Significance
Heart failure (HF) has a high morbidity and mortality and its incidence is increasing worldwide. While protein biomarkers have been established for diagnostic and prognostic evaluation of patients with HF, there is currently no systematic assessment of RNA biomarkers. We determined the composition of myocardial tissue and circulating microRNAs (miRNAs) in a large cohort of patients with stable and advanced HF and compared it to the composition of normal adult and fetal samples. The advanced HF patients underwent mechanical unloading with left ventricular assist devices and samples were collected at different postoperative time points. Our findings provide the underpinning for miRNA-based therapies and emphasize the usefulness of circulating miRNAs as biomarkers for heart injury performing similar to established diagnostic protein biomarkers.
Keywords: exRNA, body fluids, miRNA-mRNA regulation, development, cardiovascular disease
Abstract
Heart failure (HF) is associated with high morbidity and mortality and its incidence is increasing worldwide. MicroRNAs (miRNAs) are potential markers and targets for diagnostic and therapeutic applications, respectively. We determined myocardial and circulating miRNA abundance and its changes in patients with stable and end-stage HF before and at different time points after mechanical unloading by a left ventricular assist device (LVAD) by small RNA sequencing. miRNA changes in failing heart tissues partially resembled that of fetal myocardium. Consistent with prototypical miRNA–target-mRNA interactions, target mRNA levels were negatively correlated with changes in abundance for highly expressed miRNAs in HF and fetal hearts. The circulating small RNA profile was dominated by miRNAs, and fragments of tRNAs and small cytoplasmic RNAs. Heart- and muscle-specific circulating miRNAs (myomirs) increased up to 140-fold in advanced HF, which coincided with a similar increase in cardiac troponin I (cTnI) protein, the established marker for heart injury. These extracellular changes nearly completely reversed 3 mo following initiation of LVAD support. In stable HF, circulating miRNAs showed less than fivefold differences compared with normal, and myomir and cTnI levels were only captured near the detection limit. These findings provide the underpinning for miRNA-based therapies and emphasize the usefulness of circulating miRNAs as biomarkers for heart injury performing similar to established diagnostic protein biomarkers.
The clinical syndrome of heart failure (HF) is the result of heterogeneous myocardial or vascular diseases, and is defined by the insufficiency to maintain blood circulation throughout the body. Despite significant advances in the clinical management of HF, conventional therapies are ultimately ineffective in many patients who progress to advanced HF. In these cases, implantation of left ventricular assist devices (LVADs) and/or heart transplantation remain the only viable options.
MicroRNAs (miRNAs) are promising targets for drug and biomarker development (1). They represent an abundant class of regulatory 20- to 23-nt RNAs (2), and each miRNA is able to down-regulate hundreds of target mRNAs comprising partially complementary sequences to the miRNAs. Binding of miRNA-containing ribonucleoproteins (miRNPs) to mRNAs leads to recruitment of the CCR4–NOT deadenylase complex and subsequent mRNA destabilization as well as inhibition of translation (3).
Target recognition requires base pairing of the miRNA 5′-end nucleotides (seed sequence) to complementary target mRNA regions located typically within the 3′UTR (2). The recent detection of miRNPs in body fluids (4) pointed toward their value as biomarkers for tissue injury (5, 6), and they have also been discussed as paracrine and endocrine regulators of gene expression (4, 7).
The function of miRNAs has been widely studied in animal models of HF. The muscle-specific miR-1/206 and miR-133a/b and the heart-specific miR-208a/b and miR-499 (also referred to as myomirs) (Fig. S1) were shown to contribute to muscle or myocardial function (8, 9). miRNAs and other classes of RNA have been profiled in failing human myocardium (10–17), and a selected subset was also investigated as circulating biomarkers in HF (18–24). However, in these studies heart tissue and circulating miRNA abundance changes were not acquired simultaneously to correlate changes in tissue versus circulating miRNA composition.
Here, we report myocardial and circulating sRNA composition changes in human HF demonstrating that relative miRNA abundance changes in myocardial tissue could not be detected within the pool of circulating miRNAs. However, we show that myocardial miRNAs still serve as excellent biomarkers of heart muscle injury similar to an established protein biomarker. In heart tissues, we observed that changes in abundant miRNAs coincided with seed-dependent mRNA target responses indicative of active miRNA regulation during development and disease. This may impact development of miRNA-based therapeutic intervention.
Results
miRNA Profiles in the Left Ventricular Myocardium.
