Significance
Scientists have long debated the dynamic form of perpetual reciprocal adaptations, or coevolution, between hosts and their parasites. The two main types of antagonistic coevolution described to date are arms race dynamics, in which interaction traits escalate through time, and fluctuating selection dynamics, in which traits cycle through time. We used experimental evolution between Pseudomonas aeruginosa and a panel of its lytic phages and found the full known range of coevolutionary dynamics. We argue that the coevolutionary pattern is determined by whether phages typically adsorb directly to receptors on the bacterial outer membrane or instead use retractable type IV pili as a primary or alternative site. Our results have applications in the use of phages as therapeutics or disinfectants to control bacterial pathogens.
Keywords: phage therapy, nosocomial pathogen, fluctuating selection, arms race, type IV pili
Abstract
Many antagonistic interactions between hosts and their parasites result in coevolution. Although coevolution can drive diversity and specificity within species, it is not known whether coevolutionary dynamics differ among functionally similar species. We present evidence of coevolution within simple communities of Pseudomonas aeruginosa PAO1 and a panel of bacteriophages. Pathogen identity affected coevolutionary dynamics. For five of six phages tested, time-shift assays revealed temporal peaks in bacterial resistance and phage infectivity, consistent with frequency-dependent selection (Red Queen dynamics). Two of the six phages also imposed additional directional selection, resulting in strongly increased resistance ranges over the entire length of the experiment (ca. 60 generations). Cross-resistance to these two phages was very high, independent of the coevolutionary history of the bacteria. We suggest that coevolutionary dynamics are associated with the nature of the receptor used by the phage for infection. Our results shed light on the coevolutionary process in simple communities and have practical application in the control of bacterial pathogens through the evolutionary training of phages, increasing their virulence and efficacy as therapeutics or disinfectants.
Many host–parasite associations coevolve, and patterns in this antagonistic interaction are influenced by biology and environment (1–3). In single-species host–parasite interactions, parasite genotypes show differences in their host ranges and specificities on host genotypes, providing the basis for such coevolution (4–8). Arms race dynamics (ARD) driven by directional selection favors a broader resistance range in the host against a greater number of parasite genotypes and an increased host range in the parasite allowing more host genotypes to be infected (2, 9). In contrast, fluctuating selection dynamics (FSD), in which there is no directional change in the evolution of the host resistance range, is governed by negative frequency-dependent selection, favoring hosts that resist the most frequently encountered parasite genotypes and parasites that infect the most common host genotypes (9–11). It has been suggested that ARD predominates during the initial stages of coevolution, when adaptations to the coevolving opponent are largely cost-free, whereas FDS is more significant at later stages, when attack/defense alleles accumulate in the genome and impose costs (12). Thus, when systems shift from ARD to FSD, dynamic coevolutionary equilibria may arise, with constant numbers of attack/defense alleles at the individual level (13) and the continuous frequency-dependent (re)cycling of alleles at the population level (14).
Coevolutionary dynamics between hosts and parasites is increasingly investigated using experimental evolution (15) and more specifically time-shift assays (16–18), in which hosts or parasites from a given time point are compared with their counterparts from the past or future, enabling the examination of putative reciprocal adaptations (19, 20). Some of the prime experimental models are bacteria and their lytic phages, which exhibit rapid evolution and are amenable to time-shift tests. Recent study indicates that coevolving populations may exhibit either ARD or FSD (12, 21–24), and there is some evidence for the genetic mechanism involved [e.g., mutations at tail fiber genes (12, 25)] and for the expansion of the phage host range as coevolution proceeds (6). Most studies of bacteria–phage coevolution involve the model system Pseudomonas fluorescens SBW25 and its lytic phage ϕ2 (15). Although these studies are important for an in-depth understanding of this process, their restriction to a single host–parasite pair is unfortunate, given the immense diversity of bacteria and phages in both terrestrial and aquatic ecosystems (26–28) and the importance of many of these organisms in human health (29). Thus, the generality of previous results to other bacterial species and to different phages parasitizing a given bacterium remains an open question.
Coevolution may be particularly important in an applied context, namely when predators or parasitic organisms are used for biocontrol (30). For instance, Conrad et al. (31) recently argued that a community perspective with its ecological and evolutionary underpinnings is needed to explore the usefulness of phages as antimicrobial agents to treat cystic fibrosis patients infected with several bacteria and notably strains of Pseudomonas aeruginosa, a congeneric to the model organism P. fluorescens. Different phages naturally have different host ranges (8, 32, 33), but whether phage taxonomic origin influences impacts on P. aeruginosa is not known. Despite the study of phage mixtures to control P. aeruginosa infections (34), the nature of bacterial cross-resistance to phages other than that with which the bacterium evolved has not been addressed.
Previous studies are inconclusive regarding whether P. aeruginosa coevolves with bacteriophages (35, 36). Given the ubiquity of this model organism in natural habitats (37) and as a widespread pathogen in hospitals (38, 39), it is important to know whether phage parasitism influences P. aeruginosa population biology and adaptation, whether coevolution between these antagonists actually occurs, and, if so, whether there are general patterns shared by different phages. Here, we test coevolutionary dynamics and their consistency in a panel of lytic bacteriophages and their host P. aeruginosa PAO1. We allowed this bacterium to interact and potentially coevolve with each of six different phage isolates separately, four from the Podoviridae and two from the Myoviridae (Table S1), for a total of 10 serial transfers (∼60 generations). Using bacteria and phages isolated from different time points, we conducted time-shift assays of resistance to infer patterns of the coevolutionary process. Finally, to assess specificity, we performed a cross-resistance assay with evolved bacteria and the six ancestral phage isolates to compare resistance of the bacteria to their “own” phage and to “foreign” phages with which they had not coevolved.
Results
Evidence for Coevolution.
