detected during each data acquisition (Table S1 and Supporting Information). In all of the 21 samples with the IDH1 R132H mutation, we clearly detected 2-HG with a limit of detection estimated to be 3 μmol 2-HG/g of tissue (Fig. S4), which is below the lowest concentration of 2-HG in tissue in IDH1 mutant human gliomas as measured by HPLC-MS analysis (23).
We also observed a correlation between the concentration of tumor cells and the intensity of the 2-HG signal: Samples with low concentrations of tumor cells (<50%) had lower 2-HG levels, whereas samples with high concentrations of tumor cells (>50%) had higher 2-HG levels (Fig. S5). Although the sample set of high-density tumors (≥80% tumor cells) is relatively small, we noted that glioblastomas with mutant IDH1 generally had lower levels of 2-HG than oligodendrogliomas (Fig. S5).
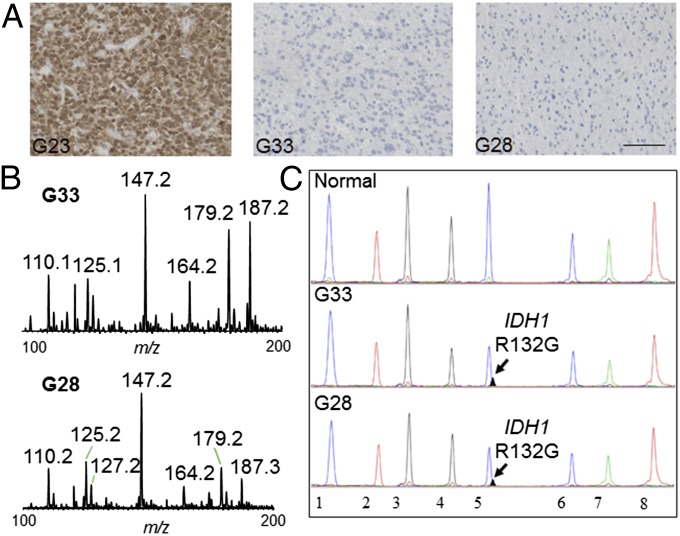
Interestingly, in two of the samples (G33 and G28) that were negative for the IDH1 R132H mutation by immunohistochemical staining (Fig. 2A), we detected 2-HG signal by DESI MS (Fig. 2B). The signal from both samples was confirmed to be from 2-HG by MS2 analysis. Because other mutations in IDH1 or IDH2 can lead to 2-HG accumulation (35–37), we performed targeted sequencing (38) for all of the major mutations in IDH1 and IDH2 that have been described in gliomas (39). This analysis revealed that both samples G33 and G28 harbored a less common but previously described IDH1 mutation that leads to substitution of the amino acid arginine with glycine at position 132 (R132G) (Fig. 2C). These results provide a clear example of how detecting 2-HG with DESI MS allows rapid and accurate determination of IDH1 status in human gliomas. Whereas the diagnostic antibody only recognizes one of the many IDH1 mutants (37), DESI MS captures the presence of 2-HG independent of the underlying genetic mutation in IDH1. Notably, our results show that DESI MS can detect 2-HG with very high sensitivity and specificity: We detected 2-HG signal in all cases with mutant IDH1 (even when the tumor concentration was as low as 5%) and did not detect 2-HG signal in any of the cases with wild-type IDH1.
Fig. 2.
Detecting 2-HG in glioblastoma with IDH1 R132G mutation. (A) Immunohistochemistry using an IDH1 R132H point mutation-specific antibody on formalin-fixed and paraffin-embedded sections from glioma samples (G23, G33, and G28). (Scale bar, 100 μm.) (B) Negative ion mode DESI mass spectra obtained using a linear ion trap mass spectrometer for samples G33 and G28 that are negative for IDH1 R132H mutant immunohistochemistry. (C) Targeted mutational profiling using SNaPshot analysis on nucleic acids extracted from gliobastoma archival specimens (G33 and G28) run in parallel with a normal genomic DNA control, as indicated. The arrows point to the IDH1 R132G (c.394C > G) mutant allele identified in both tumor samples. The assayed loci were as follows: (1) KRAS 35, (2) EGFR 2236_50del R, (3) PTEN 517, (4) TP53 733, (5) IDH1 394, (6) PIK3CA 3139, (7) NOTCH1 4724, and (8) NOTCH1 4802.
