Significance
Increasing the integrity of the bacterial envelope is necessary to allow the successful survival of bacterial pathogens within the host and allow them to counteract damage caused by membrane-targeting antibiotics. We demonstrate that components of a clustered, regularly interspaced, short palindromic repeats–CRISPR associated (CRISPR-Cas) system, a prokaryotic defense against viruses and foreign nucleic acid, act to regulate the permeability of the bacterial envelope, ultimately providing these cells with the capability to resist membrane damage caused by antibiotics. This regulation further allows bacteria to resist detection by multiple host receptors to promote virulence. Overall, this study demonstrates the breadth of function of CRISPR-Cas systems in regulation, antibiotic resistance, innate immune evasion, and virulence.
Keywords: gene regulation, innate immune evasion
Abstract
Clustered, regularly interspaced, short palindromic repeats–CRISPR associated (CRISPR-Cas) systems defend bacteria against foreign nucleic acids, such as during bacteriophage infection and transformation, processes which cause envelope stress. It is unclear if these machineries enhance membrane integrity to combat this stress. Here, we show that the Cas9-dependent CRISPR-Cas system of the intracellular bacterial pathogen Francisella novicida is involved in enhancing envelope integrity through the regulation of a bacterial lipoprotein. This action ultimately provides increased resistance to numerous membrane stressors, including antibiotics. We further find that this previously unappreciated function of Cas9 is critical during infection, as it promotes evasion of the host innate immune absent in melanoma 2/apoptosis associated speck-like protein containing a CARD (AIM2/ASC) inflammasome. Interestingly, the attenuation of the cas9 mutant is complemented only in mice lacking both the AIM2/ASC inflammasome and the bacterial lipoprotein sensor Toll-like receptor 2, but not in single knockout mice, demonstrating that Cas9 is essential for evasion of both pathways. These data represent a paradigm shift in our understanding of the function of CRISPR-Cas systems as regulators of bacterial physiology and provide a framework with which to investigate the roles of these systems in myriad bacteria, including pathogens and commensals.
Clustered, regularly interspaced, short palindromic repeats–CRISPR associated (CRISPR-Cas) systems are adaptive bacterial defenses against foreign nucleic acids derived from bacteriophages, plasmids, and other sources (1–4). Foreign nucleic acids are targeted by direct hybridization of small CRISPR RNAs (crRNAs), which act in conjunction with conserved Cas proteins to mediate cleavage of the target. Interestingly, there is evidence that CRISPR-Cas components are up-regulated in the presence of bacteriophages or due to perturbations in the cell envelope (5–7), suggesting that CRISPR-Cas systems are induced in response to envelope stresses. Despite this up-regulation, it is unknown whether CRISPR-Cas systems function to counteract the stresses occurring at the envelope.
We demonstrated a role for components of a type II-B CRISPR-Cas system, which are encoded predominantly in pathogens and commensals (8–10), in the regulation of a membrane lipoprotein produced by the intracellular pathogen Francisella novicida (11). Through the action of the RNA-directed endonuclease Cas9 and two small RNAs, tracrRNA and scaRNA, the transcript for a bacterial lipoprotein (BLP; FTN_1103) is targeted and its stability altered, resulting in a decrease in protein production (SI Appendix, Fig. S1) (11). As this is the only known direct and natural example of CRISPR-Cas–mediated endogenous gene regulation, the F. novicida type II-B CRISPR-Cas system represents an important model to understand how these common prokaryotic genetic elements can act as regulators to control microbial physiology.
F. novicida is capable of causing disease in a number of mammalian species, including humans (12–14). During infection, F. novicida must resist the action of numerous antimicrobials that are present on mucosal surfaces and within phagosomes of innate immune cells such as macrophages (15). Compared with other Gram-negative species, F. novicida is highly resistant to the effects of several antimicrobials, including cationic antimicrobial peptides that disrupt bacterial membranes causing lysis and death (16–18). These cationic antimicrobial peptides act similarly to polymyxin antibiotics which are often used as surrogates for their study, and F. novicida is also extremely resistant to polymyxins. Following phagocytosis by macrophages, F. novicida escapes the phagosome and replicates to high titers in the cytosol (19). Throughout this cycle, the macrophage employs numerous pattern recognition receptors to respond to F. novicida infection. This includes the BLP receptor Toll-like receptor 2 (TLR2), present at both the plasma membrane and in the phagosome, which initiates a proinflammatory response (20). Additionally, F. novicida can be recognized in the host cytosol by the absent in melanoma 2/apoptosis associated speck-like protein containing a CARD (AIM2/ASC) inflammasome (21–23). This protein complex triggers activation of the cysteine protease caspase-1, which mediates an inflammatory host cell death. Cell death results in the loss of the intracellular replicative niche for F. novicida, and as such, plays an important role in controlling infection. Because both TLR2 and the AIM2/ASC inflammasome are important for host defense against F. novicida infection, dampening the activation of these innate signaling pathways is critical for F. novicida pathogenesis (24–26).
