Abstract
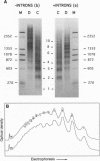
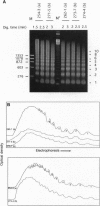
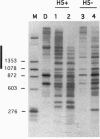
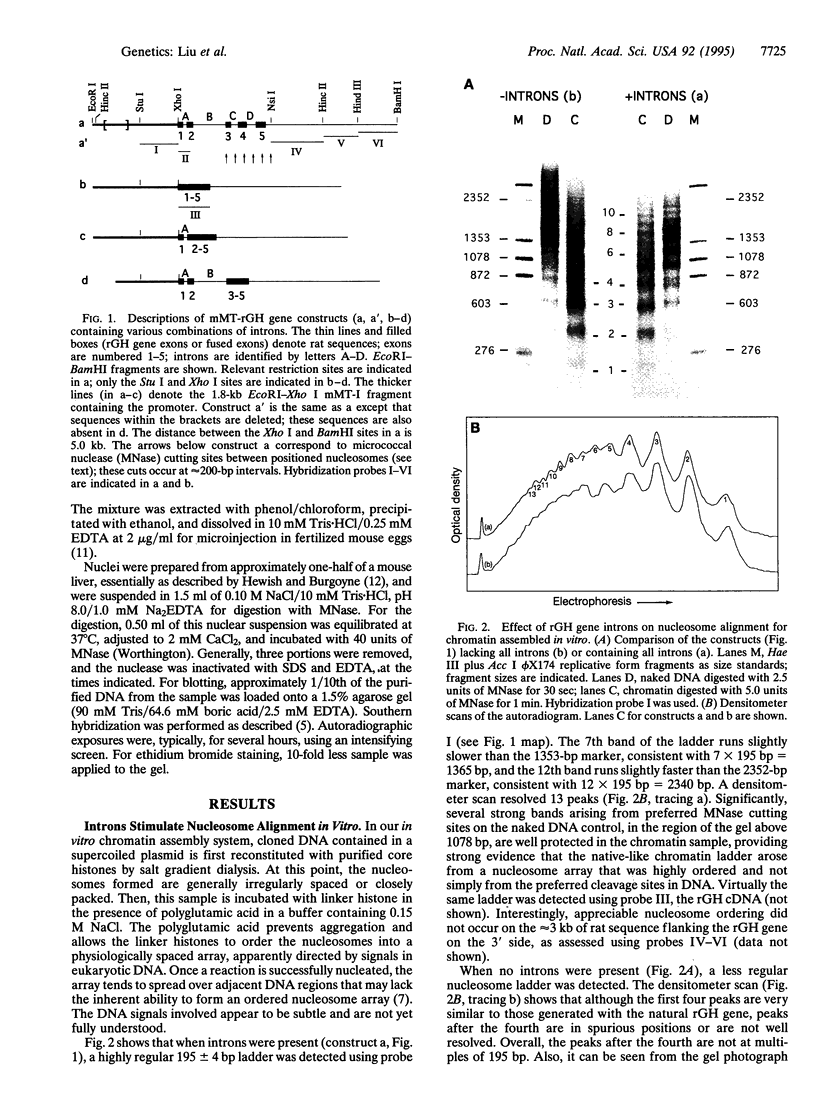
Average hepatic expression (mRNA per cell per gene) of a metallothionein-rat growth hormone (rGH) gene with its natural introns was about 15-fold higher than an intronless version when tested in transgenic mice. We examined the idea that intron removal leads to an alteration in chromatin structure that might be responsible for this effect. Using an in vitro chromatin assembly system, we observed that nucleosomes were aligned in a characteristic ordered array over the gene and promoter when all introns were present. Linker histones were necessary for this alignment to occur. In contrast, nucleosome alignment was perturbed in constructs lacking some or all of the introns. A similar disruption of nucleosome alignment was observed when comparing chromatin from livers of transgenic mice carrying rGH transgenes with or without introns. In vitro, sequences at the 3' end of the rGH gene position nucleosomes and facilitate nucleosome alignment upstream; however, nucleosome alignment does not occur on the approximately 3 kb of downstream flanking rat sequence. These observations suggest that signals present in genomic rGH DNA may serve to establish appropriate nucleosome alignment during development and, possibly, to restore nucleosome alignment to the transcribed region after disruption incurred by the passage of an RNA polymerase molecule, thereby facilitating subsequent rounds of transcription.