DESI MS 2D Imaging of 2-HG in Glioma Sections Delineates Tumor Margins.
To further validate DESI MS as a tool for monitoring 2-HG levels, we turned to 2D DESI MS imaging to study the spatial distribution of molecules across a tissue section (40). DESI MS imaging has recently been shown not to destroy a sample as it is being analyzed when a histologically compatible solvent system is used (40). This relative preservation allows the same tissue section to be stained with H&E following DESI MS data acquisition, and the spatial molecular information derived from DESI MS can then be overlaid onto the optical image of the tissue (40). As such, this approach provides a powerful way to correlate 2-HG levels with histopathology and, importantly, to validate the DESI MS observations.
As a control, we acquired 2D DESI MS data from frozen sections of human glioblastoma orthotopic xenograft models that had been implanted into the brains of immunocompromised mice (Fig. S6). A signal for 2-HG was not detected from xenografts of a glioblastoma cell line (BT329) that has wild-type IDH1 (Fig. S6A). Strikingly, however, a strong signal for 2-HG was found throughout the tissue section of the mouse brain that was diffusely infiltrated by a glioblastoma xenograft (BT116) that has the IDH1 R132H mutation (Fig. S6B), as was similarly observed in an IDH1 R132H mutated oligodendroglioma xenograft model by liquid extraction surface analysis nano electrospray ionization-MS imaging (41).
We next turned to tissue sections of human glioma specimens that had been surgically resected. Using 2D DESI MS with both an LTQ ion trap (Thermo Fisher Scientific) and an amaZon Speed ion trap (Bruker Daltonics), we observed accumulation of 2-HG within a densely cellular glioblastoma with mutated IDH1 (Fig. S7). 2-HG was absent in an area of hemorrhage abutting the tumor (Fig. S7). In tissue specimens from two additional glioma resections, we identified areas that contained regions of tumor as well as regions of brain with only scattered infiltrating glioma cells (i.e., the margin of the tumor). DESI MS revealed strong 2-HG signals in the cellular portions of these samples but weaker signals in the portions of brain with scattered infiltrating tumor cells. By validating our DESI MS results directly with tissue histopathology, we show that monitoring 2-HG levels with DESI MS can help to readily discriminate tissue with dense tumor from tissue with only scattered tumor cells. Such discriminatory capacity can help define tumor margins.
Three-Dimensional Mapping of 2-HG onto MRI Tumor Reconstructions.
MRI information is critical for planning neurosurgical procedures. During the surgery, neuronavigation systems allow the neurosurgeon to register the position of surgical instruments with preoperative plans (i.e., confirming where the tools are relative to the imaging findings). Surgeons can therefore digitally mark the site of a biopsy relative to the tumor in the MRI. We resected two IDH1 mutated gliomas in this manner, using 3D mapping, marking the positions of multiple biopsies in each case. In both cases, we measured the 2-HG content of each stereotactic specimen and normalized to its lipid signals (see Materials and Methods and Supporting Information for details). We then correlated this information with the tumor cell content of each stereotactic specimen, as determined by review of both H&E and immunostains for IDH1 R132H.
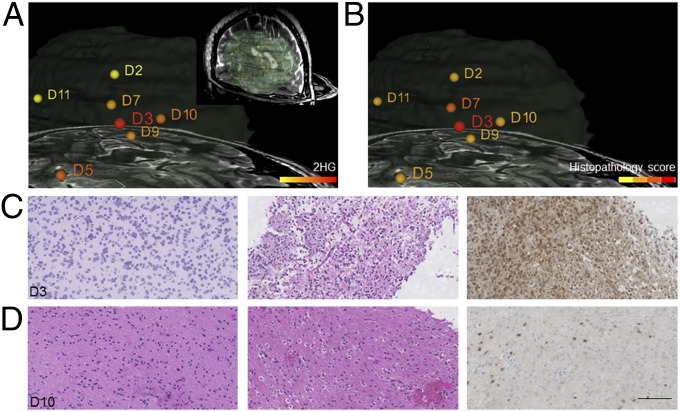
In the resection of an oligodendroglioma (Fig. 3), we identified strong 2-HG signals in the sample (D3) taken from the center of the tumor mass (Fig. 3A). This sample was composed of dense tumor (Fig. 3 B and C). Biopsies from the margins of the radiographic mass (e.g., D10, Fig. 3 B and D) contained low concentrations of infiltrating glioma cells (Fig. 3D). In such samples we detected low levels of 2-HG (Fig. 3A). Consistent with our prior findings on a large panel of glioma specimens (Fig. S4 and Table S1), these stereotactic samples demonstrate that the normalized level of 2-HG correlates with the tumor cell concentration and can help define samples that are at the infiltrating border of the tumor.