We initially sought to identify genes that allow F. novicida to resist antimicrobials, using polymyxin for these studies. Surprisingly, we identified the CRISPR-Cas gene cas9 as being required for F. novicida resistance to this membrane-targeting antibiotic. We subsequently found that tracrRNA and scaRNA, two small RNAs that function with Cas9, were also necessary for polymyxin resistance, and that this process was dependent on their ability to repress production of the FTN_1103 BLP. We further observed that this regulation was critical for the enhancement of envelope integrity, which facilitated resistance to other antimicrobials as well. This process also occurred during infection of host cells and subsequently dampened AIM2/ASC inflammasome activation. The importance of Cas9-mediated evasion of the inflammasome, as well as evasion of TLR2, in F. novicida pathogenesis was highlighted by the demonstration that the cas9 deletion mutant was rescued for virulence in mice lacking both ASC and TLR2, but not either component alone. Thus, the work presented here demonstrates that CRISPR-Cas systems are capable of enhancing the integrity of the bacterial envelope, a previously unappreciated role in bacterial physiology. This promotes resistance to antimicrobials and, during infection, facilitates the evasion of multiple innate defense pathways. This represents a previously unappreciated CRISPR-Cas function that is likely relevant to numerous bacteria, including pathogenic and commensal species.
Results
Cas9 Regulatory Axis Promotes Enhancement of Envelope Integrity.
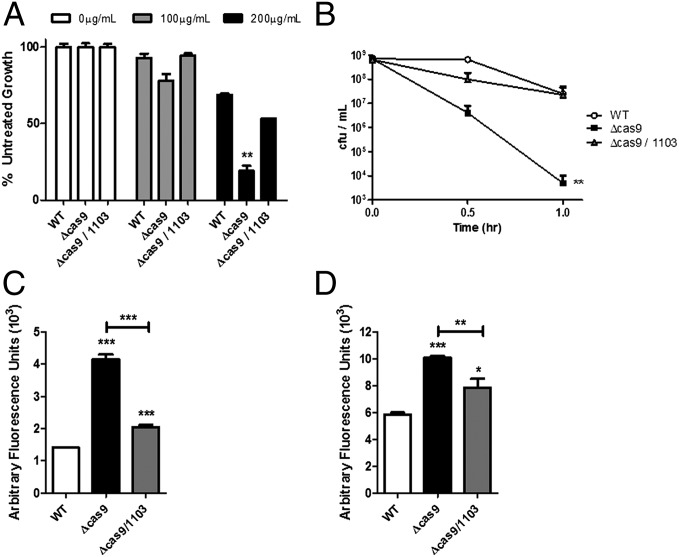
We sought to determine if CRISPR-Cas systems could enhance bacterial membrane integrity, because they are known to be up-regulated in response to nucleic acid transfer events associated with envelope stress. A genetic screen for determinants of F. novicida resistance to the membrane targeting antibiotic polymyxin B (details of which can be found in the SI Appendix, Figs. S2 and S3 and Tables S1–S3) identified the gene encoding the CRISPR-Cas endonuclease Cas9 (FTN_0757). The cas9 mutant was significantly hindered in its ability to grow in the presence of polymyxin, even at doses that had little effect on the growth of WT bacteria (Fig. 1A and SI Appendix, Fig. S4), and was also unable to resist a lethal dose of this antimicrobial (Fig. 1B). This defect could be successfully complemented by restoration of the cas9 gene to the deletion mutant (SI Appendix, Fig. S5). In contrast, mutants lacking cas1, cas2, cas4, or the CRISPR locus were not defective in their ability to survive in the presence of polymyxin B (SI Appendix, Fig. S6). Resistance to polymyxin is often mediated by alterations to the structure of lipid A in the outer membrane and an increase in surface charge. However, the cas9 deletion mutant produced a lipid A identical to WT cells (SI Appendix, Fig. S7) and had similar total surface charge (SI Appendix, Fig. S8). Together, these data clearly demonstrate the importance of Cas9 in the enhancement of resistance against a membrane-damaging antibiotic through a mechanism independent of lipid A modifications.