After 10 transfers without exposure to bacteriophages, the bacteria from control replicate lines completely lost their ability to resist any of the six phages. In fact, when 120 arbitrarily selected colonies from each of these phage-free bacterial populations (20 colonies from each of six replicates against each ancestral phage) were tested for phage resistance, not a single resistant colony was found (Fig. S1). This result indicates that resistance to phages comes at a fitness cost (40–44), because no bottlenecking could have occurred, given that ca. 108 bacterial cells were introduced to fresh microcosms at each serial transfer.
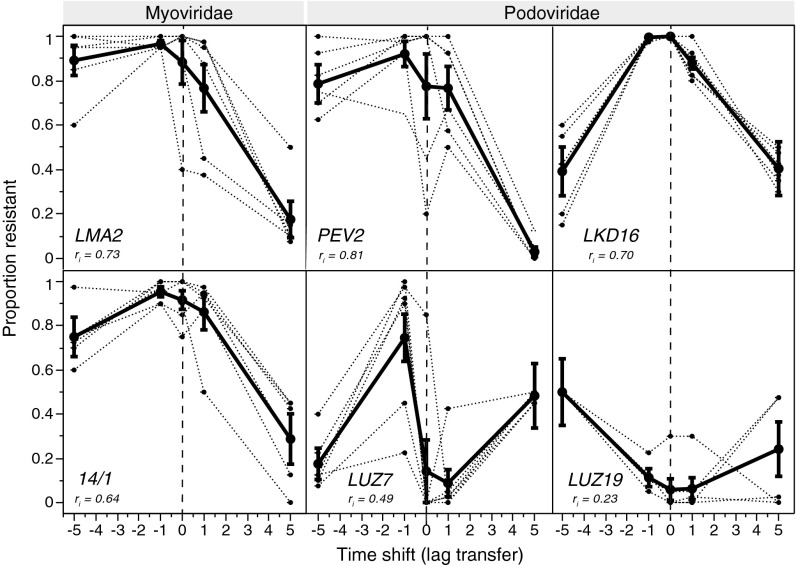
The time-shift assay revealed signatures of coevolution with all six phages. Most of the time-shift curves showed considerable nonlinearity, resulting in significant quadratic and cubic polynomial terms of the time-shift covariate (Table S2). However, the precise patterns differed among phages, as indicated by significant interactions between phage type and time-shift higher-order polynomial terms (F5, 269 > 3.2, P < 0.008, Table S2; see also Tables S3 and S4 for phage-by-phage type analyses). Namely, the curves for four of the phages describe hump-shaped patterns (Fig. 1), with more or less pronounced maximum levels of resistance when bacteria were one transfer in the future relative to the phage (lag −1). Resistance tended to decrease when bacteria were five transfers in the future (lag −5) and also when the phage was ahead in time relative to the bacteria (lag +1, lag +5). The peaks of resistance indicate a pattern of local temporal adaptation of the bacteria. In contrast, phage LUZ19 showed the reverse pattern of local temporal adaptation, having the highest infectivity when confronted with contemporary bacteria from transfer 5 (lag 0). Finally, phage LUZ7 showed a zig-zag pattern, with multiple highs and lows of resistance along the time-shift axis (Fig. 1).
Fig. 1.
Time-shift curves of resistance of P. aeruginosa PAO1 coevolving with six phage types. Each panel shows the curve for each of six replicate populations (dotted lines) and the overall mean ± SE (bold line). Time-shift lag 0 refers to contemporary combinations of bacteria and phages from transfer 5 (vertical dashed line); lag +1 and lag +5 are combinations in which phages are one or five transfers in the future relative to the bacteria; lag −1 and lag −5 are combinations in which bacteria are one or five transfers in the future relative to phages. Intraclass correlation coefficients (ri) describe the degree of similarity of trajectories among replicate populations (1 = highly correlated; 0 = uncorrelated). Phage types represent Myoviridae or Podoviridae families.
Most time-shift studies consider a relatively narrow time window and often show monotonic patterns (16, 35, 45). When we restricted our analysis to time lags of one transfer (lag +1, lag −1), we also found a general trend of increased resistance when bacteria were one transfer ahead in time (lag −1) and decreased resistance when phages were one transfer ahead in time (lag +1). Similar decreases in resistance for lag +1 combinations were seen in additional tests in which the phage was ahead in time relative to bacteria from transfers 4 and 6 (Fig. S2). Thus, over these narrow time windows, we found evidence of typical coevolutionary interactions, with selection for increased resistance in the bacteria and increased infectivity in the phage.
Following more widely used approaches (46), we also carried out statistical analyses separately for bacteria and phages. These analyses (Fig. S3 and Table S5) confirmed the main patterns detected by the combined analysis presented above. Namely, when the combined analyses detected bacterial local adaptation in time (Fig. 1), the separate analyses revealed bacterial time-shift curves with a local resistance maximum (Fig. S3); conversely, phage local adaptation in time had a concomitant local resistance minimum for the phage time-shift curve (Fig. S3).
Finally, we used intraclass correlation coefficients (ri; refs. 47, 48) to assess the synchrony of time-shift curves. All ri values were positive, indicating similarity of time-shift curves among replicate populations coevolving with the same phage (Fig. 1). The similarity was less pronounced for the LUZ19 and LUZ7 replicate populations (ri <0.5) than for the other four phages (ri >0.6).
Mean Resistance and Infectivity Ranges.