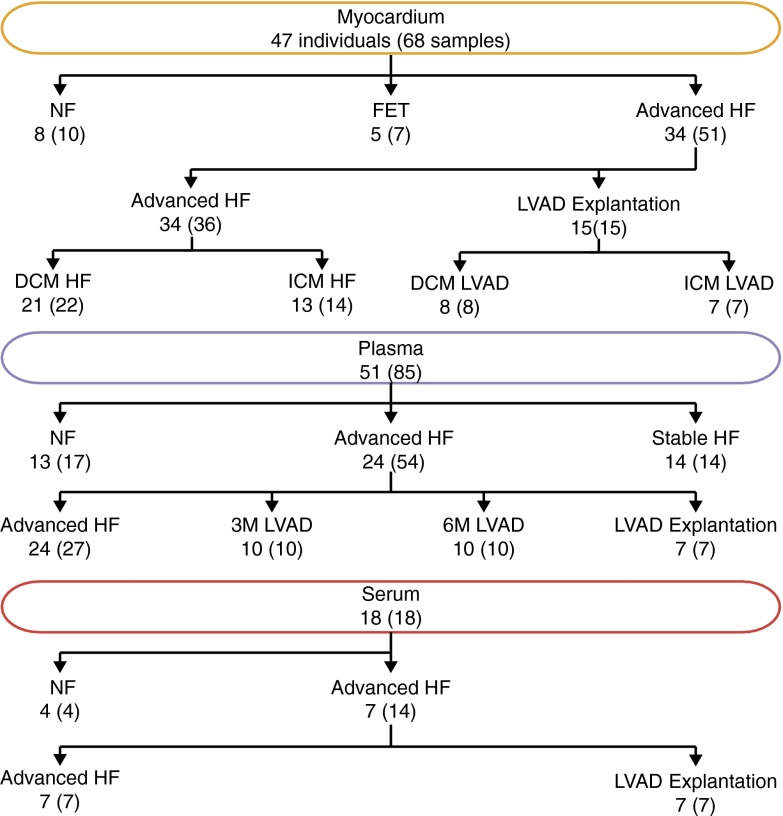
RNA was isolated from left ventricular tissue samples from a total of 47 subjects: 21 patients with advanced HF due to dilated cardiomyopathy (DCM), 13 patients with advanced HF due to ischemic cardiomyopathy (ICM), 8 individuals without heart disease (NFs), and 5 fetuses (FETs) (Fig. 1). The advanced HF samples were collected at the time of LVAD implantation (DCM/ICM HF) and LVAD explantation (DCM/ICM LVAD) during heart transplantation (Table S1, and Dataset S1). The median total RNA yield was 0.5 µg per milligram of myocardium [interquartile range (IQR) = 0.2; Fig. S2A]. We obtained a median of 4.6 million miRNA reads per cardiac tissue sample (0.4–10.6 million), representing 67–93% of the total reads. In selected noncardiac samples included for comparison, the median was 1.7 million miRNA reads (0.6–2.9 million), representing 37–99% of total reads (Dataset S1). The myocardial miRNA content was 20 fmol per microgram of total RNA (IQR = 9 fmol; Fig. S2B) and was calculated from the read ratios of all miRNA reads to spiked-in calibrator reads. The miRNA content was not significantly different between groups (Fig. S2B) and comparable with other tissues (25). We reviewed the sequence alignments of unannotated reads to the genome and found no evidence of novel highly expressed miRNA genes. We also analyzed miRNA sequence variation as described previously (25–27) and found no novel SNPs in positions known to affect miRNA biogenesis or their target mRNA regulation (Dataset S1).
Fig. 1.
Number of individuals in each group and tissue with the number of samples shown in parentheses. Advanced HF group at LVAD implantation 3 mo (3M LVAD) or 6 mo (6M LVAD) after LVAD implantation and at LVAD explantation.
To investigate myocardial miRNA expression changes, we combined the reads corresponding to miRNA genes organized in miRNA cistrons (25, 28). The cistrons are all labeled in lowercase followed by the number of the founding member and the number of cistronic miRNAs in parentheses (for details, see Fig. S1; results for individual miRNAs and miRNA families are in Dataset S1). Forty-two miRNA cistrons changed in DCM (23 up and 19 down) and 54 cistrons changed in ICM (30 up and 24 down) HF compared with NF (Fig. 2A). The majority of these differences affected miRNAs with low expression [Fig. 2A (to the left of the dashed line) and Dataset S1]. Experiments with siRNAs or antagomirs (29) showed that only highly expressed miRNAs effectively repress target mRNAs. For simplicity of data presentation and discussion, we thus focused on regulatory miRNAs that contribute to ∼85% of sequencing reads per sample (7, 25), corresponding to 15, 25, 21, and 28 miRNA cistrons in NFs, FETs, DCM HF, and ICM HF, respectively (Fig. 2 A–C and Fig. S2C). Of these highly expressed cistrons ∼20% changed in DCM HF and ICM HF compared with NFs ranging in absolute values from 1.4-fold for mir-1-1(4) to 2.9-fold for mir-221(2) (Fig. 2A). The most highly expressed cistron mir-1-1(4) in myocardial tissue changed from an average read frequency of 25% in NF to 18% (1.4-fold) and 17% (1.6-fold) in DCM HF and ICM HF, respectively. Although some of the differentially expressed cistrons were common to DCM HF and ICM HF, others were exclusive to ICM HF (Dataset S1). Interestingly, the myomirs mir-208a(1), mir-208b(1), and mir-499(1) were unaltered in either DCM HF or ICM HF. Considering less abundant miRNA cistrons and their variation across sample groups, they were typically less than fourfold, except for mir-216a(3) that increased 22-fold in DCM HF and a 47-fold in ICM HF compared with NFs (Fig. 2A). mir-216a(3) was at least 10 times higher expressed in human umbilical vein endothelial cells (HUVECs) possibly indicative of altered endothelial cell function in the heart. Finally, we did not observe significant changes in miRNA cistron expression comparing the patient-matched myocardial samples taken at the time of LVAD implantation and during explantation (Dataset S1).