Fig. 3.
Three-dimensional mapping of 2-HG over MRI volume reconstruction for surgical case 10, an oligodendroglioma grade II with extensive involvement of the right frontal lobe. (A) Normalized 2-HG signal is represented with a warm color scale as indicated by the scale bar, set from the lowest (yellow) to highest (red) levels detected from this individual case. MS data were acquired using a DESI LTQ instrument. Stereotactic positions were digitally registered to the preoperative MRI using neuronavigation (BrainLab system) in a standard operating room. (Inset) The segmented tumor in green as it relates to brain anatomy. (B) Histopathology scoring of tumor cell concentrations determined from reviewing of H&E-stained tissue sections corresponding to samples analyzed by MS. The scale is divided into four discrete binned colors corresponding to normal brain and low (1–29%), medium (30–59%), and high (60–100%) tumor cell concentrations. (C) High magnification microscopy images of H&E-stained sections of sample D3 representing high tumor cell concentration. (Left) From the MS-analyzed frozen section. (Center) From the corresponding formalin-fixed tissue section. (Right) From IHC for IDH1 R132H mutant (fixed tissue). (D) High magnification microscopy images of H&E-stained sections of sample D10 representing infiltrating tumor cells. (Left) From the MS-analyzed frozen section. (Center) From the corresponding formalin-fixed tissue section. (Right) From IHC for IDH1 R132H mutant (fixed tissue). (Scale bar, 100 μm.)
We performed a second surgical resection (Fig. S8) in the Advanced Multimodality Image Guided Operating (AMIGO) suite (42) at Brigham and Women’s Hospital that is a part of the National Center for Image-Guided Therapy. In this advanced surgical and interventional environment, MRI can be performed during the operation to see whether additional tumor remains in situ. This residual tumor can then be resected before the procedure is completed.
An oligoastrocytoma was resected in this second case. We digitally registered the location of multiple biopsy pieces to the preoperative MRI and measured 2-HG levels in each of them (Fig. S8A). The highest levels of 2-HG were detected in specimens that were taken from the center of the tumor mass, which proved to be dense cellular tumor (Fig. S8B). We took an intraoperative MRI of the patient’s brain once it seemed that the entire tumor had been removed (i.e., following an apparent gross total resection). The T2-weighted intraoperative image revealed a region that was of concern for residual tumor, and surgery for more complete resection was continued based on the MRI finding (Fig. S8C, outlined in green in the inset). Because the areas that were of concern for residual tumor were close (just anterior) to the premotor cortex, we carefully sampled them to preserve the patient’s motor function. We digitally registered two additional specimens to the intraoperative MR image, samples S60 and S61. We detected an equivocal 2-HG signal from one sample (S61) but robust 2-HG signal from the other (S60) (Fig. S8C). Microscopic review of the HE and IDH1 R132H immunostained sections revealed only scattered tumor cells in sample S61 (<5% tumor nuclei by H&E frozen section analysis) but numerous tumor cells in sample S60 (∼20% tumor by H&E frozen section analysis) (Fig. S8D). This clinical example demonstrates a scenario where surveying the resection cavity with DESI MS could eventually identify areas of residual tumor without interrupting surgery for intraoperative MRI.
Real-Time Intraoperative Detection of 2-HG.
