Significance
The ColE1 Rop protein is a paradigm of a canonical 4-α-helical bundle and an attractive model system for protein-folding studies. This work characterizes Rop and several of its loop mutants, focusing on their extreme structural plasticity. Plasticity gives rise to new helical bundle topologies and properties and establishes a variety of native-like and molten globule states that depend on nonnative disulfide bonds. This plasticity and the capacity of the Rop mutants to assemble a large variety of different hydrophobic cores add Rop to the list of proteins that are suitable for the engineering of bio-inspired materials. Additionally, our findings have implications for the theory of protein folding and for understanding certain pathogenic mechanisms and diseases.
Keywords: recurrent tertiary motifs, core packing, disulfide bonds, dimensionless Kratky plot
Abstract
The dimeric Repressor of Primer (Rop) protein, a widely used model system for the study of coiled-coil 4-α-helical bundles, is characterized by a remarkable structural plasticity. Loop region mutations lead to a wide range of topologies, folding states, and altered physicochemical properties. A protein-folding study of Rop and several loop variants has identified specific residues and sequences that are linked to the observed structural plasticity. Apart from the native state, native-like and molten-globule states have been identified; these states are sensitive to reducing agents due to the formation of nonnative disulfide bridges. Pro residues in the loop are critical for the establishment of new topologies and molten globule states; their effects, however, can be in part compensated by Gly residues. The extreme plasticity in the assembly of 4-α-helical bundles reflects the capacity of the Rop sequence to combine a specific set of hydrophobic residues into strikingly different hydrophobic cores. These cores include highly hydrated ones that are consistent with the formation of interchain, nonnative disulfide bridges and the establishment of molten globules. Potential applications of this structural plasticity are among others in the engineering of bio-inspired materials.
Recurrent motifs of tertiary structure are convenient model systems for studying protein folding and potentially also for the design of bio-inspired materials. For protein design purposes, structural plasticity is an important, although poorly understood, parameter to be considered, as it is among the main reasons that the re-engineering of proteins toward novel materials is not yet satisfactorily manageable (1, 2).
The present study focuses on the structural plasticity associated with the 4-α-helical bundle (4HB) motif. 4HBs consist of four amphipathic α-helices packed in a parallel or antiparallel fashion (3, 4). Their folding is largely determined by a repeating pattern of hydrophobic and hydrophilic residues, organized on the basis of seven-residue repeats (heptads) (5). Being the simplest tertiary motif, 4HBs have been subject to numerous protein-folding studies; attempts have been made to exploit them as building blocks for bio-inspired materials (6).
A paradigm of a highly regular 4HB is the RNA-binding ColE1 Repressor of Primer (Rop) protein (7–9), also referred to as RNA-one-modulator (ROM). Each monomer is an α-helical hairpin consisting of two antiparallel α-helices connected by a short loop. The sequence of Rop displays a heptad repeats pattern that is interrupted only in the loop region.
Structural simplicity makes Rop an attractive model system for the study of the folding of 4HBs. The loop region and the hydrophobic core have thereby attracted particular attention, as these regions are linked with the remarkable ability of Rop mutants to adopt altered topologies and properties (10–15). Striking examples of loop variants include mutant Loopless Rop (LLR), in which an uninterrupted pattern of heptad repeats is established through a five-residue deletion in the loop. In this “loopless” mutant, the α-helical hairpin of the monomer is converted into a single helix (15, 16). The complete LLR molecule is a tetramer that is completely reorganized relative to the dimeric wild-type (WT) Rop, thereby becoming a hyper-thermostable protein (16). On the other hand, establishment of an uninterrupted heptad periodicity through a two-residue insertion in the loop produces minimal changes relative to WT in terms of structure and properties (12). Thus, these two mutants with uninterrupted patterns of heptads reveal that there is a considerable structural plasticity inherent to the Rop sequence, but the relationship between heptad periodicity and the structural/physicochemical properties is complex.
Extreme structural plasticity producing completely altered 4HB topologies is also associated with point mutations in the loop region. Replacement of loop residue Ala31 by Pro (17) results in a complete reorganization of the entire protein, which is converted from the canonical left-handed, all-antiparallel form into a right-handed mixed-parallel and antiparallel 4-α-helical bundle, displaying a “bisecting U” topology that is to a large extent determined by the local conformation at residue 31 (18). Mutant A31P displays two variations of the bisecting U topology; these differ in the relative juxtaposition of the α-helices (19). These conformations crystallize in different space groups (orthorhombic and monoclinic); both space groups have been occasionally observed in the same crystallization drop, indicating the coexistence of the two forms in solution. Molecular dynamics simulations for A31P have demonstrated a potential for the interconversion between the two conformations (14).
Hydrophobic core mutants occasionally also display structural plasticity producing a new (“syn”) topology (20) that results from the “anti”-topology of WT through a 180° flip of one monomer around the dyad axis normal to the long axis of WT. The competition between the anti and syn topologies and the mixture of the two structures have been studied in detail for some Rop mutants (21, 22).
Apart from structural plasticity, a closely related issue associated with the folding of Rop is the role of Cys residues (Cys-38 and Cys-52). Both residues are buried in the hydrophobic core and are not involved in the formation of disulfide bridges in any of the known structures of WT and its mutants. Surprisingly, however, a Cys-free variant (CYSfree) that conserves the structure, stability, and in vivo activity of WT exhibits dramatically faster unfolding kinetics (23).