Fig. 2.
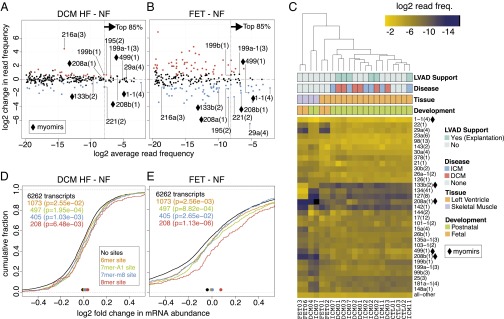
miRNA cistron abundance changes and target gene regulation in failing and fetal myocardium. (A and B) Relative changes in miRNA cistron abundance compared with nonfailing postnatal hearts. Cistrons with a false discovery rate (FDR) of <10% are colored red if up- regulated and blue if down-regulated. Cistrons contributing to the top 85% sequencing reads are to the right and residual cistrons to the left of the dotted vertical line. Labeled in both A and B for comparison are (i) cistrons consistently changed in DCM and ICM HF, (ii) myomirs, and (iii) mir-216a(3) and mir-29a(4) due to their large differences. (C) Unsupervised hierarchical clustering of miRNA cistrons representing the cumulative top 85% miRNA sequence reads with residual cistrons represented as “all–other” at the bottom of the heatmap. The row dendrogram has been omitted. The column labels at the bottom represent the unique IDs for each subject; demographic details on the individual subjects and for each sequencing sample can be found in Dataset S1 (Tables 34 and 35). (D and E) Cumulative distribution function (CDF) showing the changes in mRNA abundance (x axis) for transcripts with different target sites (colored) for family miR-1-1(3) in the 3′UTR compared with transcripts without a site in the 3′UTR (black line). Colored points at bottom of the graph indicate the median of the CDF; P values are from a one-sided Kolmogorov–Smirnov test.
In FETs, a total of 111 cistrons changed compared with NFs (54 up and 57 down; Fig. 2B). In contrast to DCM and ICM HF, 60% of the highly expressed miRNAs in FETs were differentially regulated at a higher magnitude than in failing myocardium (Fig. 2B and Dataset S1). This was particularly the case for mir-29a(4). This cistron was expressed at 0.06% read frequency in FETs and increased 90-fold to 5.6% in NFs, whereas its expression was unchanged in HF. We observed a similar large difference in mir-29a(4) expression in skeletal muscle (Fig. 2C). Considering myomir expression, levels of mir-1-1(4) were reduced in FETs versus NFs, mirroring the changes in HF described above; however mir-208a(1), mir-208b(1), and mir-499(1), all of which are located in introns of myosin genes, were lower by 2.6-, 4.0-, and 3.9-fold, respectively, and unaltered in HF.
Identification of miRNA-Guided Repression of Target mRNAs in the Myocardium.
miRNAs predominantly mediate mRNA destabilization (30). To evaluate if the observed changes in miRNA abundance between HF or FETs compared with NFs were reflected in quantifiable differences in target mRNA abundance, we performed a correlational analysis of the abundance of miRNA sequence families (sf) and miRNA seed-containing mRNAs versus residual mRNAs. mRNA levels were determined using Illumina BeadChips (n = 4 for each condition; Fig. S3 and Dataset S1). Indicative of miRNA regulation (31), we found that target mRNAs containing miRNA binding sites in their 3′UTRs were more repressed than those with sites in their coding sequence (Fig. S4 A and B) and repression increased according to the extent of seed pairing between miRNA and mRNAs (8-mer > 7-mer-m8 ≈ 7-mer-A1 > 6-mer; Fig. 2 D and E and Fig. S4). Target mRNAs for sf-miR-1-1(3) detected with a probing signal intensity above the median and comprising at least one 8-mer site in the 3′UTR were elevated on average by 1.03-fold in DCM or ICM HF (P = 0.006 and 0.002, respectively), and 1.09-fold (P = 1.13 × 10−6) higher in FETs compared with NFs. mRNAs detected by signal intensities in the top 25% range were 1.05-fold (P = 0.02) and 1.04-fold (P = 0.002) higher in DCM and ICM HF, respectively and 1.2-fold (P = 2.13 × 10−5) higher in FETs compared with NFs (Fig. S4 C and D). This observation indicates that higher-expressed target mRNAs respond better to miRNA regulation or are better quantifiable by hybridization expression analysis. The target analysis gave similar responses for other dysregulated miRNA families (Fig. S4 E–M). Collectively, these findings support bona fide miRNA regulation in heart tissues under normal and disease conditions for well and differentially expressed miRNAs.
