Significance
This is the first report, to our knowledge, of the absence of lipofuscin, “aging pigment,” in a setting of motor neuron neurodegenerative disease, mutant SOD1-linked ALS. Although the initial hypothesis was that this must be due to a block in the autophagy/lysosome pathway, instead studies of autophagy markers and of ALS mice treated with chloroquine suggest the opposite, that there is hyperactivity of the autophagy/lysosome pathway, at least in part mediated through MTORC1.
Abstract
Lipofuscin, or aging pigment, is accreted as red autofluorescence in the lysosomes of motor neuron cell bodies in the ventral horn of WT mice by 3 mo of age. Strikingly, in two presymptomatic ALS mouse strains transgenic for mutant human Cu/Zn superoxide dismutase (SOD1), G85R SOD1YFP and G93A SOD1, little or no lipofuscin was detected in motor neuron cell bodies. Two markers of autophagy, sequestosome 1 (SQSTM1/p62) and microtubule-associated protein 1 light chain 3 (LC3), were examined in the motor neuron cell bodies of G85R SOD1YFP mice and found to be reduced relative to WT SOD1YFP transgenic mice. To elucidate whether the autophagy/lysosome pathway was either impaired or hyperactive in motor neurons, chloroquine was administered to 3-mo-old G85R SOD1YFP mice to block lysosomal hydrolysis. After 2 wk, lipofuscin was now observed in motor neurons, and SQSTM1 and LC3 levels approached those of WT SOD1YFP mice, suggesting that the autophagy/lysosome pathway is hyperactive in motor neurons of SOD1-linked ALS mice. This seems to be mediated at least in part through the mammalian target of rapamycin complex 1 (MTORC1) pathway, because levels of Ser757-phosphorylated Unc-51-like kinase 1 (ULK1), an MTORC1 target, were greatly reduced in the G85R SOD1YFP motor neurons, correspondent to an activated state of ULK1 that initiates autophagy.
Lipofuscin, referred to as aging pigment, is an accumulation of protein, lipid, and carbohydrate within the lysosomes of postmitotic cells, typically neurons, that exhibits autofluorescence (1). This material is thought to comprise remnant products of lysosomal hydrolysis. For example, in retinal pigment epithelial cells, products of retinal, the cofactor of opsin, are a principal component (2, 3). In the aging brain, a variety of proteins/lipids are accreted. Additionally, in a set of inherited lipofuscinoses, deficiency of various lysosomal enzymes leads to massive accumulation of particular lipofuscin species. For example, in infantile lipofuscinosis, deficiency of palmitoyl protein thioesterase prevents cleavage of thiol-linked fatty acids from protein species, leading to accumulation of sphingolipid activating proteins A and D (4). By contrast, in late infantile and juvenile forms of lipofuscinosis, the accumulated material is largely composed of subunit c of the mitochondrial F1/F0 ATPase (5). Here we observe an opposite behavior in a pathologic setting, the specific lack of lipofuscin accumulation in motor neurons of mice affected by an ALS neurodegenerative state. Our observations point to a hyperactivity of the autophagy/lysosome pathway as responsible for turning over products that would otherwise accumulate as lipofuscin and implicate mammalian target of rapamycin complex 1 (MTORC1) and Unc-51-like kinase 1 (ULK1) in regulating this state.
Results
Lack of Autofluorescent Lipofuscin in Motor Neurons from Mutant Cu/Zn Superoxide Dismutase-Associated ALS Mice.
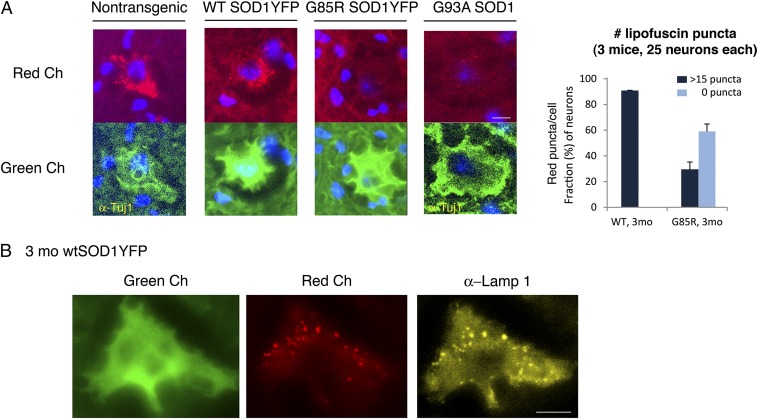
During inspection of spinal cord sections taken from 3-mo-old nontransgenic B6/SJL mice and WT Cu/Zn superoxide dismutase (SOD1)YFP transgenic mice (6), we observed numerous perinuclear red-autofluorescent puncta in motor neuron cell bodies in the ventral horn of the gray matter (Fig. 1A). These puncta were costainable with anti-LAMP1 antibody (Fig. 1B), a lysosomal marker, indicating that the red autofluorescence comes from lipofuscin, an “aging pigment” previously observed to localize within lysosomes and to comprise undegraded protein and lipid products of lysosomal hydrolysis (1). In these two mouse strains, such puncta were absent at 1 mo of age, present at 3 mo of age (Fig. 1A), and increased in number and intensity at 13 mo of age (Fig. S1). Surprisingly, when similar spinal cord sections were examined from transgenic G85R SOD1YFP ALS mice (6) at 3 mo of age, a presymptomatic point in a typically 5- to 6-mo course to paralysis, we observed that most cell bodies of the motor neurons exhibited no such autofluorescence (Fig. 1A). Similarly, at the time of paralysis at 5 mo of age, little or no lipofuscin was detected in the remaining intact motor neurons in the ventral horn (Fig. S2). We then examined spinal cord motor neurons from the classically studied G93A SOD1 mouse at 3 mo of age in a ∼5-mo course to paralysis (7). Here also, most motor neuron cell bodies lacked lipofuscin fluorescence (Fig. 1A).