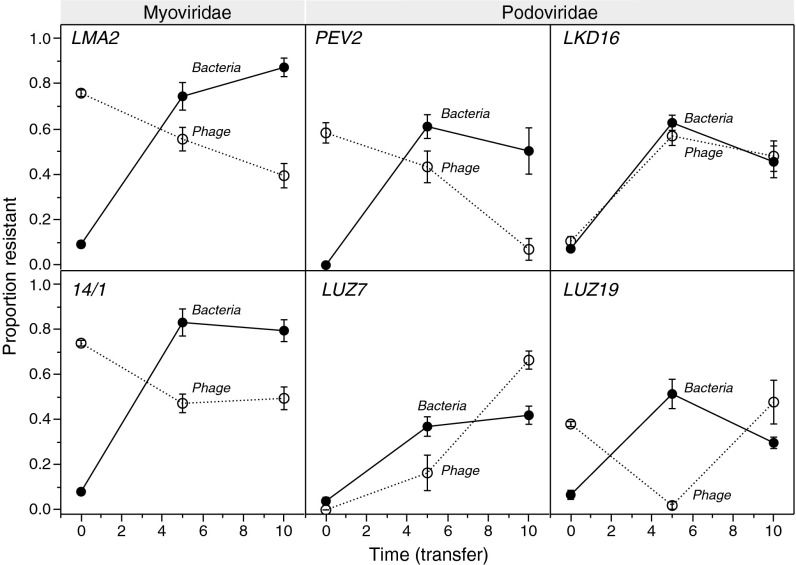
Changes in resistance/infectivity ranges can provide further information on the nature of the coevolutionary dynamics, whereby directional selection in an escalating arms race should result in a cumulative increase in the mean resistance and infectivity over evolutionary time (15). To test this notion, we measured the overall mean resistance of bacteria from three time points (transfers 0, 5, and 10) to all phages from these same time points. Overall, initially low ancestral resistance (<15%) increased during the experiment. However, although resistance to the two Myoviridae phages (14/1 and LMA2) increased to very high levels (80%), resistance to the four Podoviridae phages appeared to saturate at intermediate levels (∼50%; significant effect of phage type: F5, 30 = 6.2, P = 0.0005) (Fig. 2 and Table S6). This saturating pattern is reflected by a significant quadratic term for the time covariate (F1, 273 > 70.1, P < 0.0001) (Table S6).
Fig. 2.
Mean resistance of P. aeruginosa PAO1 and mean infectivity of six phage types at three time points during the experiment. Means (± SE) were calculated from a cross-infection matrix using all combinations between bacteria and phages from transfers 0, 5, and 10. Phage types represent Myoviridae or Podoviridae families.
Changes in mean phage infectivity varied significantly among phage types (significant time × phage type interactions: F5, 273 > 6.0, P < 0.0001; Table S7). In four cases, infectivity increased between transfers 0 and 5, mirroring the increases in bacterial resistance. In contrast, two initially successful phages (LUZ7 and LKD16) lost much of their general infection capacity at the end of the experiment (Fig. 2).
Specificity and Cross-Resistance.
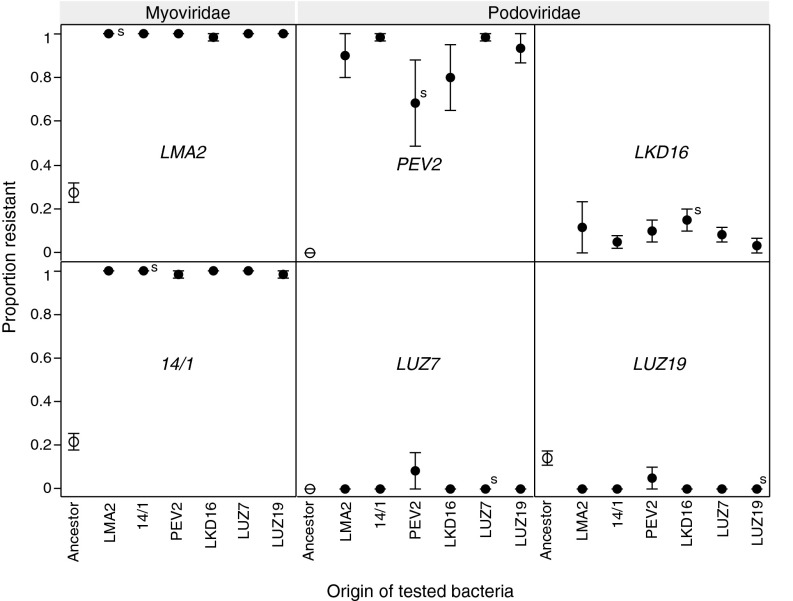
To assess patterns of cross-resistance, we measured resistance of evolved bacteria from transfer 10 against all six ancestral phage isolates. Even though the bacteria origin × phage interaction was significant (F25, 60 = 2.5, P = 0.0017; Table S8), we found no evidence for specialization (Fig. 3) that would make bacteria more resistant to the ancestors of the phages with which they had coevolved. There was no significant overall difference between own vs. foreign combinations of bacteria and phages [F1, 5 < 1], nor were any of the own–foreign comparisons statistically significant when analyzed separately for each bacterial origin (all P > 0.06).
Fig. 3.
Mean (± SE) resistance of evolved P. aeruginosa PAO1 to six ancestral phage types. “Origin of tested bacteria” indicates the phage with which the bacteria had coevolved for 10 transfers. The letter “s” (= sympatric) indicates combinations of coevolved bacteria and phage. “Ancestor” indicates the resistance of the ancestral bacterial strain (P. aeruginosa PAO1). Phage types represent Myoviridae or Podoviridae families.
Observed levels of resistance depended mainly on the identity of the ancestral phage isolate against which the bacteria were tested (F5, 25 = 200, P < 0.0001; Table S8). In fact, bacteria collectively either showed reduced resistance (<20%) or gained very high levels of resistance, in particular to the two Podoviridae phages (∼100%; Fig. 3).