The present study focuses on the role of the loop region and Cys residues in the structural plasticity of Rop. To explore the conversion of the WT anti-topology into the bisecting U topology of A31P, the three double mutants D30P/A31G (PG), D30G/A31P (GP), and D30P/A31P (PP) have been constructed for loop positions 30 and 31. These mutations combine the effects of the most constrained amino acid (Pro) and of the least constrained one (Gly). In addition, the potential role of Cys residues in Rop folding is explored by following the effects of reducing agents.
Results
Combining Pro and Gly Residues in the Loop Does Not Necessarily Alter the Anti-Topology.
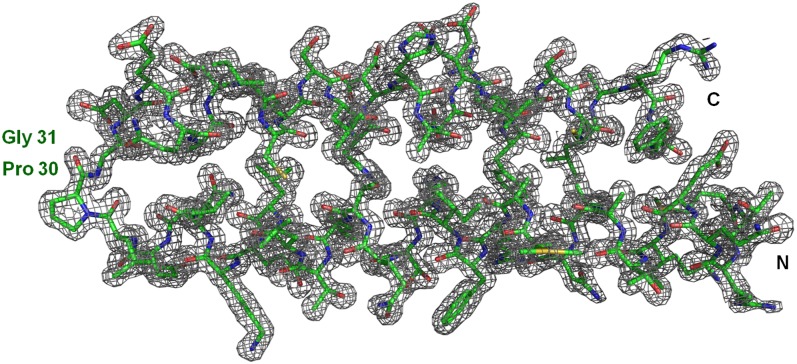
X-ray crystallography was used to determine the 3D structures of loop mutants PG, GP, and PP. Data quality crystals could be obtained only for PG (24), the structure of which was determined at a resolution of 1.4 Å (Table S1). The electron density map (Fig. 1) illustrates the high quality of the PG crystal structure; a schematic representation is given in Fig. S1. The PG dimer is an all-antiparallel 4HB, consisting of two nearly identical subunits with an rmsd to each other of 0.66 Å for the Cα atoms (Table S2). In WT the two subunits are identical, being related to each other by a crystallographic (i.e., an exact) dyad. PG adopts the anti-topology, exhibiting a high overall similarity to WT; considerable deviations exist relative to A31P, reflecting the topological differences between the two proteins. The PG loop adopts an intermediate conformation between the loops of WT and A31P (Fig. S2). Relative to WT, it is extended from the side of the first helix by one residue (Leu-29) that adopts a preproline conformation (Table S3) (25). The topological similarity to WT can be, to a large extent, attributed to the conformation of Gly31, which is WT-like, thus placing the helices of the 4HB in positions corresponding to the anti-topology (Table S3 and Fig. S2). The small size of Gly31 contributes to the formation of a water-filled pocket, leading to an increased accessibility of the hydrophobic core residues at the end of the 4HB (Fig. S3).
Fig. 1.
Crystal structure of PG. A 2Fo-Fc electron density map for the PG monomer (chain A) contoured at 1.1 σ. The mutated residues Pro30 and Gly31 and the N and C termini are labeled.
Reducing Agents Strongly Affect the Chromatographic Properties of Rop and Its Mutants.
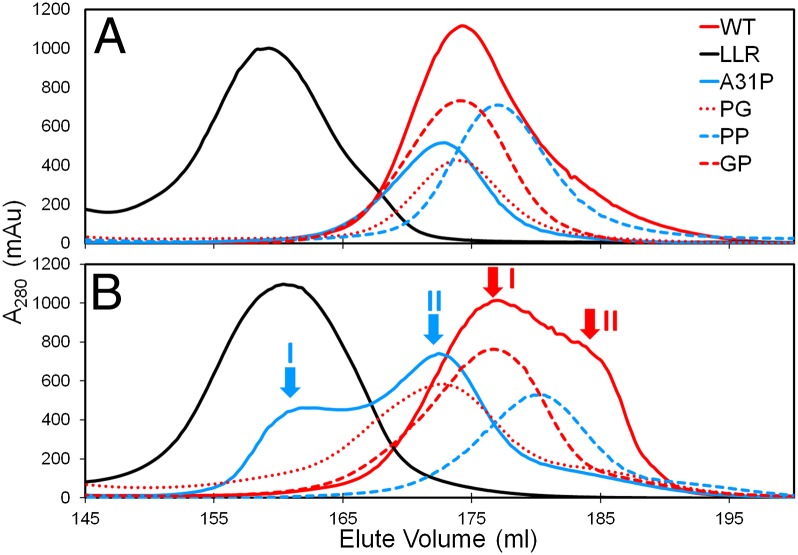
Although no disulfide bridges are found in the known structures of Rop and its mutants, size exclusion chromatography (SEC) experiments of the proteins studied reveal an unexpected dependence of the elution profiles on reducing agents. As shown in Fig. 2A, the proteins elute in the presence of the reducing agent β-mercaptoethanol (β-met) as single peaks that are consistent with largely monodisperse populations of protein molecules. On the other hand, in the absence of β-met (Fig. 2B), the elution profiles change, exhibiting either two distinguishable peaks (mutant A31P, peaks I and II), peak broadening (mutants PP, PG, and GP) or a broad profile with a main peak (I) followed by a pronounced shoulder (II) for WT. These profiles suggest a tendency toward heterodispersity. Only LLR is not affected by the reducing agent because its elution profiles are nearly identical, reflecting monodispersity under all conditions.
Fig. 2.
SEC elution profiles for Rop and its mutants. (A) Elution profiles in the presence of β-met. (B) Elution profiles in the absence of β-met with the two elution peaks for A31P (cyan arrows) are labeled as I and II (in cyan), and the broad peak and its shoulder of WT (red arrows) are labeled as I and II (in red), respectively.