Full text
PDF

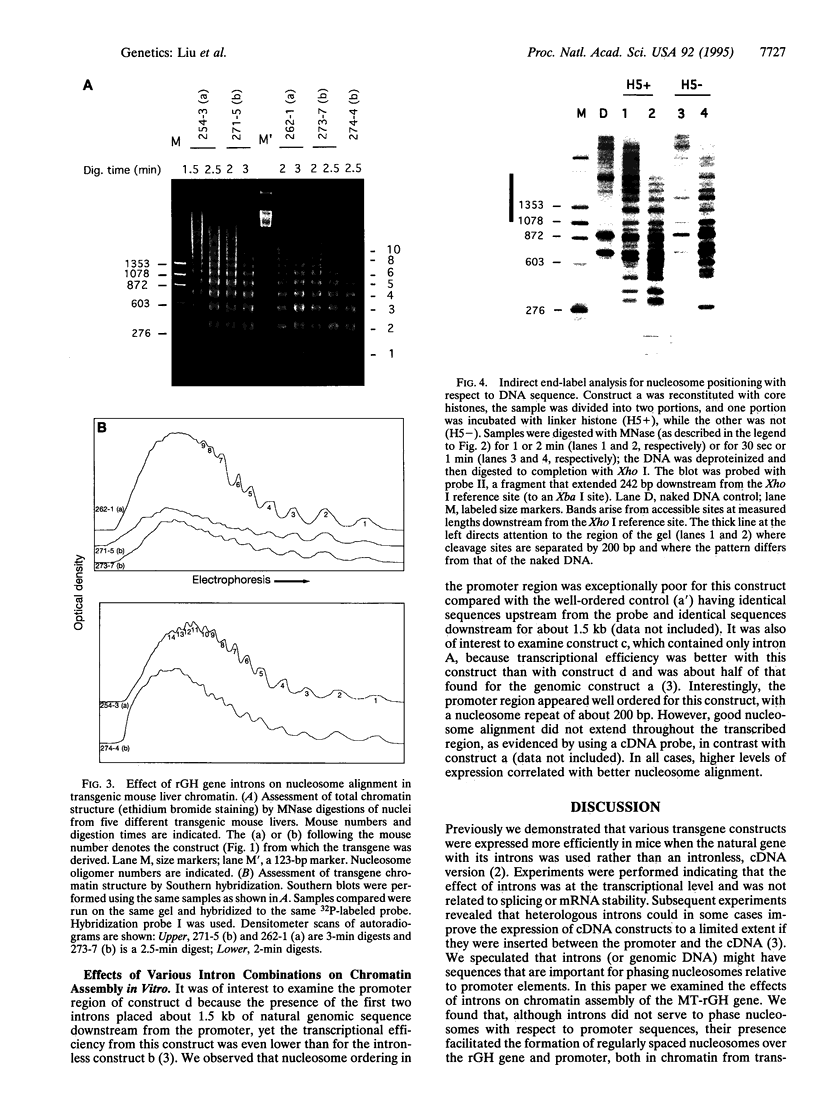

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bacolla A., Wu F. Y. Mung bean nuclease cleavage pattern at a polypurine.polypyrimidine sequence upstream from the mouse metallothionein-I gene. Nucleic Acids Res. 1991 Apr 11;19(7):1639–1647. doi: 10.1093/nar/19.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta A., Richards R. I., Baxter J. D., Shine J. Primary structure and evolution of rat growth hormone gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4867–4871. doi: 10.1073/pnas.78.8.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R. L., Allen J. M., Behringer R. R., Gelinas R. E., Palmiter R. D. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988 Feb;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R. L., Chen H. Y., Trumbauer M. E., Yagle M. K., Palmiter R. D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G., Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993 Dec;12(12):4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer D. H. Metallothionein. Annu Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Jeong S. W., Lauderdale J. D., Stein A. Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J Mol Biol. 1991 Dec 20;222(4):1131–1147. doi: 10.1016/0022-2836(91)90597-y. [DOI] [PubMed] [Google Scholar]
- Jeong S. W., Stein A. DNA sequence affects nucleosome ordering on replicating plasmids in transfected COS-1 cells and in vitro. J Biol Chem. 1994 Jan 21;269(3):2197–2205. [PubMed] [Google Scholar]
- Lauderdale J. D., Stein A. Introns of the chicken ovalbumin gene promote nucleosome alignment in vitro. Nucleic Acids Res. 1992 Dec 25;20(24):6589–6596. doi: 10.1093/nar/20.24.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Lauderdale J. D., Stein A. Signals in chicken beta-globin DNA influence chromatin assembly in vitro. Mol Cell Biol. 1993 Dec;13(12):7596–7603. doi: 10.1128/mcb.13.12.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Palmiter R. D., Brinster R. L. Germ-line transformation of mice. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Sandgren E. P., Avarbock M. R., Allen D. D., Brinster R. L. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter R. D., Sandgren E. P., Koeller D. M., Brinster R. L. Distal regulatory elements from the mouse metallothionein locus stimulate gene expression in transgenic mice. Mol Cell Biol. 1993 Sep;13(9):5266–5275. doi: 10.1128/mcb.13.9.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky V. M., Clark D. J., Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994 Jan 28;76(2):371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]