The Circulating Small RNA Pool Consists of miRNAs and Fragments of tRNAs and Small Cytoplasmic RNAs.
To determine whether myocardial miRNA expression changes translated into measurable changes in the circulating miRNA fraction, we isolated total RNA from potassium–EDTA-treated plasma as well as serum samples from three cohorts representing different clinical stages of HF (Fig. 1 and Table S2): Cohort 1 included healthy controls (NF, n = 13); cohort 2 included patients with advanced HF with samples collected at LVAD implantation (advanced HF, n = 24) and during routine outpatient visits after 3 mo (3M LVAD, n = 10) or 6 mo (6M LVAD, n = 10), and at the time of LVAD explantation (n = 7). Twelve of the 24 advanced HF samples were procured from patients for whom we generated myocardial miRNA profiles. Cohort 3 included ambulatory patients with highly reduced left ventricular function stabilized with conventional pharmacologic therapy (stable HF, n = 14). We sequenced sRNA cDNA libraries prepared from plasma total RNA from all patients of the three cohorts as well as libraries prepared from serum total RNA from 18 individuals of cohorts 1 and 2 (serum–plasma pairs).
The median recovery of total RNA was 30 ng/mL (IQR = 17 ng/mL) for plasma and 69 ng/mL (IQR = 70 ng/mL) for serum (Fig. S5 A and B). The circulating sRNA content was mainly miRNAs, and fragments of small cytoplasmic RNAs (scRNAs) and tRNAs. The average plasma and serum tRNA composition differed 47-fold and was 0.6% (IQR 0.9%) in plasma and 28% (IQR 33%) in serum, whereas the scRNA content remained stable (Fig. S5 C and D). EDTA treatment has been shown recently to destabilize tRNA-containing RNPs in plasma (32) without affecting scRNAs (33).
The serum samples had a median of 0.9 million miRNA reads (80,000 to 6 million) and the plasma samples 1.4 million (40.000 to 14 million; Dataset S1). The median miRNA content was 51 fmol/µg total RNA (IQR = 26 fmol/µg) in serum, and due to the lower tRNA concentration, higher in plasma with 116 fmol/µg total RNA (IQR = 119 fmol/µg; Fig. S5 E and F). This value was 5- to 10-fold higher than previously noted in plasma samples (7), and likely the result of a different RNA isolation protocol here that avoided column-based RNA isolation.
The Circulating miRNA Profile in HF.
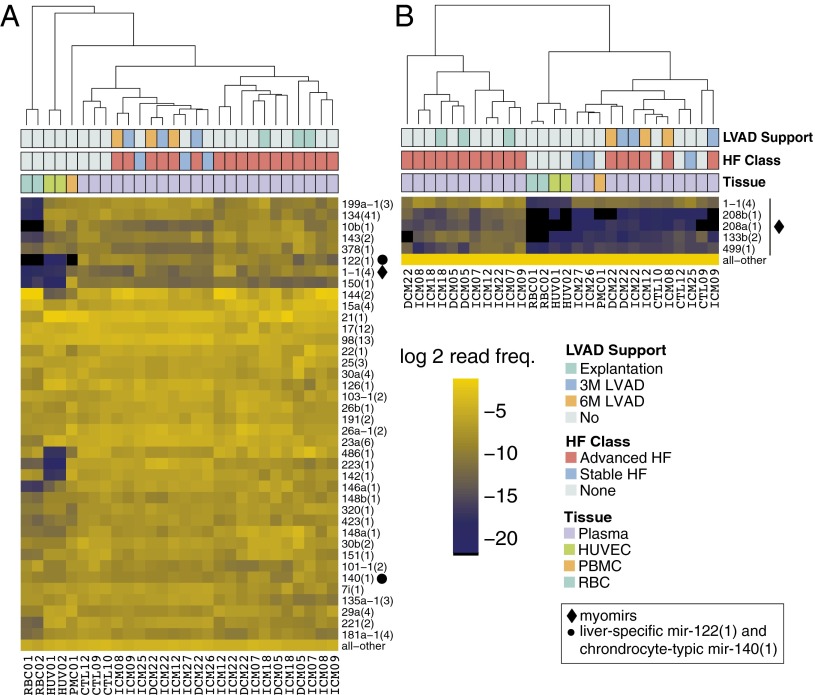
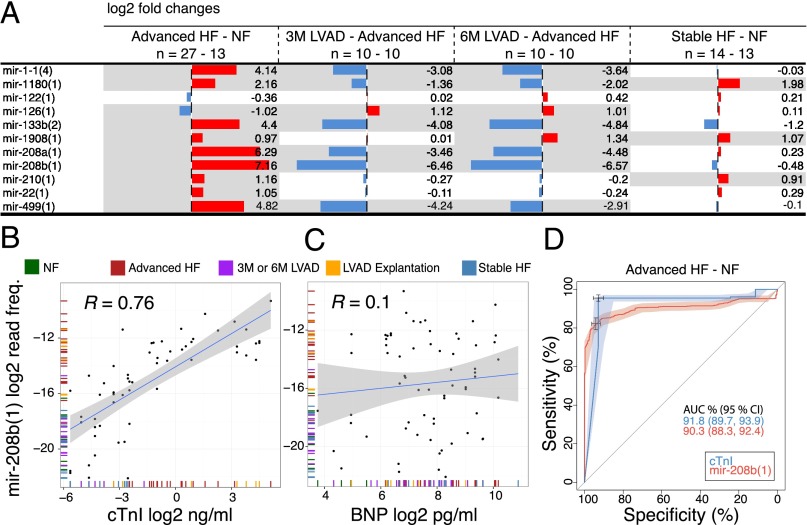
The most abundant circulating miRNAs in healthy individuals probably originate from circulating blood cells (7) and endothelial cells, where they are highly expressed (Fig. 3A and Fig. S5G). In healthy individuals, only a few miRNAs known to be specifically expressed in solid tissues were among the top 85% sequence reads, including mir-122(1) from liver and mir-1-1(4) from muscle, but not the cardiac-specific myomirs. The combined myomir abundance in