Based on the calibration of the SEC column, WT, PG, GP, PP, and A31P in the presence of reducing agents elute as dimers and LLR as a tetramer (Fig. 2A) in agreement with the known 3D structures. In the absence of reducing agents (Fig. 2B), the heterodispersity of the eluting proteins is occasionally associated with changes in the apparent molecular weights: Small shifts are observed for WT between peaks I and II; for A31P, peak II elutes at an apparent molecular weight that is consistent with a dimer, whereas peak I elutes at a molecular weight that is comparable to that of a tetramer. This major change is probably linked to the nonglobular characteristics of peak I as suggested by circular dichroism (CD) and small angle X-ray scattering (SAXS) experiments (see below).
Looser Structural Packing and Disorder Effects Are Associated with the Presence of Pro at Position 31.
The dependence of the properties of Rop and its mutants from reducing agents was further characterized by SAXS at the SWING beamline/SOLEIL synchrotron in which sample delivery is performed via high-performance liquid chromatography (HPLC) (26). In the absence of reducing agents, WT and all variants (except LLR) were found to be polydisperse for a considerable fraction of their elution peaks from the HPLC column. Polydispersity produced a strong variation of the observed radii of gyration (Rg). SAXS data were thus analyzed only for the fraction of each elution peak that was characterized by constant Rg. In contrast, for those proteins (WT, LLR, and A31P) measured in the presence of a reducing agent, constant Rg values, i.e., monodispersity, were observed over a considerably larger fraction of the elution profiles, thus confirming also at the level of SAXS that reducing agents do affect the properties of Rop and most of its mutants. WT and GP exhibit the most tightly packed structures on the basis of their Rg values (21.0 ± 0.1 Å), followed by PG (22.1 ± 0.2 Å). On the other hand, the Rg values for PP (23.0 ± 0.2 Å) and A31P (24.1 ± 0.4 Å) suggest less efficient packing. The Rg value of LLR (32.9 ± 0.1 Å) reflects the bigger size and the elongated shape of this protein in agreement with the crystallographically determined structure (15).
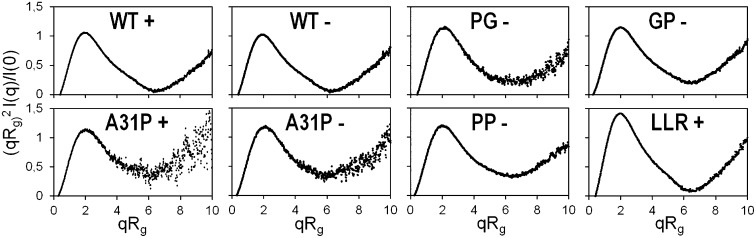
Dimensionless Kratky plots (27) were calculated to judge the state of folding for the proteins studied (Fig. 3). The bell-shaped curves for WT and, to a lesser extent, for GP and PG are consistent with compact, well-folded proteins. In addition, the maximum values of their plots are close to 1.2 at qRg ∼ 1.9 [with q = 4πsin(θ)/λ being the momentum transfer, 2θ the scattering angle, and λ = 1.03 Å the wavelength of X-rays]. These values confirm the presence of fully folded, single-domain globular proteins. On the other hand, the characteristics of the dimensionless Kratky plots for A31P and PP (maximum values of 1.35 at qRg ∼ 2.1) indicate a looser structural packing or partial disorder (27); these characteristics become more pronounced in the absence of a reducing agent. Mutants A31P, PP, and GP provide strong evidence that, among the Rop variants studied, the presence of Pro at position 31 is a necessary but not sufficient condition for loose structural packing and disorder effects. The well-defined, bell-shaped dimensionless Kratky plot of LLR (maximum value close to 1.7 at qRg ∼ 2) is consistent with the elongated, nonglobular shape of LLR and indicates a compact, fully folded protein (28).
Fig. 3.
Dimensionless Kratky plots for Rop and its variants. For WT, PG, GP, A31P, PP, and LLR. Plus (“+”) and minus (“−”) indicate the presence and the absence of β-met, respectively.
Molten Globule States Are Established for Some Mutants in the Absence of Reducing Agents.
CD was used to validate the integrity of the secondary structure of WT and its mutants as a function of temperature. At 10 °C far-UV CD scans (195–260 nm) exhibited the two characteristic minima (at 208 and 222 nm) of highly α-helical proteins. The stability of the proteins reflected in the loss of secondary structure was followed by the gradual loss of the characteristic minima at increasingly higher temperatures (Fig. 4A1–10). For the thermal unfolding transition, melting curves were obtained from the temperature dependence of the CD signal at 222 nm (Fig. 4B1–10 and Fig. S4). In the presence of reducing agents, the shape of the melting curves is sigmoid, corresponding to a two-state transition between the folded and the unfolded state. Melting temperatures (Tm) were calculated from the maximum value of the first derivative of the melting curves, confirming that LLR is hyper-thermostable (Tm = 92 °C) in accordance with previous studies (15, 16) and surpasses the stability of WT (Tm = 58 °C). Mutants GP (Tm = 45 °C), PG (Tm = 40 °C), PP (Tm = 38 °C), and A31P (Tm = 36 °C) are considerably less stable. The presence of an isodichroic point at 203 nm (shown for WT, Fig. 4A1–3) is also consistent with a two-state unfolding transition. In the absence a reducing agent, the melting curves of mutants A31P from peak I in Fig. 2 (A31P-peak I) and PP (Fig. 4B5,10) monotonically increase, a property that characterizes molten globule states (29), whereas A31P-peak II (Fig. 4B6) shows a two-state unfolding transition, thus being comparable to A31P in the presence of the reducing agent (Fig. 4B4). In both cases the melting temperature is approximately Tm = 37 °C, whereas no melting temperatures are defined for the molten globule states with their monotonically increasing melting curves. Singular value decomposition (SVD) analyses (30) of the far-UV CD spectra confirm (with the exception of A31P-peak I and PP without reducing agents) the presence of two-state transitions, as they produce only two significant species of linearly independent CD spectra (Fig. S5). The first significant species corresponds to the α-helical state, the second one to random coil. In the absence of reducing agents, however, the SVD analysis of the CD spectra of A31P-peak I and PP shows a more complex pattern with more than two states (Fig. S5), thus reflecting the presence of molten globule states.
