Abstract
Salmonella FliR and FlhB are membrane proteins necessary for flagellar export. In Clostridium a fliR-flhB fusion gene exists. We constructed a similar Salmonella fusion gene which is able to complement fliR, flhB, and fliR flhB null strains. Western blotting revealed that the FliR-FlhB fusion protein retains the FlhB protein's cleavage properties. We conclude that the FliR and FlhB proteins are physically associated in the wild-type Salmonella basal body, probably in a 1:1 ratio.
The bacterial flagellum, a rotary motor, is the organelle responsible for the motility of numerous bacteria, including Salmonella enterica serovar Typhimurium. The flagellum consists of a basal body and motor structure, a flexible hook, and a filament. Assembly begins with the formation of the MS ring, which is imbedded in the inner membrane (14). The MS ring houses the export apparatus which is responsible for the translocation of the majority of the flagellar proteins that are external to the cell membrane. Export substrates are translocated across the cytoplasmic membrane, travel through a narrow (ca. 3-nm) channel in the nascent structure (17, 25), and then assemble at the distal end of the growing flagellum (2, 9). The flagellar export apparatus is a member of the type III secretion system family (15), which is also responsible for virulence factor secretion in pathogenic bacteria. In Salmonella an apparatus distinct from yet homologous to the flagellar export machinery is present for the secretion of virulence factors (8).
The flagellar export apparatus consists of six integral membrane proteins (FlhA, FlhB, FliO, FliP, FliQ, and FliR) and three associated cytoplasmic proteins (FliH, FliI, and FliJ) (14, 15). Together these proteins are responsible for the export of rod-type, hook-type, and filament-type export substrates (21, 23). FliI is a flagellum-specific ATPase (3) and is regulated by FliH (7, 20, 24). FliJ is a putative general chaperone (5, 18). This note focuses on two of the membrane components, FliR and FlhB.
The membrane components are likely located in a patch of membrane within the central pore of the MS ring (6, 29). There is direct evidence for the presence of FliP and FliR in the basal body (4) and intergenic suppression evidence for the presence of FlhA, which probably interacts with the MS ring protein FliF (12). Very little is known about the functions of FliO, FliP, FliQ, FliR, and FlhA, but their presence is required for flagellum-specific protein export (21). Although FliO, FliP, FliQ, and FliR are extremely hydrophobic, FlhA and FlhB both have large cytoplasmic domains that interact with FliH, FliI, and FliJ (23). FlhB, along with the hook-length control protein FliK, is responsible for substrate specificity switching from rod- and hook-type export to filament-type export (22). Minamino and Macnab showed that FlhB can be divided into two domains: FlhBTM, comprising the four-membrane-spanning N-terminal domain, and FlhBC, the cytoplasmic domain (22). The ability to switch is related to the cleavage of FlhBC into FlhBCN and FlhBCC (22). Fraser et al. have shown that a noncleaving mutant, FlhBN269A, does not switch from rod- and hook-type to filament-type export (6). FlhBC binds to numerous export substrates (23) as well as FlhAC and itself (34). Mutations in FlhBC are able to suppress a FliK null background (32).
In many bacterial genomes, fliR and flhB are positioned as consecutive genes within the same operon (Fig. 1). In Salmonella the two genes are separated by about 55 kb (16). A database search for FlhB homologs found that Clostridium acetobutylicum (26) and Clostridium tetani (1) have a gene that is equivalent to a fusion of fliR and flhB. In the present study we show that an engineered Salmonella fusion protein has the ability to complement fliR and flhB null strains of Salmonella as well as a fliR flhB double-null strain. This strongly suggests that FliR and FlhB are in close proximity to each other within the basal body. In addition, these findings clarify the predicted membrane topology of FliR, which had previously been unclear (27), and firmly establish the presence of FlhB in the basal body.
FIG. 1.
The genes coding for the integral membrane proteins of the flagellar export apparatus. The genomes of Salmonella (16) and Bacillus subtilis (13) are representative of the two most common arrangements. In Salmonella, flhA and flhB are located in a separate operon approximately 55 kb from the operon containing fliO, fliP, fliQ, and fliR. In Bacillus these genes are contained within the same operon, with flhB immediately following fliR. The arrangement of genes in Clostridium (1, 26) is similar to that in Bacillus, but a gene homologous to both fliR and flhB is present in place of the separate genes.