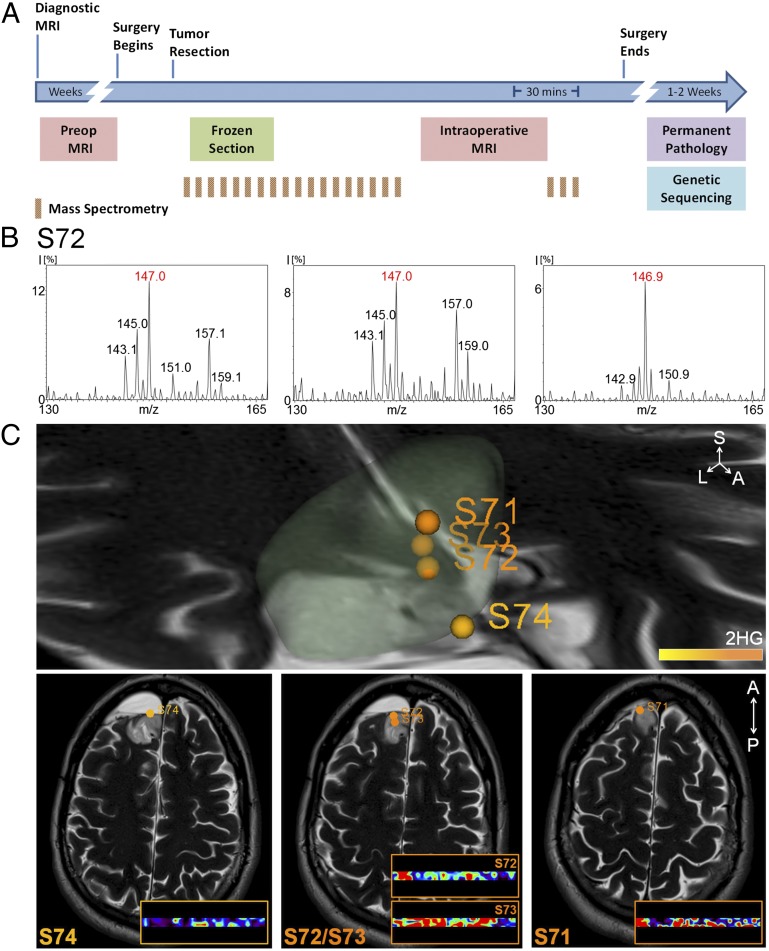
Successfully implementing DESI MS in the operative setting requires that we demonstrate the feasibility of immediately detecting 2-HG in the operating room from tissue biopsies. In Fig. 4A we outline the standard work flow for brain surgery in the AMIGO suite using current methodologies and the increased sampling that is possible with DESI MS. To test our ability to measure 2-HG in this setting, we installed a complete DESI MS system in the AMIGO suite and monitored 2-HG levels from multiple biopsies as they were resected from two patients.
In one case, a patient had had an oligoastrocytoma [World Health Organization (WHO) grade II] resected 6 y earlier. Upon recurrence of the tumor, we reoperated on the patient in our AMIGO suite. Interestingly, subsequent IDH1 molecular testing showed that the tumor lacked the R132H mutation by immunohistochemistry (IHC) testing (Fig. S9A) but had an R132C mutation as detected by targeted sequencing (Fig. S9B). This information was unknown to us at the time of surgery. We sampled the tumor biopsies in two ways: We applied miniscule amounts of biopsy material to a standard glass slide either with a swab (the ones used for swab cultures) or by smearing a tiny tissue fragment between two glass slides (i.e., a standard smear preparation) (Fig. S9C). Within minutes, from both preparations, we clearly detected a peak that corresponded to 2-HG (m/z 147.0) (e.g., data from sample S72 are shown in Fig. 4B) and confirmed this with tandem MS (Fig. S9D). After the operation, in our laboratory outside of the AMIGO suite, we confirmed the presence of 2-HG in these biopsies with DESI MS imaging in samples taken from the center of the tumor (S73) as well as those taken from along the tumor edge (S71 and S74) (Fig. 4C). Similarly, in a resection of a recurrent anaplastic oligoastrocytoma (WHO grade III) in our AMIGO suite we rapidly detected 2-HG on smear preparations from multiple biopsies (Fig. S10). IHC performed days later as part of the routine clinical analysis confirmed the presence of the IDH1 R123H mutation.
Fig. 4.
(A) Time course and work flow of patient care associated with a typical 5-h neurosurgery in the AMIGO, MRI-equipped, operative suite at Brigham and Women’s Hospital. See Supporting Information for additional description. (B) Negative ion mode DESI mass spectra obtained using an amaZon Speed ion trap from m/z 130–165 (Bruker Daltonics) from a swab (Left), a smear (Center), and a section (Right) for sample S72. (C) Normalized 2-HG signal is represented with a warm color scale as indicated by the scale bar, set from the lowest (yellow) to highest (orange) levels detected from this individual case. Stereotactic positions were digitally registered to the preoperative MRI using neuronavigation (BrainLab system) in a standard operating room. The 3D tumor volume is shown (Upper). Classification results of samples S74, S72, S73, and S71 are further visualized on axial sections (Lower).