Fig. 4.
Thermal unfolding for Rop and its variants, recorded by CD. (A) Smoothed far-UV CD spectra recorded for a range of temperatures. Isodichroic points are indicated by black circles. (B) Melting curves monitoring the variation of the CD signal at 222 nm are shown with the temperature noted. Sigmoid melting curves suggest cooperative unfolding transitions, whereas monotonically varying curves (B5, B10) suggest molten-globule–like states. In all cases, the CD signal has been transformed into mean residue ellipticity. Subscripts in A and B designate the following cases: 1, WT in the presence of β-met; 2, WT-peak I, no β-met; 3, WT-peak II, no β-met; 4, A31P in the presence of β-met; 5, A31P-peak I, no β-met; 6, A31P-peak II, no β-met; 7, LLR in the presence of β-met; 8, LLR, no β-met; 9, PP in the presence of β-met; and 10, PP, no β-met.
Establishment of Molten Globule States Is Reversible.
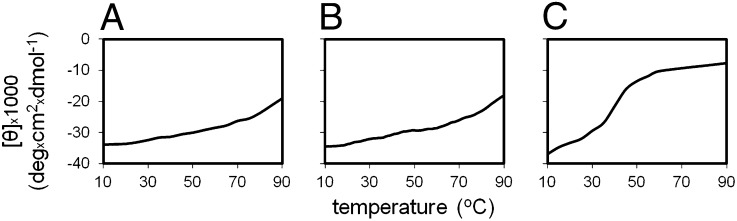
Mutant A31P was purified in the absence of reducing agents and showed the typical thermal unfolding curve corresponding to a molten globule state (Fig. 5A). Half of the protein sample was subsequently stored at 4 °C (sample 1), and β-met was added to the other half of the sample and was also stored at 4 °C (sample 2). Thermal unfolding curves for samples 1 and 2 were recorded after 7 d, revealing that sample 1 conserves its molten globule properties (Fig. 5B), whereas, in sample 2, the molten globule features are largely lost, and a melting curve corresponding to a two-state transition was established (Fig. 5C).
Fig. 5.
Test for the reversibility of the molten globule state for mutant A31P by CD (A) Melting curve for A31P produced in the absence of β-met. (B) Melting curve of a sample from the original protein after a 7-d storage at 4 °C. (C) Melting curve obtained after a 7-d storage at 4 °C of a sample from the original protein to which β-met has been added before storage. The CD signal is transformed into mean residue ellipticity.
Discussion
Properties of Rop and Its Mutants Suggest the Involvement of Nonnative Disulfide Bonds in Their Folding Pathways.
A striking result of our studies is the dependence of the properties of Rop and its loop mutants (with the exception of LLR) on reducing agents. In the presence of reducing agents, the elution peaks of all Rop variants indicate considerable monodispersity, whereas in their absence they show an increased polydispersity (except for LLR), which manifests itself in SEC experiments as broadening of elution peaks, as a clear shoulder of an elution peak, or even as two well-separated elution peaks (Fig. 2). This suggests a role for one or both Cys residues of Rop in the folding of the 4HB, although in all known structures both Cys residues exist in the reduced form.
It is known from various proteins of prokaryotic (31) or eukaryotic origin (32) that Cys residues can affect protein-folding pathways due to improper formation and reshuffling of transient disulfide bonds (33–35). Our findings for Rop and its variants make a strong case for the formation of nonnative disulfides in the absence of reducing agents; these appear to play a role in directing folding of these proteins toward a variety of different states, including the molten globule state for mutants A31P and PP. The insensitivity of LLR to reducing agents suggests that its folding does not involve transient disulfides.
Native, Native-Like, and Molten Globule States Depend on Loop Conformations, 4HB Topologies, and the Capacity of Nonnative Disulfide Formation.
The structural states identified by our experiments differ considerably in their properties. Thermal unfolding in the presence of reducing agents always produces sigmoidal melting curves reflecting cooperative transitions between the folded and the unfolded state (Fig. 4). The SVD analysis of the CD spectra (Fig. S5) and the presence of an isodichroic point (Fig. 4A1) confirm that only these two states are present to any significant extent. Because the crystallization of Rop and most of its mutants has been performed in the presence of reducing agents, we conclude that under these conditions both Cys residues must exist in the reduced form in the 4HB core, in agreement with all known crystal structures of these proteins. We thus define the state established for WT and its loop mutants in the presence of reducing agents (Fig. 2A) as the native state; the available crystal structures correspond to this state.
In the absence of reducing agents, the situation becomes more complex: From the Tm values calculated from the corresponding melting curves of WT (peaks I and II) (Fig. 4B2,3), PG and GP, it can be concluded that these proteins retain the stability of their respective native states. Furthermore WT, PG, and GP also conserve the sigmoidal shapes of their native-state melting curves (Fig. S4), which suggests cooperative two-state unfolding. We thus term the states adopted by these proteins as “native-like” to account for their overall similarity to the native state, despite slight differences between the two states in their SEC elution profiles. For LLR, the melting curve and the SEC behavior in the absence of a reducing agent conserve the properties of native LLR (Fig. 2 and Fig. 4B8).