Fig. 1.
The Cas9 regulatory axis is necessary for polymyxin resistance. (A) WT, cas9, or cas9/1103 deletion mutants were grown overnight in broth culture containing the indicated concentration of polymyxin B. Percent growth compared with untreated cultures is plotted (n = 3). (B) 109 cfu of WT, cas9, or cas9/1103 deletion mutants were treated with 800 µg/mL of polymyxin B, and cfu were enumerated at the indicated times to quantify antimicrobial killing. (C and D) WT, cas9, or cas9/1103 deletion mutants were grown to midlog phase, washed, and stained with (C) propidium iodide or (D) ethidium bromide, and fluorescence was measured (n = 3). *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.001.
Because F. novicida Cas9 functions with two small RNAs (tracrRNA and scaRNA; the three components are together referred to as the Cas9 regulatory system) to regulate an endogenous transcript encoding a membrane-localized BLP (FTN_1103) (11), we tested whether mutants lacking these small RNAs had a diminished ability to grow in the presence of polymyxin. tracrRNA and scaRNA deletion mutants exhibited an increase in susceptibility, similar to the cas9 deletion strain (SI Appendix, Fig. S9 A and B). Furthermore, deletion of FTN_1103 from the cas9, tracrRNA, or scaRNA mutants restored their resistance to polymyxin to near WT levels (Fig. 1A and SI Appendix, Fig. S9 A and B). In addition, we observed that the Cas9 regulatory axis mutants displayed a modest increase in susceptibility to the nonionic surfactant, Triton-X, but not hydrogen peroxide (SI Appendix, Figs. S10 and S11). We further found that these strains were more susceptible to streptomycin and kanamycin, first-line choices for treatment of Francisella infection (27) (SI Appendix, Figs. S12 and S13), in a manner dependent on overproduction of FTN_1103. These surprising observations suggest that the regulatory action of these CRISPR-Cas components promotes resistance to multiple antimicrobials through regulation of FTN_1103.
Because we observed a marked defect in antimicrobial resistance, we sought to address whether Cas9, tracrRNA, and scaRNA promoted resistance by enhancing the integrity of the bacterial envelope. We therefore directly analyzed the permeability of the cas9 deletion mutant by measuring its uptake of propidium iodide (PI), which fluoresces when bound to nucleic acid. The cas9 deletion mutant demonstrated a limited, yet significant, increase in fluorescence compared with WT bacteria, indicating that it is more permeable to PI (Fig. 1C). Importantly, similar levels of colony-forming units were recovered from the mutant and WT bacteria during this experiment (SI Appendix, Fig. S14), and we observed no significant difference in the ability of the strains to grow in rich or minimal media (SI Appendix, Fig. S15), together indicating that although envelope permeability was altered, bacterial viability was unaffected. As a further proof of principle, we performed similar experiments with the nucleic acid-staining dye ethidium bromide (EtBr) and observed a near-identical increase in fluorescence in the cas9 mutant (Fig. 1D). Comparable effects were observed in both the tracrRNA and scaRNA deletion mutants (SI Appendix, Fig. S16), which also did not display an observable defect during growth in broth (SI Appendix, Fig. S15). Furthermore, the increased permeability of all three mutant strains could be restored to near WT levels through deletion of FTN_1103 (Fig. 1 C and D and SI Appendix, Fig. S16), demonstrating that overproduction of this envelope lipoprotein results in decreased envelope integrity. Thus, the Cas9 regulatory axis acts to directly enhance envelope integrity in part through regulation of a BLP and thereby mediates resistance to multiple antimicrobials.
Cas9 Regulatory Axis Promotes Enhanced Bacterial Integrity During Intracellular Infection.