Bacterial strains and plasmids are listed in Table 1. Luria broth and soft tryptone agar plates were prepared as previously described (23). T7 medium contains, per liter, 20 g of tryptone, 10 g of yeast extract, 5 g of NaCl, and 8.7 g of K2HPO4 (pH 7.2). Ampicillin was added to a final concentration of 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| NovaBlue | Recipient for cloning experiments | Novagen |
| BL21(DE3)/pLysS | Used for overproduction of proteins from pET-based vectors | Novagen |
| Salmonella | ||
| JR501 | Used for converting E. coli plasmids to Salmonella compatibility | 28 |
| SJW1103 | Wild type | 33 |
| SJW170 | fliR null | 33 |
| MKM50 | flhB null strain | 6 |
| MKM65 | fliR null strain | This study |
| MKM58 | fliR flhB double-null strain | This study |
| Plasmids | ||
| pET19b | N-terminally His-tagged T7 expression vector | Novagen |
| pET19bFH | N-terminally His-FLAG-tagged T7 expression vector | 4 |
| pTrc99A-FF4 | Modified pTrc expression vector | 27 |
| pMM26 | pTrc99A-FF4/FlhB | 22 |
| pRMK3000 | pET19b/N-His-FliR | This study |
| pGFBN269A | pet19b/N-His-FlhBN269A | 6 |
| pJSV003 | pET19bFH/N-His-FLAG-FliR-FlhB | This study |
| pJSV004 | pTrc99A-FF4/FliR-FlhB | This study |
| pJSV013 | pET19bFH/N-His-FLAG-FliR-FlhBN269A | This study |
| pJSV014 | pTrc99A-FF4/FliR-FlhBN269A | This study |
| pJSV024 | pTrc99A-FF4/FliR + FlhB | This study |
Construction of the fliR-flhB fusion gene.
The fliR-flhB fusion gene was constructed by a method adapted from Toker et al. (30). Complementary primers coding for the 3′ end of fliR and the 5′ end of flhB were used with outside primers to create an NdeI site 5′ to fliR and a BamHI site 3′ to flhB. Our fusion protein has a short sequence of four amino acids (GAGA; italicized in the sequence) linking the otherwise wild-type sequence in order to maximize the possibility of a functional protein (FliR-SEMPINNNP-GAGA-MAEESDDDKTEAPTP-FlhB), although there is no such spacer in the Clostridium protein (FliR-KLVPIGLIFADSDKTEEATP-FlhB) (26).
The fliR and flhB portions of the fusion gene were amplified from plasmids pRMK3000 and pMM26, respectively, by using Pfu Turbo DNA polymerase (Stratagene). These PCR products were then used as templates for a second round of PCR to construct the fusion gene. Amplified DNA was purified by using the QiaQuick gel extraction kit (QIAGEN Inc.), digested with NdeI and BamHI, and cloned into pUC18. The fusion gene was then subcloned into the pTrc99A-FF4 and pET vectors for use in protein expression studies. The construction of the fusion gene fliR-flhBN269A used the same protocol, but pGFBN269A was used in place of plasmid pMM26. As a control, we also constructed a plasmid containing fliR and flhB as separate but consecutive genes by cloning the fliR gene with NdeI and BamHI and the flhB gene with XbaI and HindIII into pTrc99A-FF4.
Construction and complementation of null strains.
The fliR and flhB null strains were constructed as previously described (11). The fliR flhB double-null strain was constructed by transferring the flhB null genotype to SJW170, a fliR mutant strain containing a 10-bp deletion resulting in a frameshift after amino acid 89 and having 27 foreign amino acids before reaching a stop codon. Cells were tested for their swarming ability on soft tryptone agar plates as described by Fraser et al. (5).