Discussion
In theory, coevolutionary dynamics in communities can be complex (49) and influenced by numerous factors, including community structure and diversity and habitat productivity (50). Despite an increased understanding of single-species coevolutionary interactions, little is known about variation within interaction networks of functionally similar species (8, 32). Here we investigated the coevolutionary dynamics between a focal host and individual bacteriophage isolates.
The combined analyses of time shift and of mean resistance and infectivity for a panel of six phages indicate antagonistic coevolution with the bacterium P. aeruginosa PAO1. The pace of coevolutionary change was substantial, occurring on time scales of one or a few serial transfers (ca. six bacterial generations per transfer). In contrast to other systems (3), variation persisted throughout the experiment, with neither resistance nor infectivity reaching either 0% or 100% in the majority of replicates. Additionally, we present the first example, to our knowledge, of a host species showing varying degrees of ARD and FSD coevolution depending on the parasite with which it interacts. The generally high consistency of these results across replicate populations (see ri values in Fig. 1) suggests a certain level of repeatability and determinism in the coevolutionary processes underlying the observed patterns. That independent replicate populations can evolve the same molecular innovations has been shown recently for bacteriophages evolving on Escherichia coli (51).
Coevolutionary Arms Race or Fluctuating Selection?
ARD and FSD (also known as “Red Queen dynamics”) are the two main concepts used to describe host–parasite coevolution (2, 15). Under ARD, directional selection predominates, and successive selective sweeps lead to the accumulation of infectivity and resistance genes so that parasites are capable of infecting all past host genotypes, and hosts are able to resist all past parasites (6, 52). In contrast, FSD is characterized by temporal fluctuations in infectivity and resistance genes rather than their accumulation (9), resulting on average in unchanged infectivity and resistance ranges. Our analyses revealed signatures of ARD-like dynamics, as indicated by monotonic increases in infectivity of the phages or large resistance ranges of the bacteria. These patterns were most evident for bacteria coevolving with the phages LMA2 and 14/1 and, to a lesser degree, PEV2 (Fig. 2). These observations are consistent with previous work reporting ARD-like coevolution in the well-studied interaction between P. fluorescens SBW25 and its phage ϕ2 (12, 15).
Our main result, however, was the nonmonotonic coevolutionary dynamics with five of the six phages tested. Nonmonotonic patterns, with local maxima or minima of resistance in time, indicate deviations from the pure directional antagonistic selection typical of ARD (10). Only coevolution with LMA2 did not differ statistically from pure ARD. Local temporal minima or maxima of resistance in the five other phages (Fig. 1) are more consistent with an FSD coevolutionary process, based on frequency-dependent selection (20). Thus, bacteria resisted phages from the present or very recent past particularly well, but were less resistant against phages from the more distant past or distant future. Such patterns are predicted to arise under frequency-dependent selection, with the parasite or host adapting to the most common genotype of the antagonist (10, 19). Our data further support an important prediction from the model of Gandon et al. (10), showing that ARD and FSD can give identical patterns for narrow time-shift windows. These identical patterns are seen because adaptation to the most common genotype represents a temporarily restricted selective sweep that is indistinguishable from a selective sweep under ARD. In other words, time-shift assays may need to be conducted sufficiently far into the past or future to detect FSD reliably. Patterns similar to ours have been detected in time-shift experiments on bacterial parasites in natural populations of Daphnia (17, 53).
Results from the P. fluorescens–ϕ2 system suggest that ARD may predominate in the coevolutionary process for more than 100 bacterial generations and give way to FSD only subsequently, when the accumulation of resistance or infectivity genes becomes costly (12). In contrast, we detected a strong signature of FSD early in coevolution, during the first 60 generations. A possible explanation is that ARD predominates in coevolutionarily naive systems and that several of the ancestral phages we used already had adaptive signatures to the PAO1 ancestor. This conjecture is consistent with the observation of the virtually 100% susceptibility of the P. fluorescens SBW25 ancestor to the ϕ2 ancestor (figure 1 in ref. 16) and our finding that the ancestral bacteria had significant levels of resistance (14–27%; Fig. S1) against the ancestors of four of our six phages. Another possible explanation is that the genetic architecture could differ in different phage–bacteria associations (see also below). For example, the FSD systems may involve reciprocal selection of matching-allele type of infectivity and resistance, whereas ARD dynamics have an underlying gene-for-gene type of interaction (9). Evidence from the P. fluorescens–ϕ2 system indicates that genotypic change is consistent with ARD or FSD, depending on the observation period in the coevolutionary process (12). For example, declining gene di-versity in experimentally coevolving populations of Caenorhabditis elegans and Bacillus thuringiensis may have resulted from ARD dynamics or from FSD dynamics with large amplitudes (54).
Mechanistic Basis of Coevolution.
Selection for resistance and infectivity may act at several stages in the infection cycle (3, 55). The first barrier to infection is receptors on the bacterial outer cell membrane, to which lytic phages adsorb and inject nucleic acids into the cell. Many phages attach to LPS receptors, and also to type IV bacterial pili, which must retract to bring the phages into contact with membrane receptors (56, 57). We hypothesize that the relative use of LPS and pili as primary receptors contributed to the range of coevolutionary dynamics observed for our phage isolates. First, under our experimental conditions (agitated liquid culture and rapid bacterial growth), bacteria likely produce few pili (58) and/or retract them infrequently (59). This sparsity of pili can reduce adsorption by pili-dependent phages and thereby lessen selection for resistance relative to phages using LPS as primary receptors. Second, resistance to phages mediated by loss or nonretraction of pili can have substantial fitness costs, because it impairs bacterial swimming ability (41, 60). Such costly adaptations therefore may be impeded over evolutionary time, thus facilitating the action of frequency-dependent selection (9). These pilus-specific effects may explain why our four Podoviridae phages, all putatively using pili to some extent in infection (Table S1), produced FSD-like coevolutionary patterns and did not cause the same ARD-like increases in resistance ranges seen for the two Myoviridae phages, both of which are LPS-receptor specialists.