A very different situation exists for mutant A31P in the absence of reducing agents: Whereas the melting curve of A31P-peak II is sigmoidal (Fig. 4B6) with a Tm value close to that of the native state, the melting curve for A31P-peak I (Fig. 4B5) is indicative of a molten globule state (29). Similarly, the melting curve of the PP protein that elutes in the absence of reducing agents as a broadened peak (Fig. 2B) is also suggestive of a molten globule state (Fig. 4A10). The CD spectra of A31P-peak I and PP in the absence of a reducing agent (Fig. 4A5, 10) show that even at high temperatures the α-helical secondary structure is not completely lost, a property characterizing the molten globule state (36).
The establishment of the above states has been also confirmed by SAXS, as the highest Rg values correspond to PP and A31P in the absence of reducing agents, indicating a looser packing compared with the native or native-like states of WT, GP, and PG. Dimensionless Kratky plots (27, 28, 37) show that WT and LLR are fully folded under all conditions studied, whereas PG and GP in the native-like state are slightly less tightly packed. PP and A31P in the absence of reducing agents correspond to less compact and partially unfolded structures in agreement with the molten-globule–like characteristics elucidated by CD and predicted on the basis of molecular dynamics simulations (14). Importantly, as it has been shown for A31P, the molten globule characteristics can be largely reversed (Fig. 5) through the addition of a reducing agent. These observations support the concept of a possible role of Cys38 and Cys52 in the establishment of native-like and molten globule states through the formation of nonnative disulfides.
Overall, the similarities between A31P-peak I and PP in the absence of reducing agents suggest that they adopt comparable folding states, i.e., molten globules. At the level of sequence and conformational similarities, both mutants share the conformationally constrained residue Pro in the critical loop position 31, i.e., the residue that is responsible not only for a new loop conformation (Table S3), but also for the drastic rearrangement of A31P relative to WT and the establishment of the bisecting U topology. It is noteworthy that, in the A31P loop region, residues Leu29 and Asp30 adopt a conformation that can be described as preproline-like (25). It can be thus reasonably assumed that mutant PP with Pro residues in loop positions 30 and 31 will adopt a similar loop conformation to A31P with its two residues in positions 29 and 30 adopting also a preproline conformation. The similarities in the loop conformations that ultimately determine the overall topology of the 4HB imply for A31P and PP similar topologies (bisecting U) and properties, a fact supported by the establishment of molten globule states for both proteins. Still, as demonstrated by GP, the presence of Pro31 is not alone sufficient for the establishment of molten globule characteristics. Possibly, the intrinsic structural flexibility of Gly compensates for the effects of Pro, restricting the accessible states for GP to native or native-like one. In mutant PG, the effects of the mutations are localized relative to WT, affecting only the conformations of Pro30 and Leu29, whereas Gly31 adopts a WT-like conformation (Table S3), which is associated as already stated with a native-like (anti) topology (Fig. S1) and native-like properties.
Consequently, the exact position and the order of Pro and Gly residues are critical for the local conformation of the loop and the overall topology of the 4HB and thus for the accessible folding states, which may include native-like and molten globule states linked to nonnative disulfides.
Crystal Structures Provide Insights into the Capacity for the Formation of Nonnative Disulfides and the Molten Globule State.
For all known structures of WT and its loop mutants, the distances between Cys residues are incompatible with the formation of disulfide bonds. However, in one A31P structure (termed hereafter “A31P-A”) associated with the monoclinic crystal form [Protein Data Bank (PDB) ID code 1GMG, chain A, dimer formed via a crystallographic dyad], the Sγ atoms of Cys38 and Cys52 residues from different chains of the dimer approach each other within a distance of ∼2.9 Å; this distance is reduced to ∼2.4 Å with a stereochemistry that is favorable for an intersubunit disulfide bridge if a different rotamer is used for the side chain of Cys52 (38). A31P-A thus provides a structural model for a state of A31P capable of establishing nonnative disulfides between Cys38 and Cys52 upon folding/assembly of the dimer without requiring major structural changes. The properties of the A31P-A structure are consistent with the dependence of the various folding states observed on reducing agents and provide a basis for understanding the CYSfree variant with its dramatically faster unfolding kinetics compared with WT (23). In contrast, in other structures, e.g., WT or A31P from the orthorhombic crystal form (PDB ID code 1B6Q), the closest interchain distances between the Sγ atoms from Cys residues are in the range 4.2–7.6 Å and thus are incompatible with the formation of disulfide bridges.
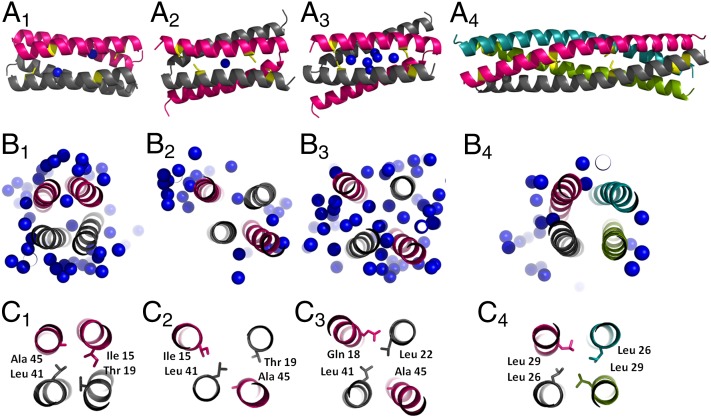
The structure A31P-A is also consistent with the molten globule properties of A31P-peak I. In fact, comparison of A31P-A with the structures of WT, the orthorhombic form of A31P, or LLR shows that A31P-A possesses a much looser structure and a highly hydrated core (Fig. 6B1–4), which are typical characteristics of the molten globule state (36).