Discussion
We have previously demonstrated that many tumor types can be discriminated based on their lipid profile. Here, using gliomas with IDH1 mutations as an example, we show that a single metabolite, which was monitored during surgery with ambient MS techniques, rapidly provides highly relevant information: tumor classification (i.e., 2-HG–expressing CNS tumors are nearly always gliomas), genotype information (i.e., 2-HG–expressing tumors carry mutations in IDH1 or IDH2), and prognostic information (i.e., 2-HG–expressing tumors have a more favorable outcome) (43–46)—all with excellent sensitivity and specificity.
Because about 80% of grade II and grade III gliomas as well as the majority of secondary glioblastomas contain IDH1 or IDH2 mutations (43), monitoring 2-HG with intraoperative MS could conceivably become routinely used for surgeries of primary brain tumors, first to classify the tumor and then, if 2-HG is present, to guide optimal resection. In tumors lacking 2-HG, surgical guidance would require monitoring lipid signatures or other metabolites. Presumably, the approach we describe here could be applicable for the resection of all 2-HG–producing tumors including chondrosarcoma and cholangiocarcinoma. Used in conjunction with lipid profiles, a detailed understanding of the tumor could be generated rapidly.
Unlike more time-consuming HPLC MS approaches that are standard for quantifying 2-HG, ambient MS techniques enable rapid data acquisition and are therefore compatible with the rigorous time constraints of surgery. Because of this, the approach described in our work can provide the intraoperative guidance needed to guide the iterative process of optimizing a resection—discriminating tumor from normal brain tissue—a distinction that is of utmost importance in neurosurgery for improving patient outcomes [increased survival (29, 31, 32) and decreased morbidity (47)]. Note that the typical spatial resolution of DESI MS is ∼200 µm (3, 7), which is ample for evaluating surgical biopsies, which are often 2 mm or more in size.
Although MRI is an important intraoperative tool (48) it does have limitations. MRI is an indirect measure of the presence of a tumor; it does not definitively reveal the type of tumor that is being operated on and can sometimes not discriminate tumor from reactive adjacent tissue; each intraoperative MRI scan requires 1 h or longer to perform and interpret; MRI is not an iterative process (i.e., generally only one scan can be performed during a procedure); and the surgeon needs to extrapolate what is learned from the MRI to judge how much more tissue needs to be removed (without being able to ask specifically and directly whether the exact tissue area in question in the surgical field is truly tumor tissue). Moreover, deformation of brain structures occurring following craniotomy (i.e., “brain shift”) renders preoperative images inaccurate, as seen for the mapping of sample biopsy sites for S92 and S95 in Fig. S9B. Importantly, performing an MRI is a major interruption to the surgical procedure. Moreover, each operating room that contains an MRI machine costs over $10 million, so intraoperative MRIs are found in only the most advanced operating rooms and access to these important technologies is somewhat restricted for many surgeons and patients alike. DESI-MS platforms could be implemented in essentially any operating room facility at a very small fraction of the costs. It is clear how characterizing 2-HG with DESI MS could play an important role in neurosurgery.
Other metabolites such as succinate and fumarate, which accumulate in specific tumor types (49), may similarly prove to be valuable metabolite markers for guiding surgery with MS approaches. As metabolomic discovery efforts intensify, the cadre of useful metabolite markers will expand significantly. This will undoubtedly increase the breadth of applications and the diagnostic utility of MS-based approaches that could use DESI technologies or other ambient ionization methods (2, 50–52). Fluidly assessing molecular information, in a rapid timeframe, should allow more accurate determination of tumor margins informed by molecular cues (i.e., “molecular margins”), enhancing the likelihood of achieving optimal tumor resection. The low tissue requirements for our methods also raise the possibility of detection in fine-needle aspirations, core-needle biopsies, or bone-marrow biopsies of a wide range of tumor types in both surgical and nonsurgical settings, and some preliminary data supporting this claim are available (53, 54).
To date, surgery remains the first and most important treatment modality for patients suffering from brain tumors. Because of the potential that we describe here, metabolite-imaging MS is a new tool with broad and powerful clinical and research applications that could transform the surgical care of patients with brain and other solid tumors.
Materials and Methods
Tissue Samples.
The tissue samples used in this study were obtained from the Brigham and Women’s Hospital/Dana–Farber Cancer Institute Neurooncology Program Biorepository collection as previously described (3) or from stereotactic surgical cases as described in Fig. 4 and Fig. S10. For additional details see Supporting Information. Genetic analysis was performed as described in Supporting Information.