The view that resistance to pili specialists can be costly is supported by a study on P. aeruginosa PAO1 and a pilus-dependent phage (PP7), in which a reduction in the phage-encounter rate coincided with a drop in bacterial resistance and a change in population composition from predominantly resistant and less motile variants to susceptible swimmer variants (41). Moreover, this notion is consistent with our observed breakdown of resistance to the pilus-specialist LUZ19 at transfer 5, accompanied by a local temporal infectivity maximum of the phage. Interestingly however, we show that such loss of resistance is reversible and that populations can reestablish resistance within a small number of generations (observed at transfer 10). As illustrated by phage LUZ7, these FSD-like dynamics even may lead to zig-zag patterns, with alternating temporal maxima of bacterial resistance and phage infectivity.
Further experimentation is required to test experimentally and statistically our hypothesis that the site of phage adsorption affects coevolutionary dynamics. Clearly, selection also may involve changes in genes such as CRISPR-Cas that mediate within-cell interactions between bacteria and phages (61). Furthermore, it may be important to consider the influence of evolutionary history. For example, the two LUZ phages were sampled from the same hospital environment (62), and thus adaptation to a common bacterial genetic background or environment may explain why these two phages produced time-shift curves so consistently different from those of the other phages. We nevertheless demonstrate that different coevolutionary dynamics may occur between phage types and that patterns are consistent within phage types.
Applications.
Our results are relevant to the use of phages in combating nosocomial infections or for disinfecting surfaces in hospitals. Provided a sufficient time window between applications, our finding that coevolution may follow FSD suggests that to some extent bacteria will lose recently gained resistance to such phages. Thus the ability of three of the ancestral phages to overcome the resistance of any of the final coevolved bacteria could have useful applications in the control of P. aeruginosa PAO1, and the same phenomenon may apply to other bacterial pathogens. On the other hand, our data suggest that cross-resistance may be a significant problem in the practical application of certain phages such as 14/1, LMA2, and PEV2, the first of which has been selected for use in phage mixtures (34, 63).
Recent study has shown that, as a strategy to control P. aeruginosa PAO1, the evolutionary “training” of phages through passaging can increase phage efficacy in reducing ancestral bacterial populations (36). Our experiment represents a case of coevolutionary passaging and shows variable outcomes in infectivity and resistance depending on phage identity (open circles, Fig. S3), with certain phages (LMA2, 14/1, PEV2, LKD16; Fig. S3) becoming more infective against bacteria from previous time points, and others (LUZ7, LUZ19; Fig. S3) losing such efficacy. Moreover, certain trained phages (LKD16, LUZ7; Fig. S3) were found to be less infectious to coevolved bacteria than the same phages from earlier passages. These findings suggest that coevolution as a phage-specific process introduces complexity that should be considered in the use of phages as therapeutics and disinfectants.
Our experiments were conducted in vitro and with a nonmucoid strain of P. aeruginosa, and thus we urge caution in interpreting our findings for in situ applications. Moreover, because different bacteriophages can exhibit contrasting coevolutionary dynamics with P. aeruginosa PAO1, it is possible that other P. aeruginosa strains also demonstrate different dynamics with their phages or do not coevolve at all. Whether the patterns observed here generalize to other clinical isolates of P. aeruginosa and other bacterial pathogens requires further investigation.
Materials and Methods
Isolates and Bacteriophage Characteristics.
We used an isogenic sample of P. aeruginosa PAO1 (referred to as the “ancestor”) and stored stock solutions of ca. 107 cfu/mL at −80 °C. Of the six bacteriophage strains used (Table S1), four (LKD16, PEV2, LUZ19, and LUZ7) were from the Podoviridae, and two (14/1 and LMA2) were from the Myoviridae (64–66). LUZ7 and PEV2 have 70-nm icosahedral capsids and short tails, typical of the Podoviridae. PEV2 is capable of infecting PA01 both aerobically and anaerobically, an important consideration for clinical applications in which infections can be anaerobic in thick mucus (65). LUZ19 and LKD16 are slightly smaller, with head diameters of 65 and 60 nm, respectively. LKD16 shows a weak, reversible attachment to P. aeruginosa PA01, but both LUZ19 and LKD16 have a broad infectivity range on clinical and environmental isolates of P. aeruginosa (62, 67). 14/1 and LMA2 have long, inflexible tails of ∼140 nm and isometric capsids with a 74-nm diameter (62, 66). All six ancestral phage strains used were stored as stocks of ca. 108 pfu/mL at 4 °C.
Experimental Conditions.
All experiments were conducted in 6-mL microcosms of King’s B (KB) culture medium (68) in 30-mL Falcon tubes (BD Biosystems), incubated at 37 °C and 200 rpm orbital agitation (Bülher compact shaker, model KS-15 Control). For each of the six ancestral phage types, six replicate lines were initiated in separate microcosms. Each microcosm was inoculated with 5 μL from the ancestral phage stock (ca. 5 × 105 phage particles) together with 20 μL of P. aeruginosa (ca. 2 × 105 cells) from an overnight (24 h of growth) population initiated from a single ancestral P. aeruginosa PAO1 colony. For each of the 36 microcosms, a 100-μL sample containing both bacteria and phages was transferred to a new microcosm with fresh KB medium every 24 h. We carried out a total of 10 such transfers. At each transfer a 500-μL sample was taken and stored in 50% glycerol at −80 °C for subsequent analysis.
Measuring Bacteriophage Infection Capacity and Bacterial Resistance.
Phage infection capacity was measured by streaking, from a given replicate, a sample of 20 arbitrarily chosen bacterial colonies through a line formed by 30 μL of a purified bacteriophage population (16). After incubation for 24 h at 37 °C, a bacterial colony was considered resistant if its growth continued uninterrupted by the line of phage.