Fig. 6.
Plasticity and hydration of hydrophobic cores. (A) Schematic representations of structures indicated by the following subscripts in A–C: 1, WT; 2, A31P-orthorhombic form, 3, A31P-monoclinic form; and 4, LLR. The individual subunits of the 4HBs are colored separately. Cys residues are colored in yellow, and water molecules located in the hydrophobic core are shown as blue spheres. (B) The central parts of the hydrophobic cores of the molecules shown in A. Water molecules from the immediate neighborhood and those hydrating the core are also shown. View is down the long axis of the helical bundle. (C) Packing of the hydrophobic residues in the central part of the helical bundles.
The capacity of Rop mutants with altered hydrophobic core residues to switch between the syn and the anti topologies by thermal fluctuations (21) suggests the presence of additional degrees of freedom in the structures of these 4-α-helical bundles that also could lead to the formation of nonnative disulfides, much like the predicted spontaneous switch between the different conformations of A31P (14) through which A31P-A also becomes accessible. This structural plasticity of the 4HBs formed by Rop and its mutants provides a mechanism to access conformations that are compatible with either the reduced form of Cys residues or the formation of intersubunit disulfides. The formation of nonnative disulfides occurs at the level of dimers because the native-like or the molten globule states are always associated with the dimeric state, as suggested by the SEC experiments.
Plasticity of the Hydrophobic Core Enables the Establishment of Different Helical Bundle Topologies.
The hydrophobic cores of proteins usually represent highly conserved elements of their structures (38). For protein cores, we have systematically quantified the structural destabilization associated with the loss of hydrophobic contacts and shown that volume changes of core residues exceeding one methyl group act in a very destabilizing way (39).
Surprisingly, the cores of loop mutants studied in the present work reveal an extreme plasticity in the way in which the hydrophobic residues from heptads of the Rop sequence (5) rearrange themselves to form a variety of cores; these cores belong to 4HBs that may drastically differ in their topologies (anti, bisecting U) and oligomerization states (tetramers or dimers). Extremely rearranged cores relative to WT are those of A31P with its bisecting U topology of structures and of the tetrameric LLR mutant. At sequence level both mutants deviate from WT only in the loop region; the hydrophobic residues are fully conserved, but extensively rearranged, as shown in Fig. 6C1–4, resulting in strikingly different protein cores and structures (Fig. 6A1–4).
Conclusions.
Our results indicate that changes in the loop region may drastically alter the structures, the hydrophobic cores, and the folding states of Rop mutants. Replacement of one or more loop residues by Pro (A31P, PP) appears to be a special case capable of drastically altering the loop conformation and the topology of the 4HB and of inducing the establishment of molten globule states that depend on nonnative disulfide bridges. On the other hand, when combined with Pro in the loop, Gly residues may in part compensate for the most drastic effects of Pro, producing native-like structures (mutant PG) and native-like properties (GP and PG). The extreme structural plasticity of Rop and its mutants also enables the formation of nonnative disulfide bonds, making the properties of these molecules dependent on reducing agents. This plasticity, and in particular the capacity for the assembly of a large variety of hydrophobic cores, provides the basis for the engineering of different modes of assembly/association of Rop variants. A better understanding of the molecular assembly puzzle of Rop could create a potentially powerful approach to engineer bio-inspired materials exploiting the plasticity in the assembly of subunits derived from Rop variants. Additionally, the prevalence of coiled-coils and helical bundles in secretion systems of pathogenic bacteria (40) or in members of epigenetic protein families that represent new frontiers for drug discovery (41) makes our results relevant to the understanding of the structural basis of various diseases and to the development of new drugs.
Materials and Methods
Mutagenesis, Expression, and Purification.
Loop mutants were obtained using the QuikChange II site-directed mutagenesis kit (Stratagene). For the attachment of a C-terminal His6 tag, the genes encoding the proteins studied were cloned into the pET-26b(+) vector (Novagen) and transformed into the Escherichia coli strain BL21(DE3). Protein expression and purification followed a protocol published earlier (24) with yields of ∼30 mg pure protein per 10 g of cell paste.
X-Ray Crystallography.
The crystallization of PG has been reported earlier (24). No data-quality crystals were obtained for PP and GP. For PG X-ray diffraction, data extending to a resolution of 1.4 Å were collected at 100 K at the EMBL/DESY synchrotron. The structure was determined by molecular replacement using PHASER (42), the WT monomer as search model (loop residues 27–34 were omitted), and data from the resolution range 50–4 Å. Rigid body refinement, followed by restrained refinement, was performed with Refmac5 (43). Six extensively disordered C-terminal residues (58D-63E) and the His6 tag were omitted from the model; water molecules were added conservatively in the C-terminal region due to the presence of noninterpretable electron density from the disordered residues. The final model of PG was deposited with the Protein Data Bank (PDB ID code 4DO2).
SAXS.
SAXS data were collected using the SWING beamline at the SOLEIL synchrotron, which integrates HPLC with a SAXS instrument (26). Dimensionless Kratky plots (27) were used to assess the globular nature/folding state of the proteins examined. More details are provided in SI Materials and Methods.