Tissues were sectioned and immunostained as previously described (3). For details see Supporting Information.
Identification of 2-HG by DESI MS.
To determine whether 2-HG could be detected directly from glioma tissue sections by DESI MS imaging, we analyzed human glioma samples by DESI MS in the negative ion mode using either an LTQ ion trap (Thermo Fisher Scientific) or an amaZon speed ion trap (Bruker Daltonics). For additional details see Supporting Information.
Supplementary Material
Acknowledgments
We thank Marian Slaney, Sebastian Valentin, and Terri Woo for assistance with histology and immunohistochemistry and Revaz Machaidze for assistance with the project. We thank Dr. Rebecca Folkerth and the Brigham and Women's Hospital Neuropathology Division and Dr. Bill Richards and the Brigham and Women's Tissue Bank for facilitating access to archived tissue. This work was in part funded by the James McDonnell Foundation (N.Y.R.A. and R.G.C.). N.Y.R.A. is supported by the US Army Medical Research/Center for Integration of Medicine and Innovative Technology Grant 2010A052245, the Daniel E. Ponton Fund for the Neurosciences, and the National Institutes of Health (NIH) Director’s New Innovator Award (Grant 1DP2OD007383‐01). Support to K.L.L. was provided by NIH Grant R01 RO1CA170592, the Sontag Foundation, and the Ivy Foundation. S.S. is supported by NIH Grant K08NS064168, the V Foundation, and the Jared Branfman Sunflowers for Life Fund. The National Center for Image Guided Therapy Grant P41RR019703 provided support (to A.J.G. and N.Y.R.A.). R.G.C. is supported by NIH Grant 1R21EB009459‐01.
Footnotes
Conflict of interest statement: S.S. and N.Y.R.A. are scientific advisors to BayesianDx.
This article is a PNAS Direct Submission.
See Commentary on page 10906.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404724111/-/DCSupplemental.
References
- 1.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: Molecular snapshots of biochemical systems. Nat Methods. 2007;4(10):828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 2.Harris GA, Galhena AS, Fernández FM. Ambient sampling/ionization mass spectrometry: Applications and current trends. Anal Chem. 2011;83(12):4508–4538. doi: 10.1021/ac200918u. [DOI] [PubMed] [Google Scholar]
- 3.Eberlin LS, et al. Classifying human brain tumors by lipid imaging with mass spectrometry. Cancer Res. 2012;72(3):645–654. doi: 10.1158/0008-5472.CAN-11-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eberlin LS, et al. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc Natl Acad Sci USA. 2013;110(5):1611–1616. doi: 10.1073/pnas.1215687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberlin LS, et al. Molecular assessment of surgical-resection margins of gastric cancer by mass-spectrometric imaging. Proc Natl Acad Sci USA. 2014;111(7):2436–2441. doi: 10.1073/pnas.1400274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calligaris D, et al. Mass spectrometry imaging as a tool for surgical decision-making. J Mass Spectrom. 2013;48(11):1178–1187. doi: 10.1002/jms.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiseman JM, Ifa DR, Venter A, Cooks RG. Ambient molecular imaging by desorption electrospray ionization mass spectrometry. Nat Protoc. 2008;3(3):517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- 8.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306(5695):471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 9.Agar NY, et al. Development of stereotactic mass spectrometry for brain tumor surgery. Neurosurgery. 2011;68(2):280–289, discussion 290. doi: 10.1227/NEU.0b013e3181ff9cbb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27(8):836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borger DR, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17(1):72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amary MF, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224(3):334–343. doi: 10.1002/path.2913. [DOI] [PubMed] [Google Scholar]
- 15.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koivunen P, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 21.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andronesi OC, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4(116):ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi C, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18(4):624–629. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elkhaled A, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):ra5. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pope WB, et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197–205. doi: 10.1007/s11060-011-0737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andronesi OC, et al. Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest. 2013;123(9):3659–3663. doi: 10.1172/JCI67229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62(4):753–764, discussion 264–266. doi: 10.1227/01.neu.0000318159.21731.cf. [DOI] [PubMed] [Google Scholar]