Fig. 1.
Lipofuscin is strongly reduced or absent in spinal cord motor neurons from two mouse models for ALS, G85R SOD1YFP, and G93A SOD1. (A, Upper Left) Representative images of motor neuron cell bodies in transverse lumbar spinal cord sections from nontransgenic, WT SOD1YFP, G85R SOD1YFP, and G93A SOD1 mice observed in the “red” channel (see Materials and Methods for specific filter sets). (Lower Left) The cell bodies corresponding to those in the top row observed in the “green” channel after staining with anti-Tuj1 and Alexa Fluor 488 secondary antibody for the nontransgenic and G93A SOD1 samples, or by virtue of the fluorescence of the transgenic WT SOD1YFP or G85R SOD1YFP fusion protein in large ventral horn neurons of the other two. In images for both channels, DAPI fluorescence is overlaid in blue. (Right) Auto-fluorescent lipofuscin puncta were counted in images similar to those on the left of 25 motor neuron cell bodies from each of three 3-mo-old mice from each SOD1YFP genotype (Materials and Methods). Cells were binned as having >15 puncta or no visible puncta. Data in the bar graph are presented as the means for the three mice, with the SDs indicated by the error bars. The majority of the WT SOD1YFP cells had >15 puncta. Conversely, >50% of the G85R SOD1YFP cell bodies had no visible puncta. (Scale bar in A, 10 μm.) (B) Lipofuscin is present in LAMP1–positive compartments. A motor neuron cell body in a spinal cord section from a 3-mo-old WT SOD1YFP mouse was imaged in the green channel for YFP (Left) and the red channel for lipofuscin auto-fluorescence (Center), and its position on the slide was recorded. The slide was then treated with copper sulfate (Materials and Methods) to eliminate the auto-fluorescence and reimaged to confirm its absence. The slide was then stained for LAMP1, a lysosomal membrane protein, using an anti-LAMP1 antibody and an Alexa Fluor 568 secondary antibody. The cell initially imaged was relocated and imaged in the red channel (Right, false-colored yellow). Note the coincidence of many of the red lipofuscin puncta with the yellow LAMP1-positive structures, confirming the lysosomal localization of lipofuscin in motor neurons. (Scale bar, 10 μm.)
A quantitative analysis was carried out, determining the number of red-fluorescent puncta in 25 large ventral horn motor neurons of three transgenic animals of each of the G85R SOD1YFP and WT SOD1YFP transgenic strains at 3 mo of age (Fig. 1A, Right). In WT SOD1YFP mice, >90% of the neurons contained >15 puncta per cell body. In contrast, a similar examination of G85R SOD1YFP transgenic mice revealed that only ∼30% of ventral horn neurons contained >15 puncta per cell body and ∼60% of cell bodies were devoid of puncta. Similar quantitation of fluorescent puncta was observed in two G93A animals at 3 mo of age.
In contrast to the observations in the ventral horn, when neuron cell bodies in the dorsal regions of the spinal cord gray matter were examined, both the G85R SOD1YFP and WT SOD1YFP, as well as nontransgenic animals, exhibited red fluorescent puncta, at equal levels. Thus, the selective absence of red autofluorescence in the ventral horn neurons seems to reflect behavior indigenous to mutant motor neurons, which exhibit strong expression of mutant SOD1YFP that is subject to misfolding and inclusion body formation, behaviors associated with cell death. Was absence of lipofuscin owing to impaired production of these autofluorescent products of lysosomal metabolism because of a block of autophagy (or endocytosis pathways that deliver products to the lysosome), or was it owing to accelerated clearance of material transiting this pathway? Concerning endocytosis, two endocytic proteins, EEA1 and RAB9, seemed unaffected in steady-state levels (Fig. S3), indicating that there was no block in this pathway. We next carried out further examination of the autophagy/lysosome pathway, inspecting two components, microtubule-associated protein 1 light chain 3 (LC3) and sequestosome 1 (SQSTM1), in WT and mutant SOD1YFP spinal cord sections (8, 9).