Circular Dichroism.
CD spectra were obtained using a J-810 CD spectropolarimeter (Jasco Inc.). Thermal denaturation was analyzed by collecting full far-UV CD spectra (260–195 nm) in the range of 10–90 °C (30–110 °C for LLR) in steps of 5 °C and monitoring the change of the typical α-helical minima at 208 and 222 nm. Melting curves were obtained from the CD signal at 222 nm for temperatures 10–90 °C with a temperature increase of 80 °C/h and a waiting time of 2 s for stabilization. SVD analysis of the far-UV CD spectra was performed with the program SVD1 (44). More details are provided are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Professor Anna Mitraki for carefully reading the manuscript and helpful advice. This work was supported by funds from the European Union–Regional Potential Programme (Project InnovCrete, Contract 316223), by a grant in the framework of the Greece–France Joint Technology Research Programmes, and by the SOLEIL synchrotron. M.A. was supported by an IRAKLITOS grant from the Greek Ministry of Education and by a short-term European Molecular Biology Organization fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4DO2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322065111/-/DCSupplemental.
References
- 1.Boyle AL, Woolfson DN. De novo designed peptides for biological applications. Chem Soc Rev. 2011;40(8):4295–4306. doi: 10.1039/c0cs00152j. [DOI] [PubMed] [Google Scholar]
- 2.Khoury GA, Smadbeck J, Kieslich CA, Floudas CA. Protein folding and de novo protein design for biotechnological applications. Trends Biotechnol. 2014;32(2):99–109. doi: 10.1016/j.tibtech.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos P, Rossmann MG, Johnson JE. A four-helical super-secondary structure. Biochem Biophys Res Commun. 1977;75(1):83–86. doi: 10.1016/0006-291x(77)91292-x. [DOI] [PubMed] [Google Scholar]
- 4.Cohen C, Parry DA. Alpha-helical coiled coils and bundles: How to design an alpha-helical protein. Proteins. 1990;7(1):1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 5.Paliakasis CD, Kokkinidis M. Relationships between sequence and structure for the four-alpha-helix bundle tertiary motif in proteins. Protein Eng. 1992;5(8):739–748. doi: 10.1093/protein/5.8.739. [DOI] [PubMed] [Google Scholar]
- 6.Hill RB, Raleigh DP, Lombardi A, DeGrado WF. De novo design of helical bundles as models for understanding protein folding and function. Acc Chem Res. 2000;33(11):745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castagnoli L, et al. Genetic and structural analysis of the ColE1 Rop (Rom) protein. EMBO J. 1989;8(2):621–629. doi: 10.1002/j.1460-2075.1989.tb03417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banner DW, Kokkinidis M, Tsernoglou D. Structure of the ColE1 rop protein at 1.7 A resolution. J Mol Biol. 1987;196(3):657–675. doi: 10.1016/0022-2836(87)90039-8. [DOI] [PubMed] [Google Scholar]
- 9.Eberle W, Pastore A, Sander C, Rösch P. The structure of ColE1 rop in solution. J Biomol NMR. 1991;1(1):71–82. doi: 10.1007/BF01874570. [DOI] [PubMed] [Google Scholar]
- 10.Munson M, O’Brien R, Sturtevant JM, Regan L. Redesigning the hydrophobic core of a four-helix-bundle protein. Protein Sci. 1994;3(11):2015–2022. doi: 10.1002/pro.5560031114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Predki PF, Agrawal V, Brünger AT, Regan L. Amino-acid substitutions in a surface turn modulate protein stability. Nat Struct Biol. 1996;3(1):54–58. doi: 10.1038/nsb0196-54. [DOI] [PubMed] [Google Scholar]
- 12.Vlassi M, et al. Restored heptad pattern continuity does not alter the folding of a four-alpha-helix bundle. Nat Struct Biol. 1994;1(10):706–716. doi: 10.1038/nsb1094-706. [DOI] [PubMed] [Google Scholar]
- 13.Shukla RT, Baliga C, Sasidhar YU. The role of loop closure propensity in the refolding of Rop protein probed by molecular dynamics simulations. J Mol Graph Model. 2013;40:10–21. doi: 10.1016/j.jmgm.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Glykos NM, Kokkinidis M. Structural polymorphism of a marginally stable 4-alpha-helical bundle. Images of a trapped molten globule? Proteins. 2004;56(3):420–425. doi: 10.1002/prot.20167. [DOI] [PubMed] [Google Scholar]
- 15.Glykos NM, et al. Loopless Rop: Structure and dynamics of an engineered homotetrameric variant of the repressor of primer protein. Biochemistry. 2006;45(36):10905–10919. doi: 10.1021/bi060833n. [DOI] [PubMed] [Google Scholar]
- 16.Lassalle MW, et al. Dimer-to-tetramer transformation: Loop excision dramatically alters structure and stability of the ROP four alpha-helix bundle protein. J Mol Biol. 1998;279(4):987–1000. doi: 10.1006/jmbi.1998.1776. [DOI] [PubMed] [Google Scholar]
- 17.Kokkinidis M, et al. Correlation between protein stability and crystal properties of designed ROP variants. Proteins. 1993;16(2):214–216. doi: 10.1002/prot.340160208. [DOI] [PubMed] [Google Scholar]
- 18.Levy Y, Cho SS, Shen T, Onuchic JN, Wolynes PG. Symmetry and frustration in protein energy landscapes: A near degeneracy resolves the Rop dimer-folding mystery. Proc Natl Acad Sci USA. 2005;102(7):2373–2378. doi: 10.1073/pnas.0409572102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glykos NM, Cesareni G, Kokkinidis M. Protein plasticity to the extreme: Changing the topology of a 4-alpha-helical bundle with a single amino acid substitution. Structure. 1999;7(6):597–603. doi: 10.1016/s0969-2126(99)80081-1. [DOI] [PubMed] [Google Scholar]