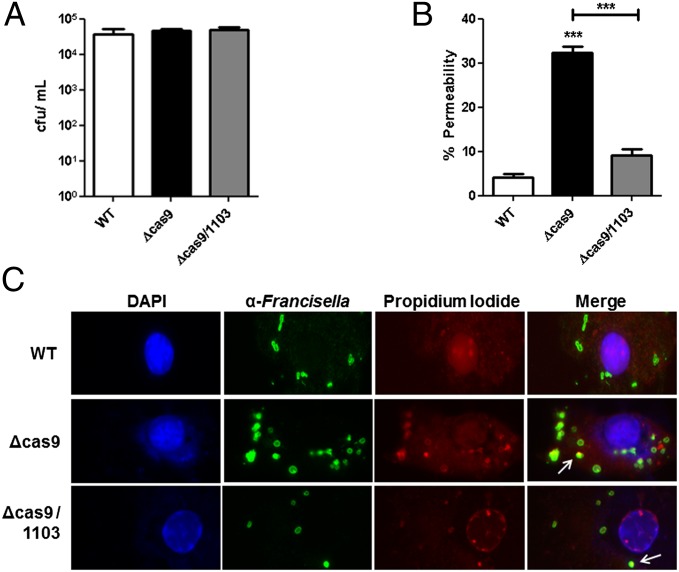
Because these data demonstrated a role for CRISPR-Cas components in enhancing envelope integrity during growth in broth culture, we examined whether they were necessary for a similar function during infection of macrophages, an important replicative niche for F. novicida. Importantly, Cas9 regulatory axis mutants and double mutants lacking FTN_1103 survived and replicated to WT levels in macrophages (Fig. 2A and SI Appendix, Fig. S17A). However, during intracellular infection we observed that cas9, tracrRNA, and scaRNA deletion mutants displayed an almost 10-fold increase in PI staining, a measure of membrane permeability (Fig. 2 B and C and SI Appendix, Fig. S17 B and C). Additionally, intracellular permeability to PI was dependent on FTN_1103, further demonstrating the importance of repression of this membrane lipoprotein for the enhancement of envelope stability during infection of host cells (Fig. 2 B and C and SI Appendix, Fig. S17 B and C).
Fig. 2.
Cas9 is necessary for enhanced envelope integrity during intracellular infection. (A) Bone marrow-derived macrophages were infected with WT, cas9, or cas9/1103 deletion mutants at a multiplicity of infection (MOI) of 20:1 (bacteria per macrophage). At 4 h postinfection, macrophages were lysed and plated to enumerate colony-forming units. (B and C) Macrophages were infected as above, and at 4 h postinfection, were permeabilized with saponin and stained with anti-Francisella antibody (green), propidium iodide (nucleic acids, red), and DAPI (DNA, blue). Colocalization was determined as no less than 50% PI overlap with anti-Francisella. One thousand bacteria were counted per strain and quantified in B. Representative fluorescence micrographs are shown in C. Arrows indicate representative PI and anti-Francisella colocalization. Data are representative of at least three independent experiments in A, whereas B and C are compiled from four independent experiments. ***P ≤ 0.001.
Cas9, tracrRNA, and scaRNA Are Required for Evasion of Inflammasome Activation.
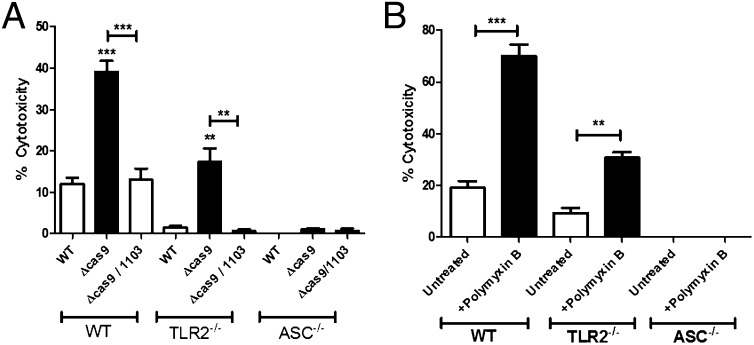
Because we observed an increase in the permeability of Cas9 regulatory axis mutants during intracellular infection, we sought to determine if the lack of enhanced membrane integrity might correlate with increased recognition of bacterial components by host cytosolic receptors that activate innate immune signaling pathways. Francisella is recognized in the cytosol by the AIM2 inflammasome, which contains the adaptor protein ASC, and is partially activated in a TLR2-dependent manner (21–23, 28). Inflammasome activation leads to an inflammatory host cell death and loss of Francisella’s intracellular replicative niche. To determine if the loss of envelope integrity in the Cas9 regulatory axis mutants could result in an inability to dampen inflammasome activation, we measured cell death following infection of bone marrow-derived macrophages. Mutants lacking cas9, tracrRNA, or scaRNA (but not other components of the CRISPR-Cas system) displayed significantly higher levels of cytotoxicity than WT bacteria (Fig. 3A and SI Appendix, Figs. S18 and S19), through a signaling pathway that was partially dependent on TLR2 and completely dependent on ASC (Fig. 3A). We further found that in the absence of FTN_1103, cytotoxicity decreased to near WT levels (Fig. 3A and SI Appendix, Fig. S18), demonstrating that dysregulation of the FTN_1103 BLP is indeed the primary factor responsible for the increased activation of ASC-dependent cell death in the Cas9 regulatory axis mutants.