Reduced LC3 and SQSTM1 in Motor Neuron Cell Bodies of G85R SOD1YFP but Not WT SOD1YFP Mice.
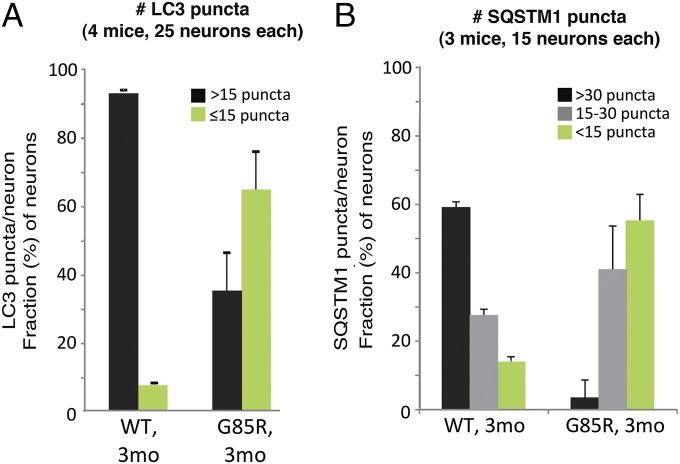
LC3, microtubule-associated protein 1 light chain 3 (MAP1LC3), is recruited to the phagophore membranes (the precursor of the autophagosome) as a phosphatidyl–ethanolamine conjugate (LC3-II) during autophagosome formation; after fusion of the autophagosome with a lysosome, the LC3 trapped inside the autophagosome is degraded (10, 11). LC3 in large motor neuron cell bodies in spinal cord sections from 3-mo-old mice was identified by staining with an anti-LC3B antibody. To avoid red fluorescent signals from the lipofuscin itself, the lipofuscin signal was ablated using copper sulfate treatment (ref. 12 and Fig. S4). In motor neurons of WT SOD1YFP transgenic animals, we observed >15 LC3 antibody-positive puncta per cell in nearly all cells examined (Fig. 2A) (see raw images in Fig. 3 and SI Text concerning colocalization with lysosomes). In contrast with the abundant LC3 puncta in the WT, the G85R SOD1YFP neurons exhibited fewer puncta, with >15 puncta observed in fewer than 40% of the cells (Fig. 2A). Thus, LC3 puncta, corresponding to autophagosomes, are less abundant in motor neurons from presymptomatic G85R SOD1YFP transgenic animals.
Fig. 2.
LC3 and SQSTM1 are reduced in G85R SOD1YFP motor neurons compared with WT SOD1YFP. Spinal cord slices were stained for autophagosome markers LC3 or SQSTM1 with the respective antibodies and Alexa Fluor 568 secondary antibody. The sections were treated with copper sulfate to remove lipofuscin auto-fluorescence then imaged in the red channel. (A) For LC3, the bar graphs show the fraction of cells of each genotype that had >15 vs. ≤15 puncta. (B) For SQSTM1, the bar graphs show the fraction of cells of each genotype that had >30 puncta, 15–30 puncta, or <15 puncta. Most WT cells had a relatively large number of LC3 and SQSTM1 puncta, whereas the majority of the mutant G85R cells had substantially fewer, indicating that autophagosomal proteins are reduced in G85R SOD1YFP motor neuron cell bodies. Data are presented as means and SD for the indicated number of mice.
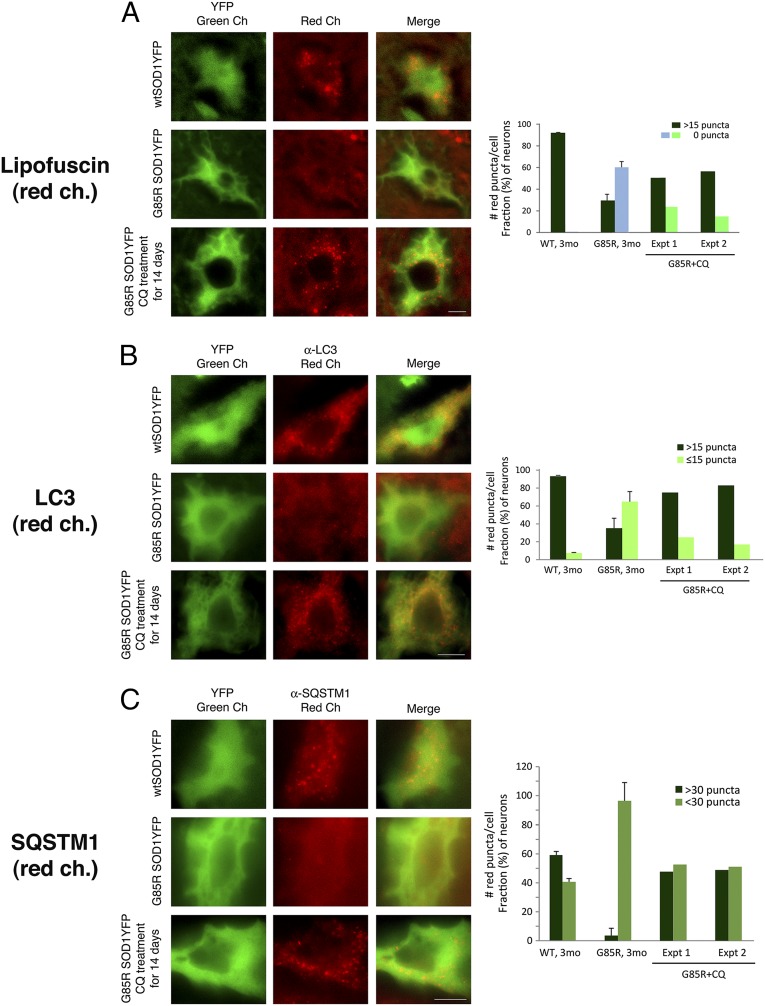
Fig. 3.