The bacterial colonies required for the test were obtained after plating diluted samples on KB agar (12 g⋅L−1 agar) and growth for 24 h at 37 °C. Bacteriophages were purified from samples containing both bacteria and phages through the addition of 100 μL chloroform to a 900-μL sample, vortexing for several seconds, centrifuging at 16,060 × g for 4 min, and then removing the supernatant of purified phages.
Time-Shift Assays: Detecting Coevolution Between Bacteria and Phages.
Based on the streaking test, a time-shift assay approach (15, 19, 20) was used to determine if bacteria and phages evolved adaptations and counter-adaptations antagonistically. Using bacteria and phages isolated from a given microcosm at different time points during the experiment, bacterial resistance is measured against phages isolated from past, present, and future transfers. Conversely, phage infectivity is tested on bacteria from past, present, and future transfers. A monotonous increase of resistance (infectivity) from the past to the present to the future is considered a signature of an escalating coevolutionary arms race. Nonlinear trajectories, in particular local maxima or minima of resistance and infectivity, indicate fluctuating coevolutionary dynamics, which can be driven by FSD (e.g., Red Queen dynamics).
Following a standard time-shift scheme, we tested bacteria from transfer 5 against their contemporary phages (transfer 5), phages from the past (transfers 0 and 4), and phages from the future (transfers 6 and 10). In the same way, phages from transfer 5 were tested against contemporary, past, and future bacteria (Fig. S1). This process yielded contemporary bacteria–phage combinations (lag 0 transfers), combinations in which the phage was from the future relative to the bacteria (lag +1 and lag +5 transfers), and combinations in which the bacteria were from the future relative to the phage (lag −1 and lag −5 transfers). Under arms race coevolution, resistance should decrease the more the phage is ahead in time relative to the bacteria (lag +1 and lag +5) and should increase the more the bacteria are ahead in time relative to the phage (lag −1 and lag −5). These time-shift assays (nine lags each) were performed for all 36 replicate populations, giving a total of 324 streaking tests. In additional time-shift assays of phage evolution, we tested bacteria from transfer 4 against phage from transfers 3, 4, and 5 and bacteria from transfer 6 against phage from transfers 5, 6, and 7 (Fig. S2).
In addition to the time-shift assay, we assessed the changes in overall resistance and infectivity ranges over the course of the 10 transfers. To this end, we performed additional streaking tests so that all bacteria from transfers 0, 5, and 10 were tested against all phages from these three transfers. This complete infection matrix consisted of 324 streakings. Monotonic increases of resistance and infectivity with transfer time would be consistent with arms race coevolution.
Cross-Resistance.
After (co)evolution with a given phage type, bacteria from the final transfer 10 were evaluated for resistance to the ancestor of the phage type with which it had coevolved (“own” phage type), and with the ancestors of the other phage types (“foreign” phage types). To this end, three bacterial replicate populations that had evolved with each phage type (three lines × six test phage types = 18 lines) were chosen arbitrarily. Each line then was confronted with the six ancestral phages, giving a total of 108 streaking tests.
Statistical Analyses.
We used General Linear Models [logistic regression with binomial error structure; JMP 10 (69)] to analyze variation in the proportion of resistance (number of resistant colonies out of 20 streakings). We fitted time shift (lag) as a continuous variable, with linear and nonlinear (quadratic, cubic) polynomial terms. Similarly, analyses of mean resistance/infectivity ranges included time (transfer number) as a continuous factor. Models further had phage type and replicate population (nested within phage type) as factors. Analysis of cross-infection data included evolved bacterial origin (as defined by the phage type with which they had evolved), bacterial replicate population, and ancestral phage type as factors.
Supplementary Material
Acknowledgments
We thank J.-P. Ceyssens for providing the phage isolates; M. A. Brockhurst for providing the ancestral strain of P. aeruginosa PAO1; and Clara Torres-Barcelo, Marie Vasse, Johan Ramsayer, Alison Duncan, Lori Burrows, Nuno Oliviera, and Sylvain Gandon for discussions throughout this work. This work was supported by a grant from Erasmus Mundus Masters Programme (to A.B.), by James S. McDonnell Foundation Studying Complex Systems Research Award 220020294 (to M.E.H.), and by the Agence National de la Recherche ‘EvolStress’ ANR-09-BLAN-099-01 and ‘EVORANGE’ ANR-09-PEXT-011 (to M.E.H. and O.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406763111/-/DCSupplemental.