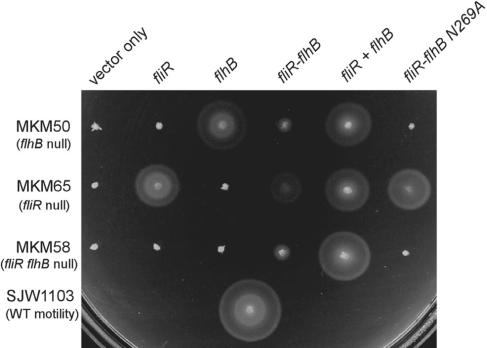
Complementation results are shown in Fig. 2. Strains MKM50 (flhB null) and MKM65 (fliR null) are complemented by plasmids pMM26 and pRMK3000, respectively. These null strains are also complemented by pJSV004 (fusion plasmid) and pJSV024 (coproducing plasmid). The fusion protein can complement MKM58 (fliR flhB double null) although providing FliR and FlhB as separate proteins (pJSV024) allows for improved swarming. This indicates that the fusion protein retains the functions of both FliR and FlhB. The complementation of MKM65 by pJSV004 is better than its complementation of the other null strains. This result could be due to the presence of wild-type FlhB (in this fliR null strain), which is not present in either of the other two null strains. This suggests that wild-type FlhB may be present in the basal body in addition to the FliR-FlhB fusion protein.
FIG. 2.
Complementation analysis in null backgrounds. The plate was incubated at 30°C for 5 h. Column 1, vector only; column 2, pRMK3000 (FliR); column 3, pMM26 (FlhB); column 4, pJS004 (FliR-FlhB fusion); column 5, pJSV024 (FliR + FlhB coproducing plasmid); and column 6, pJSV014 (FliR-FlhBN269A); WT, wild type.
Although plasmid pJSV014, containing the FliR-FlhB fusion protein with the noncleavable FlhBN269A mutation, does not complement either strain MKM50 or MKM58, it does complement strain MKM65 almost as well as plasmid pRMK3000. This finding indicates that the FliR portion of FliR-FlhBN269A is able to function in the export process although it is fused to the noncleaving FlhB variant which is locked in rod- and hook-type export. This could mean that in the fliR null strain the cleaved wild-type FlhB from the chromosome can interact with FlhBN269A of the fusion protein, enabling it to function at a level near that of the wild-type. Supporting this idea, several species have a separate gene encoding FlhBCC in addition to full-length FlhB (15). Since FlhBC can interact with itself (34), oligomerization of FlhBC may be necessary for full function.
To examine the dominant-negative effects of the fusion protein in wild-type cells, we tested the swarming of strain SJW1103 transformed with plasmids pJSV004, pJSV024, and pJSV014. In the presence of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), we saw a decrease in swarming with all three plasmids, but especially with the two fusion plasmids. This finding indicates that overproduction of these proteins can interfere with the assembly of the export apparatus (data not shown).
Purification of the FliR-FlhB fusion protein.
In order to confirm that the FliR-FlhB fusion construct remains intact and retains the cleavage properties of FlhB, we purified the fusion protein. We found it necessary to use a system yielding high levels of protein expression in order to detect the FliR-FlhB fusion protein. In Salmonella we never detected the fusion protein under a variety of culture conditions. Cultures of Escherichia coli BL21(DE3)/pLysS transformed with plasmids pJSV003 and pJSV013, coding for N-His-FLAG-FliR-FlhB and N-His-FLAG-FliR-FlhBN269A, respectively, were grown in T7 medium (3 liters) at 30°C to an optical density at 600 nm of 0.4. IPTG was then added to a final concentration of 0.2 mM, and growth continued for an additional 4 h. Cells were harvested and resuspended in 120 ml of lysis buffer (50 mM sodium phosphate [pH 8.0], 500 mM NaCl, 20% glycerol, 10 mM β-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride), sonicated, and centrifuged at 10,000 × g to pellet unbroken cells and inclusion bodies. The supernatant was then centrifuged at 100,000 × g for 45 min. Pellets were retained and resuspended by homogenization in 15 ml of solubilization buffer (lysis buffer with 6 M urea, 1% [vol/vol] Triton X-100, and 10 mM imidazole). This suspension was stirred at 4°C for 1 h and then subjected to centrifugation at 100,000 × g for 45 min. The denatured, solubilized membrane fraction was retained as the supernatant.
The supernatant was loaded by gravity flow onto a 250-μl Talon (BD Biosciences) metal affinity column, washed with 20 column volumes of wash buffer (solubilization buffer with 20 mM imidazole), and eluted in the same buffer with 250 mM imidazole.
Western blotting.
Samples to be probed were run on sodium dodecyl sulfate-15% polyacrylamide gels and transferred onto nitrocellulose membranes. Membranes were probed by using anti-FLAG M2 antibody (Sigma) and horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Bio-Rad). Immunodetection was carried out with the SuperSignal Pico detection kit (Pierce) according to the manufacturer's instructions.