healthy individuals was less than 0.1%, however, it increased to over 1% in advanced HF patients. Myomirs displayed the biggest differences in levels among the 119 significantly changed miRNA cistrons (64 up and 55 down) in advanced HF patients compared with NFs. The cardiac-specific myomirs mir-208b(1), mir-208a(1), and mir-499(1) and the muscle-specific mir-1-1(4) and mir-133b(2) were 143-, 78-, 28-, 18-, and 21-fold higher in advanced HF at LVAD implantation compared with NFs (Figs. 3B and 4A and Fig. S5H). We also noted a 25-fold increase in mir-216a(3) in advanced HF, which at first sight paralleled a similar magnitude change in cardiac tissue. However, analysis of individual–paired samples and of absolute amounts suggested that the increase in circulating mir-216a(3) in advanced HF was not directly linked to the release of cardiac myomirs. More likely, endothelial cells, which express mir-216a(3) at higher levels than whole heart tissue, released it in response to advanced HF and its clinical management. Overall, 19 cistrons differed more than fivefold in advanced HF compared with NFs.
Fig. 3.
miRNA composition in circulation. (A) Unsupervised hierarchical clustering of miRNA cistrons contributing to the cumulative top 85% sequence reads in plasma samples compared with red blood cells (RBCs), peripheral blood mononuclear cells (PBMCs), and HUVECs. (B) Unsupervised clustering of the same samples as in A but restricted to myomir cistrons with other miRNAs summarized in “all–other” at the bottom of the heatmap. The order of the annotation tracks above the heatmap and the legend are the same for A and B. The column labels at the bottom of the heatmaps are the unique IDs for each subject; demographic details on the individual subjects and for each sequencing sample can be found in Dataset S1 (Tables 34 and 35). For corresponding heatmaps with all serum and plasma samples, see Fig. S5 G and H.
Fig. 4.
Circulating miRNA dynamics in HF. (A) Changes of muscle-specific and other selected miRNA cistrons in circulation comparing the conditions as indicated in the column headings. Red bars represent higher levels and blue bars lower levels in each comparison. The gray shading marks changes that have an FDR < 10%. (B and C) Correlation of cistron mir-208b(1) with cTnI (B) and BNP (C) levels showing the fit (blue line) and its 95% confidence interval (CI) (gray shading). (D) Receiver operating characteristic curve comparing the performances for cTnI and mir-208b(1) in distinguishing patients with advanced HF at the time of LVAD implantation from healthy control individuals. Areas around the blue and red lines represent 95% CI. The cross marks represent the best threshold value, which was 0.044 ng/mL (95% CI: 0.01, 0.05) for cTnI and a relative frequency of 0.0018% (95% CI: 0.0015, 0.0019) for mir-208b(1).