- 20.Willis MA, Bishop B, Regan L, Brunger AT. Dramatic structural and thermodynamic consequences of repacking a protein’s hydrophobic core. Structure. 2000;8(12):1319–1328. doi: 10.1016/s0969-2126(00)00544-x. [DOI] [PubMed] [Google Scholar]
- 21.Gambin Y, et al. Direct single-molecule observation of a protein living in two opposed native structures. Proc Natl Acad Sci USA. 2009;106(25):10153–10158. doi: 10.1073/pnas.0904461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schug A, Whitford PC, Levy Y, Onuchic JN. Mutations as trapdoors to two competing native conformations of the Rop-dimer. Proc Natl Acad Sci USA. 2007;104(45):17674–17679. doi: 10.1073/pnas.0706077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hari SB, Byeon C, Lavinder JJ, Magliery TJ. Cysteine-free Rop: A four-helix bundle core mutant has wild-type stability and structure but dramatically different unfolding kinetics. Protein Sci. 2010;19(4):670–679. doi: 10.1002/pro.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambrazi M, et al. Purification, crystallization and preliminary X-ray diffraction analysis of a variant of the ColE1 Rop protein. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 5):432–434. doi: 10.1107/S1744309108011342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ho BK, Brasseur R. The Ramachandran plots of glycine and pre-proline. BMC Struct Biol. 2005;5:14. doi: 10.1186/1472-6807-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David G, Perez J. Combined sampler robot and high-performance liquid chromatography: A fully automated system for biological small-angle X-ray scattering experiments at the Synchrotron SOLEIL SWING beamline. J Appl Cryst. 2009;42(5):892–900. [Google Scholar]
- 27.Durand D, et al. NADPH oxidase activator p67(phox) behaves in solution as a multidomain protein with semi-flexible linkers. J Struct Biol. 2010;169(1):45–53. doi: 10.1016/j.jsb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Koch MH, Vachette P, Svergun DI. Small-angle scattering: A view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys. 2003;36(2):147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- 29.Kuwajima K, Hiraoka Y, Ikeguchi M, Sugai S. Comparison of the transient folding intermediates in lysozyme and alpha-lactalbumin. Biochemistry. 1985;24(4):874–881. doi: 10.1021/bi00325a010. [DOI] [PubMed] [Google Scholar]
- 30.Manavalan P, Johnson WC., Jr Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal Biochem. 1987;167(1):76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 31.Kim J, Robinson AS. Dissociation of intermolecular disulfide bonds in P22 tailspike protein intermediates in the presence of SDS. Protein Sci. 2006;15(7):1791–1793. doi: 10.1110/ps.062197206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissman JS, Kim PS. Reexamination of the folding of BPTI: Predominance of native intermediates. Science. 1991;253(5026):1386–1393. doi: 10.1126/science.1716783. [DOI] [PubMed] [Google Scholar]
- 33.Frand AR, Cuozzo JW, Kaiser CA. Pathways for protein disulphide bond formation. Trends Cell Biol. 2000;10(5):203–210. doi: 10.1016/s0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- 34.Mamathambika BS, Bardwell JC. Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol. 2008;24:211–235. doi: 10.1146/annurev.cellbio.24.110707.175333. [DOI] [PubMed] [Google Scholar]
- 35.Robinson AS, King J. Disulphide-bonded intermediate on the folding and assembly pathway of a non-disulphide bonded protein. Nat Struct Biol. 1997;4(6):450–455. doi: 10.1038/nsb0697-450. [DOI] [PubMed] [Google Scholar]
- 36.Mizuguchi M, Masaki K, Nitta K. The molten globule state of a chimera of human alpha-lactalbumin and equine lysozyme. J Mol Biol. 1999;292(5):1137–1148. doi: 10.1006/jmbi.1999.3132. [DOI] [PubMed] [Google Scholar]
- 37.Doniach S. Changes in biomolecular conformation seen by small angle X-ray scattering. Chem Rev. 2001;101(6):1763–1778. doi: 10.1021/cr990071k. [DOI] [PubMed] [Google Scholar]
- 38.Fadouloglou VE, Glykos NM, Kokkinidis M. Side-chain conformations in 4-alpha-helical bundles. Protein Eng. 2001;14(5):321–328. doi: 10.1093/protein/14.5.321. [DOI] [PubMed] [Google Scholar]
- 39.Vlassi M, Cesareni G, Kokkinidis M. A correlation between the loss of hydrophobic core packing interactions and protein stability. J Mol Biol. 1999;285(2):817–827. doi: 10.1006/jmbi.1998.2342. [DOI] [PubMed] [Google Scholar]
- 40.Gazi AD, Charova SN, Panopoulos NJ, Kokkinidis M. Coiled-coils in type III secretion systems: Structural flexibility, disorder and biological implications. Cell Microbiol. 2009;11(5):719–729. doi: 10.1111/j.1462-5822.2009.01297.x. [DOI] [PubMed] [Google Scholar]
- 41.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: A new frontier for drug discovery. Nat Rev Drug Discov. 2012;11(5):384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Konno T. Conformational diversity of acid-denatured cytochrome c studied by a matrix analysis of far-UV CD spectra. Protein Sci. 1998;7(4):975–982. doi: 10.1002/pro.5560070415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.