After treatment of G85R SOD1YFP mice with chloroquine, auto-fluorescent lipofuscin puncta, LC3, and SQSTM1 accumulate in motor neuron cell bodies. Two 3-mo-old G85R SOD1YFP mice were treated daily with chloroquine for 14 d as described in Materials and Methods. Spinal cord sections were prepared as described, and representative images are shown in the green channel for YFP and the red channel for (A) lipofuscin auto-fluorescence, (B) anti-LC3 immunofluorescence, and (C) anti-SQSTM1 immunofluorescence. In each set of panels, the top row shows WT SOD1YFP, the middle row shows G85R SOD1YFP, and the bottom row shows chloroquine-treated G85R SOD1YFP. For both lipofuscin and LC3, a substantial number of red puncta are present in the motor neuron cell body from the treated animal, suggesting that chloroquine, an inhibitor of lysosomal hydrolytic activity, had prevented the proteolysis of lipofuscin and LC3 components, thus permitting their accumulation. In the case of SQSTM1, a pattern of diffuse fluorescence in G85R SOD1YFP gives rise, upon treatment, to discrete puncta. The bar graphs on the right show the quantitation of the fraction of cells with different numbers of lipofuscin, LC3, or SQSTM1 puncta, respectively, in the two treated mice (two right-hand sets of bars, G85R+CQ, Exp. 1 and Exp. 2, for each mouse; >25 cells counted for each). The left-hand bars for WT and untreated G85R SOD1YFP animals are reproduced from Fig.1A in the case of lipofuscin, from Fig. 2A for LC3, and derived from Fig. 2B for SQSTM1. Note that both treated animals have an increased fraction of cells with the respective puncta. (Scale bars, 10 μm.)
Similarly, one of the “receptors” for ubiquitinated proteins, SQSTM1, that recruits substrates to the autophagy pathway and also travels in autophagosomes to the lysosome where it is degraded (13), was reduced in staining in the mutant motor neurons (Fig. 2B and Fig. S5). Whereas puncta were readily observed in the cytosol of WT transgenic spinal cord motor neuron cell bodies, with 60% exhibiting >30 puncta per cell, in the mutant cell bodies there were fewer puncta per cell, with ∼60% exhibiting fewer than 15 puncta per cell.
We conclude that two proteins normally delivered to lysosomes through the autophagy/lysosome pathway, LC3 and SQSTM1, are both diminished in ventral horn motor neurons of the G85R SOD1YFP mutant animals. Considering the apparent drop in steady-state level of these components, assuming their production is normal (verified at the transcription level; ref. 14), this might suggest that their turnover is accelerated and that the autophagy/lysosome pathway is “hyperactive.” An in vivo experiment was carried out to address this possibility.
Chloroquine Treatment of Mutant G85R SOD1YFP Mice Restores Levels of Lipofuscin, LC3, and SQSTM1 Toward Normal.
The 4-aminoquinoline compound chloroquine blocks lysosomal acidification and therefore inhibits lysosomal hydrolases, blocking lysosomal protein degradation (15). We reasoned that if lysosomes were hyperactive in motor neurons in the setting of G85R SOD1YFP, then treatment of mutant transgenic mice with chloroquine (which crosses the blood–brain barrier) should reduce the rate of clearance of both lipofuscin and components of the autophagosomal/lysosomal pathway, LC3 and SQSTM1, allowing them to accrete.