The miRNA changes in advanced HF reversed 3 and 6 mo after LVAD implantation. The levels of the myomirs mir-208a(1), mir-208b(1), mir-499(1), and mir-1-1(4) dropped as early as 3 mo after the initiation of LVAD support, approaching normal levels (Fig. 4A and Dataset S1). At LVAD explantation, the myomir levels rose again with alterations comparable in magnitude to those observed at implantation (Dataset S1).
Surprisingly, in stable HF patients the myomir levels were comparable with NFs, and the biggest differences noted were a 5.4-fold increase for mir-375(1) and a 4.5-fold drop for mir-203(1). Furthermore, there were some concordant changes in both stable and advanced HF compared with NFs: mir-210(1) was 2.2- and 1.9-fold higher in advanced HF and in stable HF, respectively. mir-1908(1) was 2.0- and 2.1-fold, and mir-1180(1) 4.5- and 4.0-fold higher in patients with advanced HF and in patients with stable HF, respectively (Fig. 4A).
Considering that myomirs showed the largest increase in the circulation of advanced HF patients compared with NF and are tissue-specifically expressed, we wanted to compare their levels with those of cardiac troponin I (cTnI) and B-type natriuretic peptide (BNP) protein levels, established biomarkers for myocardial injury and dysfunction, respectively (Dataset S1). Higher levels of the heart-specific myomirs mir-208a(1), mir-208b(1) (Fig. 4B), and mir-499(1) were positively correlated with cTnI (R = 0.75, P = 4.73 × 10−6; R = 0.76, P = 4.59 × 10−7; and R = 0.6, P = 8.86 × 10−5, respectively) but not correlated with BNP (Fig. 4C). The cTnI concentrations in the serum of NFs were below the detection limit of 0.01 ng/mL, except for one sample reaching 0.03 ng/mL, closely followed by a median of 0.04 ng/mL in 3M or 6M LVAD (IQR = 0.05), a median of 0.09 ng/mL (IQR = 0.12) in stable HF, a median of 0.5 ng/mL (IQR = 1.18) in patients with advanced HF at LVAD implantation, and maximum concentrations with a median of 9.8 ng/mL (IQR = 15.8) at LVAD explantation. In the supervised classification area under the receiver operating characteristic curve the heart-specific cistrons performed similar to cTnI (Fig. 4D). Together, these results did not support a role for circulating miRNAs as biomarkers of cardiac function beyond myocardial injury.
Discussion
We used a small RNA-sequencing (sRNAseq) protocol developed for parallel processing of large sample collections with limited amounts of input RNA (7, 34, 35) to record the miRNA composition in heart tissue and in circulation in a large cohort of HF patients and normal controls. Using the same method for miRNA profiling eliminated biases (26) otherwise affecting comparison of our data with studies, which previously profiled either tissue or circulating miRNAs in HF, but never both.
To identify changes in myocardial miRNAs abundant enough to trigger measurable differences in mRNA expression by miRNA-mediated degradation, we at first considered miRNAs contributing to the top 85% sequence reads. For these highly expressed miRNAs, the overall abundance in failing myocardium compared with normal postnatal myocardium differed not more than twofold, in agreement with a recent RNAseq tissue study by Yang et al. (17). The same miRNAs were also altered in heart development but changed up to sixfold. These alterations in miRNA abundance resulted in an average of 1.02- to 1.20-fold miRNA seed-dependent mRNA destabilization similar to observed values in mechanistic studies (31, 36).
Although miRNA expression changes in HF tissue have been studied extensively by hybridization-based methods, there is little consensus due to variation in methods and analysis (37). The miRNA signature of failing myocardium in our study comprised alterations of miRNA cistrons mir-1-1(4), mir-195(2), mir-199a-1(3), mir-199b(2), and mir-221(2). The mir-1-1(4) genes are induced during heart development and act as central regulators of muscle differentiation (38–40). The down-regulation of the myomir mir-1-1(4) therefore supports a myogenic dedifferentiation in human HF. The reported regulation in human HF and fetal myocardium, however, has been controversial, including observations of increased mir-1-1(4) expression (11, 14). Furthermore, we only detected marginal differences in miRNA expression between ICM and DCM hearts and no differences before and after LVAD support, both of which are supported by another recent RNAseq study (17), but contrasted with a microarray-based study reporting more dramatic differences in their comparison of hearts before and after LVAD treatment (14). It is conceivable that distinctive myocardial miRNA changes reflecting heterogeneous genetic or environmental etiologies as well as the myocardial response to pharmacologic and nonpharmacologic therapies exist and that these changes are detectable at early stages of HF. Tissue from early stages is hardly available at sufficient quantities and, therefore, studies on human myocardium typically focus on later stages of the disease (37).