Fig. 3.
Cas9 and enhanced envelope integrity promote evasion of inflammasome activation. (A) WT, TLR2−/−, and ASC−/− bone marrow-derived macrophages were infected with WT, cas9, or cas9/1103 deletion mutants at a multiplicity of infection (MOI) of 20:1 (bacteria per macrophage). At 5.5 h postinfection, cells were assayed for cytotoxicity using the lactate dehydrogenase release assay (n = 3). (B) WT bacteria were untreated or pretreated for 30 min with 40 µg/mL polymyxin B and subsequently used to infect macrophages, and cytotoxicity was measured as in A (n = 3). Data are representative of at least three independent experiments. **P ≤ 0.005; ***P ≤ 0.001.
To directly address whether loss of envelope integrity could lead to increased inflammasome activation, we treated WT bacteria with a sublethal dose of polymyxin B. Although this dose did not result in a loss of cellular viability (SI Appendix, Fig. S20A), it resulted in an increase in envelope permeability as measured by EtBr staining (SI Appendix, Fig. S20B), similar in magnitude to that observed in the cas9 deletion mutant (Fig. 1C). Upon infection of macrophages, WT bacteria pretreated with polymyxin B showed significantly more cytotoxicity than untreated bacteria in a manner that was partially TLR2-dependent and completely ASC-dependent (Fig. 3B), similar to the cell death elicited by the cas9 deletion mutant (Fig. 3A). Thus, these data directly show that loss of envelope integrity can lead to increased inflammasome activation. Along with both the increased permeability and cytotoxicity of Cas9 regulatory axis mutants, these data demonstrate that Cas9-dependent enhancement of envelope integrity acts to promote evasion of the inflammasome.
The cas9 Mutant Is Rescued for Virulence in ASC/TLR2-Deficient Mice.
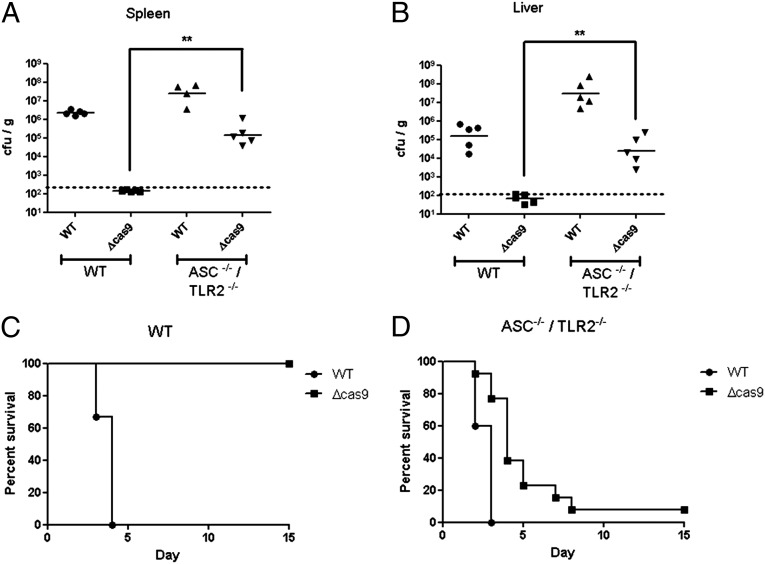
cas9 deletion mutants are severely attenuated and unable to cause lethal infection in mice (11). However, the cause of this attenuation in vivo is not clear. Because Cas9 is important for evasion of both the inflammasome and TLR2, we tested whether the cas9 mutant was rescued for virulence in the absence of these innate inflammatory pathways. Mice lacking ASC alone were able to control infection by the cas9 deletion mutant, since the bacteria were undetectable in the spleen following infection (SI Appendix, Fig. S21A) and were unable to cause morbidity in these mice (SI Appendix, Fig. S21B). Similarly, mice lacking TLR2 alone were also capable of controlling infection by the cas9 deletion mutant (SI Appendix, Fig. S21 A and C). We therefore generated mice lacking both of these innate immune proteins, and infection of macrophages derived from these mice validated that the induction of both cell death and the inflammatory cytokine response by the cas9 deletion mutant were completely abrogated (SI Appendix, Fig. S22 A and B). Strikingly, during infection of these mice, the cas9 deletion mutant was significantly rescued for survival and replication (Fig. 4 A and B and SI Appendix, Fig. S21A). The level of the cas9 mutant increased at least 3 logs in the spleen and 2–3 logs in the liver (above the limit of detection) of infected ASC/TLR2-deficient mice, reaching the levels of WT bacteria observed in WT mice (Fig. 4 A and B). This robust increase in bacterial burden correlated with mortality, because >90% of infected ASC/TLR2-deficient mice succumbed to infection with the cas9 deletion mutant (Fig. 4 C and D). This increase in virulence of the cas9 mutant in ASC/TLR2-deficient mice highlights the essential role that Cas9 plays in facilitating the evasion of two distinct and critical host innate immune receptors, providing further evidence of the important roles that CRISPR-Cas systems can play in bacterial pathogenesis.