Two mutant G85R SOD1YFP mice at 3 mo of age were treated for 2 wk with chloroquine diphosphate, 50 mg⋅kg−1⋅d−1, administered i.p. once per day. Spinal cord sections were prepared as for the earlier studies and examined for lipofuscin (Fig. 3A), LC3 (Fig. 3B), and SQSTM1 (Fig. 3C). For all three species, there was now an appearance more like that in motor neurons of untreated WT SOD1YFP mice (representative images, Left, and quantification, Right). For lipofuscin, in both treated animals (Exp.1 and 2), there was a nearly twofold increase in the number of cells with >15 puncta (Fig. 3A). For LC3, in both treated animals the level of cells with >15 puncta now approached that of the WT (75% and 85%) (Fig. 3B), and for SQSTM1 the number of cells with >30 puncta also increased to near WT levels (Fig. 3C). We conclude that autophagy/lysosome inhibition by chloroquine led to accretion of components, lipofuscin, LC3, and SQSTM1, and that these were, in the absence of the compound, rapidly turned over by a hyperactive autophagy/lysosome system in the motor neurons of the mutant G85R SOD1YFP animals.
Ser757-Phosphorylated ULK1 Is Reduced in Motor Neuron Cell Bodies of 3-Mo-Old G85R SOD1YFP Mice, Suggesting Possible Activation via the ULK1/MTORC1 Pathway.
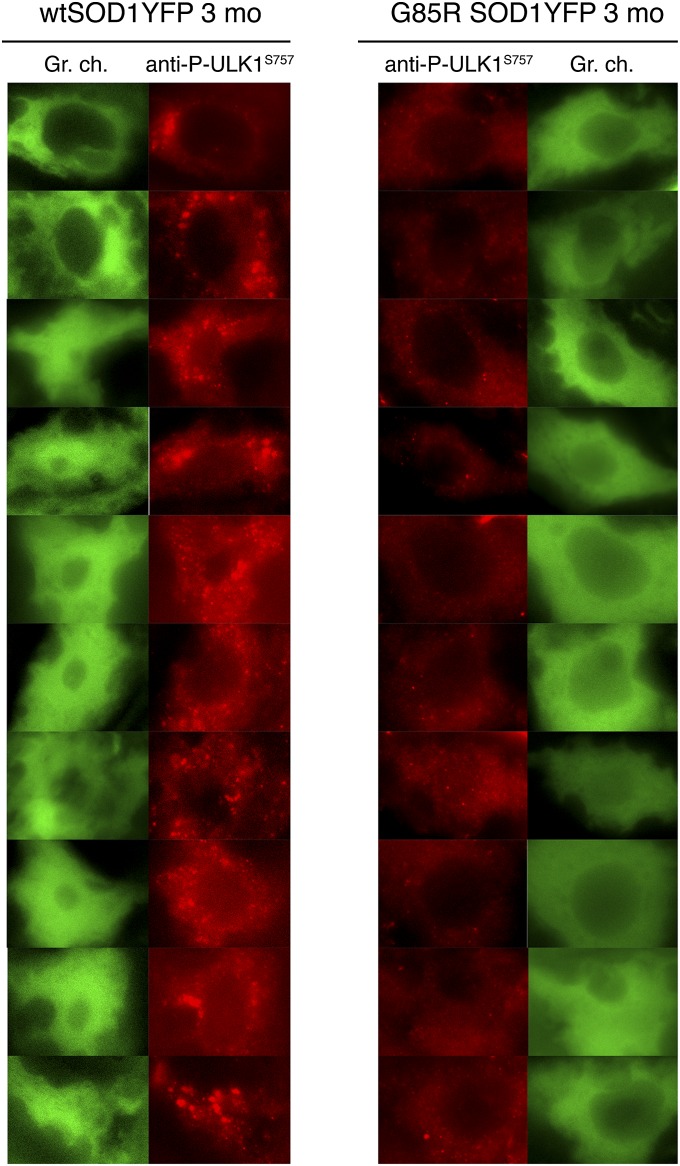
The MTORC1 complex and AMP-activated protein kinase (AMPK) play opposing roles in regulating autophagy. When a cell is “replete” (high glucose concentration), AMPK is inactive, and MTORC1 phosphorylates Ser757 of the autophagy-initiating protein ULK1, inhibiting autophagy initiation. When a cell is “starved” (low glucose), AMPK is active and inhibits MTORC1 activity (16, 17) and, hence, phosphorylation of ULK1. In addition, AMPK now phosphorylates ULK1 on Ser317 and Ser777, leading to an increase in initiation of the autophagy pathway (18, 19). We assessed the phosphorylation status of Ser757 in both WT SOD1YFP and G85R SOD1YFP motor neuron cell bodies by immunofluorescent staining with an antibody directed specifically against the Ser757-phosphorylated form of ULK1. As observed in Fig. 4, motor neuron cell bodies from 3-mo-old WT SOD1YFP mice exhibited a strong cytosolic punctate pattern, consistent with MTORC1-mediated phosphorylation of ULK1 and a basal level of autophagy. By contrast, motor neuron cell bodies from 3-mo-old G85R SOD1YFP mice exhibited minimal immunofluorescence, consistent with diminished phosphorylation at this position, likely associated with activation of ULK1 and autophagy. We were not able to assess the AMPK-mediated phosphorylation of ULK1 at Ser317 or Ser777, because an antibody available against the ULK1 Ser317 peptide, although reported to recognize the site in Western blot analysis, did not produce signals on spinal cord sections and no mouse-specific Ser777 antibody was available, leaving this arm of the pathway of activation of ULK1 to be assessed. Nevertheless, the findings related to Ser757 indicate involvement of ULK1 and MTORC1 in the pathway of activation of autophagy in the SOD1 mutant mice.