Although many of the HF-associated changes were also evident in fetal myocardium, we noted previously unreported differences—such as a the 90-, 10-, and 6-fold lower abundance of mir-29a(4), mir-22(1), and mir-195(2), respectively—in fetal but not failing myocardium compared with nonfailing myocardium. Several of these abundant miRNAs were shown to inhibit cell cycle progression in cultured cardiomyocytes (41) and in vivo (42). A similar difference in mir-29a(4) expression between fetal and adult skeletal muscle indicates a more ubiquitous antiproliferative role of mir-29a(4). Of note, HF is associated with loss of cardiomyocytes (43) and the mammalian heart has a very limited regenerative capacity (44). The reactivation of self-repair mechanisms by targeting miRNAs represents a possible approach to enhance regeneration (45).
The composition of circulating sRNAs was dominated by miRNAs that are abundant in hematopoietic cells (7) and/or the endothelium. The contribution of myomirs to all circulating miRNAs was less than 0.1% in healthy controls and patients with moderate and stable HF. However, the myomirs increased to over 1% in patients with advanced HF and reduced to nearly normal levels at 3 and 6 mo after LVAD implantation. The myomirs are subdivided into the cardiac-specific mir-208a(1), mir-208b(1), and mir-499(1) and the broadly muscle-specific mir-1-1(4) and mir-133b(2), the latter of which are responsible for the circulating myomir background levels in healthy individuals. Hence, mir-1-1(4) and mir-133b(2), which together contributed 30% of all myocardial miRNAs, showed a reduced fold increase during HF compared with cardiac-specific myomirs. The relative abundance of the three heart-specific myomirs in circulation closely followed the ratio determined in heart tissue and is consistent with it being the only source.
Increased circulation of myomirs strongly correlated with increased cTnI but not BNP protein levels; these proteins are established diagnostic heart injury and heart function markers, respectively. Increases in circulating miRNAs upon cell damage have been detected by RT-PCR–based approaches for liver, brain, and skeletal muscle (5), as well as heart (19, 46). In some instances miRNAs even performed better than established protein biomarkers (5). Our analysis indicated that heart-specific myomirs performed similar to the highly sensitive cTnI assay, but we were unable to verify the utility of any recently proposed miRNA biomarker for HF (18, 20–22).
Considering that sRNAseq experiments capture a wide spectrum of miRNAs as well as fragments of other classes of RNAs, and can be performed with low ng quantities of total RNA recoverable from 250 µL plasma or serum, this approach holds great potential for future RNA biomarker discovery. The translation of this type of RNAseq assay into clinical practice is currently limited by the time required for cDNA library preparation and sequencing, however, single-molecule direct RNAseq (47) or targeted RT-PCR assays may overcome some of these limitations.
Materials and Methods
Tissue Procurement.
Human myocardial tissue samples were obtained from the National Disease Research Interchange (Philadelphia) and from the Columbia University Medical Center, and after elective termination of pregnancy for nonmedical reasons. Serum and plasma samples were obtained from the Columbia University Medical Center.
RNA Isolation.
Total RNA from solid tissue and liquid samples was isolated with a modified TRIzol protocol and recovered by alcohol precipitation. Liquid sample RNA recovery included addition of glycogen for coprecipitation. Tissue total RNA was further purified by Qiagen RNeasy columns for bead array studies.
sRNAseq and Gene Expression Analysis.
The cDNA library preparation and annotation were done as described (27, 34, 35) with modifications for library preparations of serum and plasma samples. A calibrator cocktail of 10 22-nt synthetic oligoribonucleotides was used for quantification purposes and quality control (Fig. S6) (34). mRNA expression was assessed on Illumina HumanHT-12v4 bead arrays according to the manufacturer’s instructions.
The data were analyzed in the R statistical language (48). The functional studies testing miRNA regulation followed the approach by Grimson et al. (31). Differences in RNA quantification for unpaired samples were tested using the Kruskal–Wallis rank sum test and for paired samples using the Wilcoxon signed rank test. The differences in the cumulative distributions were tested using the one-sided Kolmogorov–Smirnov test.
For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Grant AK137-1/1 from the Deutsche Forschungsgemeinschaft (to K.M.A.); Award U19CA179564 from the National Institutes of Health (NIH) Common Fund through the Office of Strategic Coordination/Office of the NIH Director; NIH Grants K23HL095742, P30HL101272, UL1RR024156, and HL073029 and a Herbert and Florence Irving Scholar Award (to P.C.S.); NIH Grant HD068546 (to Z.W.); and, in part, by NIH Grant 8UL1TR000043. T.T. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest statement: T.T. is a cofounder of and scientific advisor to Alnylam Pharmaceuticals and a scientific advisor to Regulus Therapeutics.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE53081).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401724111/-/DCSupplemental.