Fig. 4.
A cas9 deletion mutant is rescued for virulence in mice lacking both ASC and TLR2. (A and B) WT or ASC/TLR2-deficient mice were inoculated s.c. with 105 cfu of WT or the cas9 deletion strain. Forty-eight hours postinfection, the (A) spleen and (B) liver were harvested and plated to quantify bacterial levels (n = 5). (C and D) Groups of 15 (C) WT or (D) ASC/TLR2-deficient mice were inoculated s.c. with 108 cfu of WT or cas9 deletion strains. Mice were monitored for survival over 15 d. Data are representative of at least two independent experiments in A and B; data are compiled from three independent experiments for C and D. **P ≤ 0.005.
Discussion
Here, we demonstrate that the CRISPR-Cas endonuclease Cas9, working in conjunction with tracrRNA and scaRNA, is critical for enhancing the stability of the bacterial envelope and promoting resistance to polymyxin B, as well as other antibiotics. Expression of CRISPR-Cas components can be induced by bacterial envelope stress, disruptions in envelope protein localization (5), the presence of bacteriophage (6, 7), and during infection of host cells (11, 29, 30). Taken together, this suggests that CRISPR-Cas systems are induced in response to membrane stressors, and their regulatory activity can subsequently result in the enhancement of envelope integrity to promote resistance to such stressors. It is therefore tempting to speculate that the CRISPR-Cas response to envelope stress serves two distinct purposes: (i) the activation of its canonical function as the adaptive, foreign nucleic acid restriction system and (ii) the regulation of envelope structure and content to enhance the integrity of the bacterial envelope and combat membrane stress, which represents a previously unappreciated role in bacterial physiology and a shift in the understanding of these systems.
Our data demonstrate a role for CRISPR-Cas systems in promoting antibiotic resistance, whereas previous studies have focused instead on their ability to limit this process by restricting the acquisition of mobile elements, including those which carry resistance cassettes. Studies in several bacterial species revealed a correlation between increased antibiotic resistance and nonfunctional CRISPR-Cas systems (31–33). In fact, it has been demonstrated that acquisition of resistance traits can be restricted by CRISPR-Cas systems in vivo (34). In contrast, the data presented here suggest that CRISPR-Cas systems with regulatory functions may provide bacteria with the capacity to resist certain antibiotics. Thus, loss of these systems in antibiotic-resistant species may have unappreciated regulatory effects leading to altered bacterial physiology (i.e., envelope structure) and enhanced susceptibility to certain antibiotics. Delineating the regulatory functions of CRISPR-Cas systems in diverse bacteria will be required to more broadly assess their potential roles as antibiotic resistance determinants.
During infection, the ability of CRISPR-Cas systems to enhance envelope integrity has important ramifications for the virulence of F. novicida. We demonstrate here that Cas9 regulatory axis-mediated envelope enhancement is necessary to inhibit activation of the inflammasome and host cell death. This is broadly in agreement with the idea that mutant strains with membrane defects induce increased levels of inflammasome activation (24). Furthermore, we directly demonstrate that an increase in envelope permeability induced by polymyxin B treatment leads to enhanced inflammasome activation. Because the AIM2/ASC inflammasome responds to DNA released from Francisella, it is likely that increased envelope stability serves to prevent the release of nucleic acid, thereby subverting inflammasome activation (21, 23, 24). It has been posited that the AIM2/ASC inflammasome has a low threshold for activation, perhaps requiring only a single bacterium to release DNA (24). Therefore, small changes in envelope integrity may have drastic effects on inflammasome activation, while not having any observable effects on a bacterial population’s viability as a whole. The regulation of BLP expression by the Cas9 regulatory axis thus limits the levels of this TLR2 ligand and subsequent activation of TLR2 (11), as well as promoting enhanced envelope integrity and subversion of the inflammasome. In the absence of both ASC and TLR2, the virulence of the cas9 mutant is significantly restored (Fig. 4 A, B, and D), demonstrating the importance of Cas9-mediated innate immune evasion in the ability of F. novicida to cause disease.