Fig. 4.
Ser757-phosphorylation of ULK1 measured by immunostaining with phosphospecific antibody in motor neurons of 3-mo-old G85R SOD1YFP mice is reduced relative to WT SOD1YFP. Spinal cord sections were stained with an antibody specific to Ser757-phosphorylated ULK1 and Alexa Fluor 568 secondary antibody. The sections were treated with copper sulfate to remove lipofuscin auto-fluorescence then imaged in the red channel. A gallery of images of motor neurons is shown, indicating that in most motor neuron cell bodies of the G85R SOD1YFP mice there is reduced signal for phosphor–ULK1–Ser757, relative to that in WT SOD1YFP. This parallels the apparent activation of autophagy in these cells (see text for discussion).
Discussion
A precedent for the behavior observed here has been presented in a different setting by Öst et al. (20), examining primary cultures of adipocytes from type 2 diabetes patients. Compared with cultures from nondiabetic subjects, there was little or no detectable lipofuscin (figure 3 in their publication), associated with rapid turnover of LC3. Remarkably, in this setting, accumulation of lipofuscin was restored by treatment of the cultured type 2 adipocytes with chloroquine, resembling the response observed here in motor neurons of intact ALS animals. The results here, and those of Öst et al. (20), imply that lipofuscin is not a dead-end product, but one that can be fully hydrolyzed or exported from lysosomes by a mechanism as yet unknown.
Notably, in two earlier studies of G93A SOD1 ALS mice, evidence for activation of autophagy was adduced from observation of increased spinal cord LC3-II, as measured by Western blot analysis (21, 22). Such results would agree with those presented here, although these earlier studies assessed the entire population of neurons and glia and did not distinguish the behavior of the autophagy pathway within the motor neurons themselves.
Another explanation for absence of LC3 and SQSTM1 in the motor neurons of mutant ALS animals in the present study could be a proximal block in the autophagy pathway. If this were the case, however, we would not expect chloroquine effects on lysosomal pH to restore the presence of LC3 and SQSTM1.
What is the mechanism leading to hyperactivity of the autophagy/lysosome pathway? The absence of Ser757-phosphorylated ULK1 in G85R SOD1YFP motor neuron cell bodies, relative to abundant ULK1 phosphorylation in WT SOD1YFP motor neurons from animals of the same age, suggests that MTORC1 regulation is at least partially involved. This is likely complemented by activation of AMPK, which would putatively phosphorylate ULK1 on positions Ser317 and Ser777 to initiate autophagy (18). Nevertheless, we infer from the reduced phosphorylation on ULK Ser757, and from the apparent activation of autophagy, that the motor neurons may be sensing a “starvation state.” Such a state could result from diminished energy supply, perhaps related to overactive action potential firing or to a metabolic disturbance resulting from high-level cellular expression of the misfolded SOD1YFP protein.
That MTORC1 can influence the accumulation of lipofuscin was further observed when we treated 3-mo-old WT SOD1YFP mice with rapamycin, which inhibits MTORC1 and in turn activates autophagy: We observed that a 2-wk treatment led to a substantial reduction of lipofuscin in most motor neurons of WT SOD1YFP mice (Fig. S6).
Thus, in conclusion, it seems that the absence of lipofuscin in G85R SOD1YFP motor neurons is due to hyperactivity of the autophagy/lysosome pathway in these cells. How mutant SOD1 itself dictates a starvation state or produces other metabolic effects that in turn activate ULK1 and autophagy, or whether part of the activation of the autophagic pathway could be a response to altered proteostasis in these neurons, remains to be established, but such activation might be a protective response, insofar as it could accomplish the removal of the toxic mutant SOD1 protein.
Materials and Methods
Mice.
All animal experiments were carried out according to procedures approved by the Yale University Institutional Animal Care and Use Committee. The transgenic WT SOD1YFP (607 line) and G85R SOD1YFP (737 line) strains of mice, in a B6/SJL background, have been described previously (6, 14). Mice from the transgenic G93A SOD1 strain [B6SJL-Tg(SOD1*G93A)1Gur/J; Jackson strain 002726] (7), also in a B6/SJL background, as well as nontransgenic B6/SJL mice (Jackson strain 100012) were obtained from Jackson Laboratory. When chloroquine treatment was carried out, chloroquine diphosphate (Sigma) was dissolved in sterile saline such that a dose of 50 mg/kg was contained in 100 µL, then 100 µL was injected intraperitoneally. When rapamycin treatment was carried out, rapamycin was administered at a dose of 5 mg−1⋅kg⋅d−1. Typically, 2–3 µL of an ethanol stock solution of rapamycin at 50 mg/mL was diluted into ∼30 µL of DMSO and injected i.p.