References
- 1.Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: Circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9(6):850–859. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun JE, Huntzinger E, Izaurralde E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv Exp Med Biol. 2013;768:147–163. doi: 10.1007/978-1-4614-5107-5_9. [DOI] [PubMed] [Google Scholar]
- 4.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 5.Laterza OF, et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55(11):1977–1983. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- 6.Ai J, et al. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem Biophys Res Commun. 2010;391(1):73–77. doi: 10.1016/j.bbrc.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Williams Z, et al. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc Natl Acad Sci USA. 2013;110(11):4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 9.van Rooij E, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17(5):662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13(4):486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 11.Thum T, et al. MicroRNAs in the human heart: A clue to fetal gene reprogramming in heart failure. Circulation. 2007;116(3):258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 12.Ikeda S, et al. Altered microRNA expression in human heart disease. Physiol Genomics. 2007;31(3):367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 13.Sucharov C, Bristow MR, Port JD. miRNA expression in the failing human heart: Functional correlates. J Mol Cell Cardiol. 2008;45(2):185–192. doi: 10.1016/j.yjmcc.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matkovich SJ, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119(9):1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naga Prasad SV, et al. Unique microRNA profile in end-stage heart failure indicates alterations in specific cardiovascular signaling networks. J Biol Chem. 2009;284(40):27487–27499. doi: 10.1074/jbc.M109.036541. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Leptidis S, et al. A deep sequencing approach to uncover the miRNOME in the human heart. PLoS ONE. 2013;8(2):e57800. doi: 10.1371/journal.pone.0057800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang KC, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129(9):1009–1021. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tijsen AJ, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106(6):1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 19.Corsten MF, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3(6):499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima Y, Nakanishi M, Nonogi H, Goto Y, Iwai N. Assessment of plasma miRNAs in congestive heart failure. Circ J. 2011;75(2):336–340. doi: 10.1253/circj.cj-10-0457. [DOI] [PubMed] [Google Scholar]
- 21.Goren Y, et al. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012;14(2):147–154. doi: 10.1093/eurjhf/hfr155. [DOI] [PubMed] [Google Scholar]
- 22.Dickinson BA, et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15(6):650–659. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 23.Tutarel O, et al. Circulating miR-423_5p fails as a biomarker for systemic ventricular function in adults after atrial repair for transposition of the great arteries. Int J Cardiol. 2013;167(1):63–66. doi: 10.1016/j.ijcard.2011.11.082. [DOI] [PubMed] [Google Scholar]
- 24.Kumarswamy R, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114(10):1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 25.Farazi TA, et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71(13):4443–4453. doi: 10.1158/0008-5472.CAN-11-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner M, et al. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17(9):1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown M, Suryawanshi H, Hafner M, Farazi TA, Tuschl T. Mammalian miRNA curation through next-generation sequencing. Front Genet. 2013;4:145. doi: 10.3389/fgene.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krützfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhahbi JM, et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhahbi JM, et al. 5′-YRNA fragments derived by processing of transcripts from specific YRNA genes and pseudogenes are abundant in human serum and plasma. Physiol Genomics. 2013;45(21):990–998. doi: 10.1152/physiolgenomics.00129.2013. [DOI] [PubMed] [Google Scholar]
- 34.Hafner M, et al. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods. 2012;58(2):164–170. doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farazi TA, et al. Bioinformatic analysis of barcoded cDNA libraries for small RNA profiling by next-generation sequencing. Methods. 2012;58(2):171–187. doi: 10.1016/j.ymeth.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang Z, Rajewsky N. The impact of miRNA target sites in coding sequences and in 3’UTRs. PLoS ONE. 2011;6(3):e18067. doi: 10.1371/journal.pone.0018067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda S, Pu WT. Expression and function of microRNAs in heart disease. Curr Drug Targets. 2010;11(8):913–925. doi: 10.2174/138945010791591304. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 39.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 41.Cao X, et al. MicroRNA profiling during rat ventricular maturation: A role for miR-29a in regulating cardiomyocyte cell cycle re-entry. FEBS Lett. 2013;587(10):1548–1555. doi: 10.1016/j.febslet.2013.01.075. [DOI] [PubMed] [Google Scholar]
- 42.Porrello ER, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109(6):670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whelan RS, Kaplinskiy V, Kitsis RN. Cell death in the pathogenesis of heart disease: Mechanisms and significance. Annu Rev Physiol. 2010;72:19–44. doi: 10.1146/annurev.physiol.010908.163111. [DOI] [PubMed] [Google Scholar]
- 44.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: Opportunities and obstacles. Nat Rev Drug Discov. 2012;11(11):860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji X, et al. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55(11):1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 47.Kapranov P, Ozsolak F, Milos PM. Profiling of short RNAs using Helicos single-molecule sequencing. Methods Mol Biol. 2012;822:219–232. doi: 10.1007/978-1-61779-427-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. R Core Team (2013) R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria). Available at www.R-project.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.