Although F. novicida is the only known bacterial species in which Cas9 plays a clearly demonstrated regulatory role, it is likely that Cas9-dependent regulation contributes to the virulence of other pathogens encoding this protein including Streptococcus spp., Legionella pneumophila, Listeria monocytogenes, Staphylococcus aureus, and Haemophilus parainfluenzae (8, 9, 11, 29). In fact, a role for Cas9 in controlling virulence traits has been demonstrated in Neisseria meningitidis and Campylobacter jejuni. Each has been observed to require Cas9 for both invasion and replication in eukaryotic cells (11, 35). In addition, both of these species require Cas9 to attach to host cells, further supporting the hypothesis that CRISPR-Cas systems can have effects on the bacterial envelope (11, 35). Interestingly, we have additionally observed defects in the C. jejuni envelope in the absence of Cas9. A cas9 deletion mutant in C. jejuni displays an increase in envelope permeability, similar to that observed in F. novicida (SI Appendix, Fig. S23A), and is significantly more sensitive to erythromycin, a first-line treatment for invasive campylobacteriosis (SI Appendix, Fig. S23B) (36). Therefore, although it is yet unknown how Cas9 may function as a regulator in C. jejuni, it is clear that these findings represent a broader role for Cas9 systems in modulating this important aspect of bacterial physiology.
CRISPR-Cas systems have more broadly been linked to other processes that involve the bacterial envelope and extracellular structures. For instance, the type I CRISPR-Cas system in Pseudomonas aeruginosa is capable of modulating biofilm formation (37, 38), and the type I system in Myxococcus xanthus is an essential component in regulating the development of fruiting bodies (39–41). These examples provide further support for a broader CRISPR-Cas function in the modification and regulation of the envelope and extracellular structures, extending beyond those organisms that encode Cas9. This unappreciated role for CRISPR-Cas systems would allow the myriad bacterial species encoding them to respond to envelope stresses that occur as a result of not only bacteriophage attack but also infection of host cells and exposure to other environmental conditions.
Experimental Procedures
Bacterial Manipulations.
F. novicida strain U112 and all derivatives used in this study were routinely grown at 37 °C with aeration in tryptic soy broth (TSB) supplemented with 0.2% l-cysteine (BD Biosciences), or on tryptic soy agar plates supplemented with 0.1% l-cysteine. Cas9 regulatory axis deletion mutants and complementation strains were described previously (11, 42). FTN_1254 and FTN_0109 mutants were constructed by allelic exchange as described previously (43, 44) using primers in SI Appendix, Table S3.
Polymyxin Treatments.
The indicated strains were grown overnight and subsequently diluted to an OD600 of 0.03 in Mueller-Hinton/cation-adjusted broth with 0.2% l-cysteine containing the specified doses of polymyxin B (USB Corporation). Following overnight growth at 37 °C with aeration, OD600 was measured and used to calculate the percent growth compared with the growth of the strain in media alone. For the killing assay, cultures were treated with 800 µg/mL of polymyxin B, incubated at 37 °C with aeration, and plated for colony-forming units at the indicated time points. For sublethal treatments with polymyxin, bacterial cultures were washed once and resuspended in media containing 40 µg/mL polymyxin B for 30 min. Treated cells were subsequently washed twice before preparing for infections as described below.
In Vitro Permeability.
The indicated strains were grown overnight and subsequently subcultured 1:50 in TSB and grown to an OD600 of ∼0.8–0.9. Cells were washed twice in 50 mM phosphate buffer and resuspended in 50 mM phosphate buffer containing 30 μg/mL EtBr (Fisher Scientific) or 200 μM PI (Life Technologies). Fluorescence was measured immediately in a Biotek Synergy Mx plate reader using an excitation of 250 nm and emission of 605 nm for EtBr or excitation of 534 nm and emission of 617 nm for PI, correcting with samples lacking bacteria.