Preparation of Spinal Cord Sections.
A mouse was perfused with 2% (wt/vol) paraformaldehyde (PFA) in PBS and the cord removed and postfixed overnight in 2% PFA in PBS. It was rinsed in PBS, then embedded in O.C.T. (Tissue-Tek) and rapidly frozen in 2-methylbutane cooled in a liquid nitrogen bath and stored at −80 °C. Cross-sections of 20-μm thickness were prepared on a cryostat (Leica CM3050S) and mounted on Snowcoat X-tra glass slides (Surgipath) and stored at −80 °C until used. Immunostaining was carried out as previously described (14). Briefly, slides were rapidly warmed to room temperature on silica gel dessicant, OCT was removed by washing slides in ice-cold PBS, and further postfixing was carried out with 2% PFA in PBS for 15 min, followed by three washes of 1 min in PBS. Immunodetection of LC3 (discussed below) required antigen exposure by treatment of the slide with 0.1% trypsin in 0.1% calcium chloride, pH 7.5, for 30 min at 37 °C, followed by three PBS washes of 1 min. Permeabilization was with 0.5% Triton X-100 for 15 min at room temperature, followed by three washes in PBS. Blocking was for 30 min at 37 °C in secondary antibody species-specific blocking buffer. Incubation in primary antibody was carried out first for 1 h at 37 °C, then overnight at 4 °C, followed by three washes of 3 min in PBS. Fluorescent secondary antibody was applied for 1 h at room temperature in the dark, followed by three PBS washes of 3 min. When copper sulfate treatment was used to remove lipofuscin autofluorescence that would interfere with visualization of secondary antibodies (ref. 12 and Fig. S4), slides stained with secondary antibody were dipped briefly in distilled water, followed by incubation in 10 mM CuSO4 in 50 mM ammonium acetate, pH 5.0, for 15 min at room temperature. They were then dipped in distilled water followed by a wash in PBS. Slides were coated with Vectashield containing DAPI (Vector Laboratories) and coverslips mounted.
Antibodies.
The following antibodies were used at the dilutions indicated. LC3 antibody: LC3B, mouse mAb (5F10, ALX-803-080; Enzo Life Sciences) (1:100); SQSTM1 antibody: p62, goat pAb (P-15, sc-10117; Santa Cruz Biotechnology) (1:100); Ser757 phospho-ULK1, rabbit pAb (6888; Cell Signaling) (1:100); Lamp1 antibody: LAMP-1 (CD107a) rat mAb (1D4B, MABC39; Millipore) (1:100); and Tuj1 antibody: neuron-specific β-III tubulin mouse mAb (TuJ-1, MAB1195; R&D Systems) (1:100). Immunodetection of LC3 required pretreatment with trypsin (see above). Secondary antibodies against the appropriate primary species, labeled with Alexa Fluor 488 or 568 (Molecular Probes/Life Technologies) were used at 1:1,000 dilution.
Microscopy and Images.
All images were acquired using an Olympus IX81 microscope and an Andor NEO sCMOS camera controlled by Metamorph (Molecular Devices) software. The images in Figs. 1A and 3A and Figs. S1, S2, and S6 were acquired with a 40× air objective (Olympus UPlanFLN40×, N.A. 0.75); all others were acquired with a 100× oil objective (Olympus UPlanSApo100×, N.A. 1.40). DAPI was detected with a Semrock DAP-5060C filter set (excitation, 352–402 nm; emission, 417–477 nm; dichroic, 409 nm). YFP and secondary antibodies labeled with Alexa Fluor 488 were detected with an Olympus U-MYFPHQ filter set (excitation, 490–500 nm; emission, 515–560 nm; dichroic, 505 nm); in the figures this is called the green channel. Lipofuscin and secondary antibodies labeled with Alexa Fluor 568 were detected with a custom filter set from Semrock (excitation, 562.5–587.5 nm, FF02-575/25–25; emission, 589–625 nm, FF01-607/36–25; dichroic, 593 nm, FF593-Di03), designed to avoid interference from the intense YFP fluorescence in cells from SOD1YFP transgenic animals; this is called the red channel. In general, a z-stack of images was acquired, and a single plane for all channels was selected as representative. These images were exported as TIFF files to ImageJ software (National Institutes of Health). Brightness and contrast were adjusted and the images cropped to produce final images for display. Any gamma adjustment applied is noted in the individual figure legends.
Counting and Statistics.