References
- 1.Thompson J. Geographic Mosaic of Coevolution. Chicago, IL: The University of Chicago Press; 2005. [Google Scholar]
- 2.Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32(4):569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- 3.Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014 doi: 10.1111/1574-6976.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carius HJ, Little TJ, Ebert D. Genetic variation in a host-parasite association: Potential for coevolution and frequency-dependent selection. Evolution. 2001;55(6):1136–1145. doi: 10.1111/j.0014-3820.2001.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 5.Thrall PH, Burdon JJ. Evolution of virulence in a plant host-pathogen metapopulation. Science. 2003;299(5613):1735–1737. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- 6.Poullain V, Gandon S, Brockhurst MA, Buckling A, Hochberg ME. The evolution of specificity in evolving and coevolving antagonistic interactions between a bacteria and its phage. Evolution. 2008;62(1):1–11. doi: 10.1111/j.1558-5646.2007.00260.x. [DOI] [PubMed] [Google Scholar]
- 7.Scanlan PD, Hall AR, Lopez-Pascua LDC, Buckling A. Genetic basis of infectivity evolution in a bacteriophage. Mol Ecol. 2011;20(5):981–989. doi: 10.1111/j.1365-294X.2010.04903.x. [DOI] [PubMed] [Google Scholar]
- 8.Poisot T, Lepennetier G, Martinez E, Ramsayer J, Hochberg ME. Resource availability affects the structure of a natural bacteria-bacteriophage community. Biol Lett. 2011;7(2):201–204. doi: 10.1098/rsbl.2010.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agrawal A, Lively C. Infection genetics: Gene-for-gene versus matching-alleles models and all points in between. Evol Ecol Res. 2002;4(1):79–90. [Google Scholar]
- 10.Gandon S, Buckling A, Decaestecker E, Day T. Host-parasite coevolution and patterns of adaptation across time and space. J Evol Biol. 2008;21(6):1861–1866. doi: 10.1111/j.1420-9101.2008.01598.x. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki A. Host-parasite coevolution in a multilocus gene-for-gene system. Proc Biol Sci. 2000;267(1458):2183–2188. doi: 10.1098/rspb.2000.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14(7):635–642. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 13.Frank SA. Coevolutionary genetics of plants and pathogens. Evol Ecol. 1993;7(1):45–75. [Google Scholar]
- 14.Holub EB. The arms race is ancient history in Arabidopsis, the wildflower. Nat Rev Genet. 2001;2(7):516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- 15.Brockhurst MA, Koskella B. Experimental coevolution of species interactions. Trends Ecol Evol. 2013;28(6):367–375. doi: 10.1016/j.tree.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Buckling A, Rainey PB. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci. 2002;269(1494):931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decaestecker E, et al. Host-parasite ‘Red Queen’ dynamics archived in pond sediment. Nature. 2007;450(7171):870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 18.Thrall PH, et al. Rapid genetic change underpins antagonistic coevolution in a natural host-pathogen metapopulation. Ecol Lett. 2012;15(5):425–435. doi: 10.1111/j.1461-0248.2012.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaba S, Ebert D. Time-shift experiments as a tool to study antagonistic coevolution. Trends Ecol Evol. 2009;24(4):226–232. doi: 10.1016/j.tree.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Blanquart F, Gandon S. Time-shift experiments and patterns of adaptation across time and space. Ecol Lett. 2013;16(1):31–38. doi: 10.1111/ele.12007. [DOI] [PubMed] [Google Scholar]
- 21.Koskella B, Lively CM. Evidence for negative frequency-dependent selection during experimental coevolution of a freshwater snail and a sterilizing trematode. Evolution. 2009;63(9):2213–2221. doi: 10.1111/j.1558-5646.2009.00711.x. [DOI] [PubMed] [Google Scholar]
- 22.King KC, Delph LF, Jokela J, Lively CM. The geographic mosaic of sex and the Red Queen. Curr Biol. 2009;19(17):1438–1441. doi: 10.1016/j.cub.2009.06.062. [DOI] [PubMed] [Google Scholar]
- 23.Gómez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332(6025):106–109. doi: 10.1126/science.1198767. [DOI] [PubMed] [Google Scholar]
- 24.Koskella B. Phage-mediated selection on microbiota of a long-lived host. Curr Biol. 2013;23(13):1256–1260. doi: 10.1016/j.cub.2013.05.038. [DOI] [PubMed] [Google Scholar]
- 25.Paterson S, et al. Antagonistic coevolution accelerates molecular evolution. Nature. 2010;464(7286):275–278. doi: 10.1038/nature08798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser C, Alm EJ, Polz MF, Spratt BG, Hanage WP. The bacterial species challenge: Making sense of genetic and ecological diversity. Science. 2009;323(5915):741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Brito B, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4(6):739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- 28.Martiny JBH, Riemann L, Marston MF, Middelboe M. Antagonistic coevolution of marine planktonic viruses and their hosts. Annu Rev Mar Sci. 2014;6:393–414. doi: 10.1146/annurev-marine-010213-135108. [DOI] [PubMed] [Google Scholar]
- 29.Kutter E, et al. Phage therapy in clinical practice: Treatment of human infections. Curr Pharm Biotechnol. 2010;11(1):69–86. doi: 10.2174/138920110790725401. [DOI] [PubMed] [Google Scholar]
- 30.Holt RD, Hochberg ME. When is biological control evolutionarily stable (or is it)? Ecology. 1997;78(6):1673–1683. [Google Scholar]
- 31.Conrad D, et al. Cystic fibrosis therapy: A community ecology perspective. Am J Respir Cell Mol Biol. 2013;48(2):150–156. doi: 10.1165/rcmb.2012-0059PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weitz JS, et al. Phage-bacteria infection networks. Trends Microbiol. 2013;21(2):82–91. doi: 10.1016/j.tim.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Wichels A, et al. Bacteriophage diversity in the north sea. Appl Environ Microbiol. 1998;64(11):4128–4133. doi: 10.1128/aem.64.11.4128-4133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall AR, De Vos D, Friman V-P, Pirnay J-P, Buckling A. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Environ Microbiol. 2012;78(16):5646–5652. doi: 10.1128/AEM.00757-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brockhurst MA, Buckling A, Rainey PB. Spatial heterogeneity and the stability of host-parasite coexistence. J Evol Biol. 2006;19(2):374–379. doi: 10.1111/j.1420-9101.2005.01026.x. [DOI] [PubMed] [Google Scholar]