- 29.Sanai N, Polley MY, McDermott MW, Parsa AT, Berger MS. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg. 2011;115(1):3–8. doi: 10.3171/2011.2.jns10998. [DOI] [PubMed] [Google Scholar]
- 30.Smith JS, et al. Role of extent of resection in the long-term outcome of low-grade hemispheric gliomas. J Clin Oncol. 2008;26(8):1338–1345. doi: 10.1200/JCO.2007.13.9337. [DOI] [PubMed] [Google Scholar]
- 31.Beiko J, et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro-oncol. 2014;16(1):81–91. doi: 10.1093/neuonc/not159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Snyder LA, et al. The impact of extent of resection on malignant transformation of pure oligodendrogliomas. J Neurosurg. 2014;120(2):309–314. doi: 10.3171/2013.10.JNS13368. [DOI] [PubMed] [Google Scholar]
- 33.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capper D, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20(1):245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chi AS, et al. Prospective, high-throughput molecular profiling of human gliomas. J Neurooncol. 2012;110(1):89–98. doi: 10.1007/s11060-012-0938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dias-Santagata D, et al. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: Diagnostic and therapeutic implications. PLoS ONE. 2011;6(3):e17948. doi: 10.1371/journal.pone.0017948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 38.Dias-Santagata D, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2(5):146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weller M, Wick W, von Deimling A. Isocitrate dehydrogenase mutations: a challenge to traditional views on the genesis and malignant progression of gliomas. Glia. 2011;59(8):1200–1204. doi: 10.1002/glia.21130. [DOI] [PubMed] [Google Scholar]
- 40.Eberlin LS, et al. Nondestructive, histologically compatible tissue imaging by desorption electrospray ionization mass spectrometry. ChemBioChem. 2011;12(14):2129–2132. doi: 10.1002/cbic.201100411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navis AC, et al. Increased mitochondrial activity in a novel IDH1-R132H mutant human oligodendroglioma xenograft model: In situ detection of 2-HG and α-KG. Acta Neuropathol. 2013;1(18) doi: 10.1186/2051-5960-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jolesz FA. Intraoperative imaging in neurosurgery: Where will the future take us? Acta Neurochir Suppl (Wien) 2011;109:21–25. doi: 10.1007/978-3-211-99651-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leu S, et al. IDH/MGMT-driven molecular classification of low-grade glioma is a strong predictor for long-term survival. Neuro-oncol. 2013;15(4):469–479. doi: 10.1093/neuonc/nos317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim BY, et al. Diagnostic discrepancies in malignant astrocytoma due to limited small pathological tumor sample can be overcome by IDH1 testing. J Neurooncol. 2014;118(2):405–412. doi: 10.1007/s11060-014-1451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunn GP, Andronesi OC, Cahill DP. From genomics to the clinic: Biological and translational insights of mutant IDH1/2 in glioma. Neurosurg Focus. 2013;34(2):E2. doi: 10.3171/2012.12.FOCUS12355. [DOI] [PubMed] [Google Scholar]
- 47.Sanai N, Martino J, Berger MS. Morbidity profile following aggressive resection of parietal lobe gliomas. J Neurosurg. 2012;116(6):1182–1186. doi: 10.3171/2012.2.JNS111228. [DOI] [PubMed] [Google Scholar]
- 48.Black PM, et al. Development and implementation of intraoperative magnetic resonance imaging and its neurosurgical applications. Neurosurgery. 1997;41(4):831–842, discussion 842–845. doi: 10.1097/00006123-199710000-00013. [DOI] [PubMed] [Google Scholar]
- 49.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: A metabolic disease. Nat Rev Urol. 2010;7(5):277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ifa DR, Wu C, Ouyang Z, Cooks RG. Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst (Lond) 2010;135(4):669–681. doi: 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- 51.Nemes P, Vertes A. Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. TrAC - Trends Analyt Chem. 2012;34:22–33. [Google Scholar]
- 52.Van Berkel GJ, Pasilis SP, Ovchinnikova O. Established and emerging atmospheric pressure surface sampling/ionization techniques for mass spectrometry. J Mass Spectrom. 2008;43(9):1161–1180. doi: 10.1002/jms.1440. [DOI] [PubMed] [Google Scholar]
- 53.Liu J, Cooks RG, Ouyang Z. Biological tissue diagnostics using needle biopsy and spray ionization mass spectrometry. Anal Chem. 2011;83(24):9221–9225. doi: 10.1021/ac202626f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elhawary H, et al. Intraoperative real-time querying of white matter tracts during frameless stereotactic neuronavigation. Neurosurgery. 2011;68(2):506–516, discussion 516. doi: 10.1227/NEU.0b013e3182036282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.