Cell bodies to be imaged in spinal cord sections were selected based on their position in the ventral motor columns in the section, their large size (>25–30 µm in diameter), and their relatively bright YFP fluorescence. For nontransgenic and G93A SOD1 transgenic animals, immunostaining for neuron-specific Tuj1 was used to assist identification. Previous studies have shown that such cell bodies are ventral horn motor neurons, based on staining with anti-ChAT antibodies. Cell bodies containing large, intensely fluorescent SOD1YFP aggregates were not selected. All of the identified cell bodies in an individual section (∼10–15 per ventral horn) were evaluated. For each cell, a single-plane image that seemed to have the most puncta was selected for initial counting; planes above and below were then inspected for noncoincident puncta and any observed added to the count of the main plane. Counting of at least 25 cells for each genotype, protein, or treatment was performed manually, and cells were binned according to the number of puncta as indicated in the figures: lipofuscin, 0 or >15; LC3, ≤15 or >15; SQSTM1. <15, 15–30, or >30. The fraction of cells in each bin for each condition was averaged for three or four mice as indicated in the legends, and SDs calculated based on n = 3 or 4. Because only two animals were evaluated in the chloroquine treatment experiment (Fig. 3), data from each of these are presented individually. Note that when both lipofuscin and LC3 (and/or SQSTM1) were counted, different sections from the same animal were used for each test.
For the lipofuscin autofluorescence intensity comparison between Fig.1 (3 mo) and Fig. S1 (13 mo), ventral horn motor neurons were identified with anti-Tuj1 staining in spinal cord sections from one 3-mo-old and one 13-mo-old nontransgenic mouse, and single image planes were selected as above. Imaging was carried out sequentially for sections from the two animals of different ages, using the same acquisition parameters (exposure time, neutral density filter setting, etc.) for each. The images were imported into ImageJ and the cell bodies outlined with the freeform drawing tool. From the Analyze menu in ImageJ, Area, Integrated Density, and Mean Gray Value were selected in Set Measurements. Then Measure was selected from the Analyze menu to produce a table containing these data for the selected area. The area outline was moved to a region of the ventral horn that did not contain a motor neuron and the measurement repeated to give corresponding background values. This process was repeated for 25 cells in slices from each animal, and the data were compiled in an Excel spreadsheet. Backgrounds were subtracted from the corresponding motor neuron values, and these corrected data were summed and averaged to give the integrated density and the SDs calculated.
Supplementary Material
Acknowledgments
We thank The Ellison Medical Foundation and Howard Hughes Medical Institute for supporting this work.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1409314111/-/DCSupplemental.
References
- 1.Seehafer SS, Pearce DA. You say lipofuscin, we say ceroid: Defining autofluorescent storage material. Neurobiol Aging. 2006;27(4):576–588. doi: 10.1016/j.neurobiolaging.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Eldred GE, Lasky MR. Retinal age pigments generated by self-assembling lysosomotropic detergents. Nature. 1993;361(6414):724–726. doi: 10.1038/361724a0. [DOI] [PubMed] [Google Scholar]
- 3.Sparrow JR, et al. The bisretinoids of retinal pigment epithelium. Prog Retin Eye Res. 2012;31(2):121–135. doi: 10.1016/j.preteyeres.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyynelä J, Palmer DN, Baumann M, Haltia M. Storage of saposins A and D in infantile neuronal ceroid-lipofuscinosis. FEBS Lett. 1993;330(1):8–12. doi: 10.1016/0014-5793(93)80908-d. [DOI] [PubMed] [Google Scholar]
- 5.Palmer DN, et al. Ovine ceroid lipofuscinosis. The major lipopigment protein and the lipid-binding subunit of mitochondrial ATP synthase have the same NH2-terminal sequence. J Biol Chem. 1989;264(10):5736–5740. [PubMed] [Google Scholar]
- 6.Wang J, et al. Progressive aggregation despite chaperone associations of a mutant SOD1-YFP in transgenic mice that develop ALS. Proc Natl Acad Sci USA. 2009;106(5):1392–1397. doi: 10.1073/pnas.0813045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurney ME, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264(5166):1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menzies FM, Moreau K, Rubinsztein DC. Protein misfolding disorders and macroautophagy. Curr Opin Cell Biol. 2011;23(2):190–197. doi: 10.1016/j.ceb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schnell SA, Staines WA, Wessendorf MW. Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem. 1999;47(6):719–730. doi: 10.1177/002215549904700601. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 14.Bandyopadhyay U, et al. 2013. RNA-seq profile of spinal cord motor neurons from a presymptomatic SOD1 ALS mouse. PLoS ONE 8(1):e53575.
- 15.de Duve C, de Barsy T, Poole B, Trouet A, Tulkens P, et al. Commentary. Lysosomotropic agents. Biochem Pharmacol. 1974;23(18):2495–2531. doi: 10.1016/0006-2952(74)90174-9. [DOI] [PubMed] [Google Scholar]
- 16.Inoki K, Zhu T, Guan K-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 17.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Öst A, et al. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med. 2010;16(7-8):235–246. doi: 10.2119/molmed.2010.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morimoto N, et al. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4(3):290–293. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.