Western blotting reveals that the FliR-FlhB fusion remains intact and that normal posttranslational processing of FlhB occurs (Fig. 3, lane 3). The band migrated at ∼60 kDa, consistent with an intact fusion protein in which the FlhB component has been normally cleaved at N269 (the cleavage product would not be seen with anti-FLAG antibodies). Furthermore, the uncleavable N269A fusion variant (lane 4) migrated at ∼75 kDa. A vector-only control (lane 1) and wild-type N-His-FLAG-FlhB (lane 2) are also shown.
FIG. 3.
Western blot of FLAG-tagged FliR-FlhB fusion constructs. Lane 1, vector only; lane 2, N-His-FLAG-FlhB; lane 3, N-His-FLAG-FliR-FlhB; and lane 4, N-His-FLAG-FliR-FlhBN269A. Mass markers (kDa) are given on the left.
FlhB is present in the basal body.
Although FlhB has been assumed to be present in the basal body due to its role in substrate specificity switching (6), attempts to locate FlhB within the basal body have been unsuccessful, probably because the detergent used in the basal body purification procedure solubilized FlhB. The present study has shown that a FliR-FlhB fusion protein can function in Salmonella although not as well as in the wild-type situation where the two proteins are separate. This result places FlhB within the basal body since it has previously been shown that FliR is located on the cytoplasmic face of the MS ring, probably within a membrane patch at the center of the ring (4).
Implications for export apparatus component stoichiometries.
The fusion protein is functional in a fliR flhB double-null background, suggesting that FliR and FlhB must be present in at least a 1:1 ratio in the wild-type basal body, in agreement with previous estimates of about 1 to 3 copies of FliR (4, 10) and about 2 copies of FlhB (34). However, the improved swarming with the fusion protein in the fliR null background compared to the flhB null background suggests that wild-type FlhB (produced from the chromosome of strain MKM65) could also be playing a role in the export process. Given this possibility, we can conclude that there is at least one FlhB for every FliR.
Topology of FliR.
The present results also address the question of the membrane topology of FliR. Previously it was predicted that FliO has a single transmembrane helix, FliP could have three or four transmembrane spans, and FliQ is probably a two-span, hairpin protein (27). However, it was difficult to predict the number of helices for FliR since its C-terminal region is almost exclusively hydrophobic and could contain five or six helices. We concluded then that the C terminus of FliR could lie either in the periplasm (five spans) or in the cytoplasm (six spans). We also knew that FliR is a very insoluble protein and can form aggregates even in harsh detergent conditions (e.g., 4% sodium dodecyl sulfate) (4), consistent with a six-transmembrane-spanning protein with very little soluble sequence. It is now known that the C terminus of FliR can be fused to the cytoplasmically located N terminus of FlhB (19) and still function in the export apparatus, suggesting that FliR has six transmembrane spans and that its C terminus lies in the cytoplasm.
The presence of natural fusion proteins in export apparatus homologs underscores the importance of genomic databases in protein homology research, suggesting interactions not readily detectable due to problems encountered in studying membrane proteins with unknown functions. In Buchnera aphidicola (31), a gene fusion homologous to both fliO and fliP exists, suggesting another potential interaction.
Acknowledgments
We thank Hedda Ferris, Tohru Minamino, and Bertha González-Pedrajo for helpful comments.
This work was supported by USPHS grant AI12202 to R.M.M.
Footnotes
This paper is dedicated to Robert Macnab, who died suddenly on 7 September 2003.