- 36.Betts A, Vasse M, Kaltz O, Hochberg ME. Back to the future: Evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evol Appl. 2013;6(7):1054–1063. doi: 10.1111/eva.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos JL, Filloux A. Pseudomonas. 3rd Ed. Vol 1. New York: Kluwer; 2007. [Google Scholar]
- 38.Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67(3):351–368. doi: 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 39.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis. 2010;51(Suppl 1):S81–S87. doi: 10.1086/653053. [DOI] [PubMed] [Google Scholar]
- 40.Bohannan B, Travisano M, Lenski RE. Epistatic interactions can lower the cost of resistance to multiple consumers. Evolution. 1999;53(1):292–295. doi: 10.1111/j.1558-5646.1999.tb05355.x. [DOI] [PubMed] [Google Scholar]
- 41.Brockhurst MA, Buckling A, Rainey PB. The effect of a bacteriophage on diversification of the opportunistic bacterial pathogen, Pseudomonas aeruginosa. Proc Biol Sci. 2005;272(1570):1385–1391. doi: 10.1098/rspb.2005.3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buckling A, Wei Y, Massey RC, Brockhurst MA, Hochberg ME. Antagonistic coevolution with parasites increases the cost of host deleterious mutations. Proc Biol Sci. 2006;273(1582):45–49. doi: 10.1098/rspb.2005.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koskella B, Thompson JN, Preston GM, Buckling A. Local biotic environment shapes the spatial scale of bacteriophage adaptation to bacteria. Am Nat. 2011;177(4):440–451. doi: 10.1086/658991. [DOI] [PubMed] [Google Scholar]
- 44.Koskella B, Lin DM, Buckling A, Thompson JN. The costs of evolving resistance in heterogeneous parasite environments. Proc Biol Sci. 2012;279(1735):1896–1903. doi: 10.1098/rspb.2011.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brockhurst MA, Morgan AD, Fenton A, Buckling A. Experimental coevolution with bacteria and phage. The Pseudomonas fluorescens—Phi2 model system. Infect Genet Evol. 2007;7(4):547–552. doi: 10.1016/j.meegid.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Friman V-P, Buckling A. Effects of predation on real-time host-parasite coevolutionary dynamics. Ecol Lett. 2013;16(1):39–46. doi: 10.1111/ele.12010. [DOI] [PubMed] [Google Scholar]
- 47.Duncan AB, Gonzalez A, Kaltz O. Stochastic environmental fluctuations drive epidemiology in experimental host-parasite metapopulations. Proc Biol Sci. 2013;280(1769):20131747. doi: 10.1098/rspb.2013.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zar JH. 1984. Biostatistical Analysis (Prentice-Hall, Englewood Cliffs, NJ), 2nd Ed.
- 49.Thompson JN. The coevolving web of life. Am Nat. 2009;173(2):125–140. doi: 10.1086/595752. [DOI] [PubMed] [Google Scholar]
- 50.Thrall PH, Hochberg ME, Burdon JJ, Bever JD. Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol Evol. 2007;22(3):120–126. doi: 10.1016/j.tree.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 51.Meyer JR, et al. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335(6067):428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salathe M, Scherer A, Bonhoeffer S. Neutral drift and polymorphism in gene-for-gene systems. Ecol Lett. 2005;8(9):925–932. doi: 10.1111/j.1461-0248.2005.00794.x. [DOI] [PubMed] [Google Scholar]
- 53.Decaestecker E, De Gersem H, Michalakis Y, Raeymaekers JA. Damped long-term host-parasite Red Queen coevolutionary dynamics: A reflection of dilution effects? Ecol Lett. 2013;16(12):1455–1462. doi: 10.1111/ele.12186. [DOI] [PubMed] [Google Scholar]
- 54.Schulte RD, Makus C, Schulenburg H. Host-parasite coevolution favours parasite genetic diversity and horizontal gene transfer. J Evol Biol. 2013;26(8):1836–1840. doi: 10.1111/jeb.12174. [DOI] [PubMed] [Google Scholar]
- 55.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 56.Bradley DE. Shortening of Pseudomonas aeruginosa pili after RNA-phage adsorption. J Gen Microbiol. 1972;72(2):303–319. doi: 10.1099/00221287-72-2-303. [DOI] [PubMed] [Google Scholar]
- 57.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56(1):289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 58.Weppelman RM, Brinton CC., Jr The infection of Pseudomonas aeruginosa by RNA pilus phage PP7: The adsorption organelle and the relationship between phage sensitivity and the division cycle. Virology. 1971;44(1):1–17. doi: 10.1016/0042-6822(71)90147-4. [DOI] [PubMed] [Google Scholar]
- 59.Conrad JC, et al. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys J. 2011;100(7):1608–1616. doi: 10.1016/j.bpj.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30(2):295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 61.Vale PF, Little TJ. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc Biol Sci. 2010;277(1691):2097–2103. doi: 10.1098/rspb.2010.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ceyssens P-J, et al. Survey of Pseudomonas aeruginosa and its phages: De novo peptide sequencing as a novel tool to assess the diversity of worldwide collected viruses. Environ Microbiol. 2009;11(5):1303–1313. doi: 10.1111/j.1462-2920.2008.01862.x. [DOI] [PubMed] [Google Scholar]
- 63.Merabishvili M, et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE. 2009;4(3):e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lammens E, et al. Representational Difference Analysis (RDA) of bacteriophage genomes. J Microbiol Methods. 2009;77(2):207–213. doi: 10.1016/j.mimet.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 65.Ceyssens P-J, et al. Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses”. Virology. 2010;405(1):26–30. doi: 10.1016/j.virol.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ceyssens P-J, et al. Comparative analysis of the widespread and conserved PB1-like viruses infecting Pseudomonas aeruginosa. Environ Microbiol. 2009;11(11):2874–2883. doi: 10.1111/j.1462-2920.2009.02030.x. [DOI] [PubMed] [Google Scholar]
- 67.Ceyssens P-J, et al. Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: Establishment of the phiKMV subgroup within the T7 supergroup. J Bacteriol. 2006;188(19):6924–6931. doi: 10.1128/JB.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44(2):301–307. [PubMed] [Google Scholar]
- 69.SAS Institute Inc 2012. JMP (SAS Institute, Cary, NC), Version 10.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.