REFERENCES
- 1.Brüggemann, H., S. Bäumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martínez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. USA 100:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerson, S. U., K. Tokuyasu, and M. I. Simon. 1970. Bacterial flagella: polarity of elongation. Science 169:190-192. [DOI] [PubMed] [Google Scholar]
- 3.Fan, F., and R. M. Macnab. 1996. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J. Biol. Chem. 271:31981-31988. [DOI] [PubMed] [Google Scholar]
- 4.Fan, F., K. Ohnishi, N. R. Francis, and R. M. Macnab. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26:1035-1046. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, G. M., B. González-Pedrajo, J. R. H. Tame, and R. M. Macnab. 2003. Interactions of FliJ with the Salmonella type III flagellar export apparatus. J. Bacteriol. 185:5546-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser, G. M., T. Hirano, H. U. Ferris, L. L. Devgan, M. Kihara, and R. M. Macnab. 2003. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol. Microbiol. 48:1043-1057. [DOI] [PubMed] [Google Scholar]
- 7.González-Pedrajo, B., G. M. Fraser, T. Minamino, and R. M. Macnab. 2002. Molecular dissection of Salmonella FliH, a regulator of the ATPase FliI and the type III flagellar protein export pathway. Mol. Microbiol. 45:967-982. [DOI] [PubMed] [Google Scholar]
- 8.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iino, T. 1969. Polarity of flagellar growth in Salmonella. J. Gen. Microbiol. 56:227-239. [DOI] [PubMed] [Google Scholar]
- 10.Jones, C. J., R. M. Macnab, H. Okino, and S.-I. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 11.Kihara, M., G. U. Miller, and R. M. Macnab. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kihara, M., T. Minamino, S. Yamaguchi, and R. M. Macnab. 2001. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J. Bacteriol. 183:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature (London) 390:249-256. [DOI] [PubMed] [Google Scholar]
- 14.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 15.Macnab, R. M. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 16.McClelland, M., K. E. Sanderson, J. Spleth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. All, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature (London) 413:852-856. [DOI] [PubMed] [Google Scholar]
- 17.Mimori, Y., I. Yamashita, K. Murata, Y. Fujiyoshi, K. Yonekura, C. Toyoshima, and K. Namba. 1995. The structure of the R-type straight flagellar filament of Salmonella at 9 Å resolution by electron cryomicroscopy. J. Mol. Biol. 249:69-87. [DOI] [PubMed] [Google Scholar]
- 18.Minamino, T., R. Chu, S. Yamaguchi, and R. M. Macnab. 2000. Role of FliJ in flagellar protein export in Salmonella. J. Bacteriol. 182:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minamino, T., B. González-Pedrajo, M. Kihara, K. Namba, and R. M. Macnab. 2003. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J. Bacteriol. 185:3983-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minamino, T., B. González-Pedrajo, K. Oosawa, K. Namba, and R. M. Macnab. 2002. Structural properties of FliH, an ATPase regulatory component of the Salmonella type III flagellar export apparatus. J. Mol. Biol. 322:281-290. [DOI] [PubMed] [Google Scholar]
- 21.Minamino, T., and R. M. Macnab. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minamino, T., and R. M. Macnab. 2000. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J. Bacteriol. 182:4906-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minamino, T., and R. M. Macnab. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35:1052-1064. [DOI] [PubMed] [Google Scholar]
- 24.Minamino, T., J. R. H. Tame, K. Namba, and R. M. Macnab. 2001. Proteolytic analysis of the FliH/FliI complex, the ATPase component of the type III flagellar export apparatus of Salmonella. J. Mol. Biol. 312:1027-1036. [DOI] [PubMed] [Google Scholar]
- 25.Morgan, D. G., C. Owen, L. A. Melanson, and D. J. DeRosier. 1995. Structure of bacterial flagellar filaments at 11 Å resolution: packing of the α-helices. J. Mol. Biol. 249:88-110. [DOI] [PubMed] [Google Scholar]
- 26.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, and J. Hitti. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohnishi, K., F. Fan, G. J. Schoenhals, M. Kihara, and R. M. Macnab. 1997. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J. Bacteriol. 179:6092-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu, J., and R. J. Hartin. 1990. Quick transformation in Salmonella typhimurium LT2. BioTechniques 8:43-44. [PubMed] [Google Scholar]
- 29.Suzuki, H., K. Yonekura, K. Murata, T. Hirai, K. Oosawa, and K. Namba. 1998. A structural feature in the central channel of the bacterial flagellar FliF ring complex is implicated in Type III protein export. J. Struct. Biol. 124:104-114. [DOI] [PubMed] [Google Scholar]
- 30.Toker, A. S., M. Kihara, and R. M. Macnab. 1996. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J. Bacteriol. 178:7069-7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Ham, R. C. H. J., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernández, L. Jiménez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, A. Valencia, F. Morán, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams, A. W., S. Yamaguchi, F. Togashi, S.-I. Aizawa, I. Kawagishi, and R. M. Macnab. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi, S., H. Fujita, K. Sugata, T. Taira, and T. Iino. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130:255-265. [DOI] [PubMed] [Google Scholar]
- 34.Zhu, K., B. González-Pedrajo, and R. M. Macnab. 2002. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry 41:9516-9524. [DOI] [PubMed] [Google Scholar]