Macrophage Culture and Infection.
Murine bone marrow-derived macrophages were prepared from WT C57BL/6 mice or the indicated knockout strains and cultured as described previously (42). Macrophages were seeded overnight and infected with overnight cultures of the indicated bacterial strains at a MOI of 20:1 bacteria per macrophage. Plates were centrifuged for 15 min at 335 × g at room temperature to promote bacterial uptake. Infected macrophages were incubated for 30 min at 37 °C and washed twice before adding DMEM containing 10 µg/mL gentamicin.
Intracellular Permeability.
WT murine bone marrow-derived macrophages were seeded onto glass coverslips and infected as above. At 4 h postinfection, macrophages were gently permeabilized for 15 min at room temperature with 0.1% saponin/3% (wt/vol) BSA in PBS. Cells were first stained with 2.6 µM PI and chicken–anti-F. novicida antibody (a kind gift from Denise Monack, Stanford University) for 12 min at 37 °C. Following washing, cells were fixed with 4% (vol/vol) paraformaldehyde and incubated with FITC-labeled anti-chicken antibody. Coverslips were mounted onto glass slides with SlowFade Gold reagent with DAPI (Life Technologies). Slides were imaged on a Zeiss Axioscope Z.1 microscope and a Zeiss Imager 2.1 camera. Images were analyzed with Volocity 5.5 software (Perkin–Elmer). Colocalization was determined by no less than 50% overlap between PI and Francisella-positive cells, and 1,000 cells were counted for each strain.
Cytotoxicity Assays.
Murine bone marrow-derived macrophages prepared from the indicated mice were infected with bacterial strains as described above. At 5.5 h postinfection, supernatants were collected and assayed for levels of lactate dehydrogenase using the nonradioactive cytotoxicity assay kit (Promega).
Murine Infections.
ASC−/− and TLR2−/− C57BL/6 mice were a generous gift from Bali Pulendran, Emory Vaccine Center, Atlanta (with much appreciated assistance from Paul Hakimpour) and were bred together to generate mice deficient in both ASC and TLR2. Mice were bred and kept under specific-pathogen free conditions in filter-top cages at Yerkes National Primate Center, Emory University, and provided food and water ad libitum. For bacterial burden assays, female WT or ASC/TLR2-deficient mice (of 8–10 wk of age) were infected s.c. with 2 × 105 cfu of the indicated bacterial strains in sterile PBS. At 48 h postinfection, liver and spleen were harvested, weighed, and homogenized in PBS, and serial dilutions were plated to enumerate colony-forming units. For survival experiments, mice were infected with 108 cfu s.c. and monitored for signs of illness. Mice were killed when they appeared moribund. All experimental procedures were approved by the Emory University Institutional Animal Care and Use Committee (Protocol #069-2008Y).
Statistics.
Two-tailed, Student t tests were performed to analyze pairs of data as indicated, excluding the experiments in Fig. 4 A and B, which were analyzed with the Mann–Whitney test.
Supplementary Material
Acknowledgments
We thank Emily Crispell, Thayer King, William Shafer, and Eric Skaar for helpful discussions and critical reading of this manuscript. This work was supported by National Institutes of Health (NIH) Grants U54-AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense, R56-AI87673, and R01-AI110701 (to D.S.W., a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease). T.R.S. was supported by the National Science Foundation Graduate Research Fellowship Program and the Achievement Rewards for College Scientists Foundation. J.Z. and P.Z. were supported by NIH R01-AI055588 and GM5-1310.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323025111/-/DCSupplemental.
References
- 1.Richter C, Chang JT, Fineran PC. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems. Viruses. 2012;4(10):2291–2311. doi: 10.3390/v4102291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: Acquisition of new information. Virology. 2012;434(2):202–209. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Rodriguez R, et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol. 2011;79(3):584–599. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quax TE, et al. Massive activation of archaeal defense genes during viral infection. J Virol. 2013;87(15):8419–8428. doi: 10.1128/JVI.01020-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young JC, et al. Phage-induced expression of CRISPR-associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus. PLoS ONE. 2012;7(5):e38077. doi: 10.1371/journal.pone.0038077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10(5):726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res, 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed]
- 10.Schunder E, Rydzewski K, Grunow R, Heuner K. First indication for a functional CRISPR/Cas system in Francisella tularensis. Int J Med Microbiol. 2013;303(2):51–60. doi: 10.1016/j.ijmm.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497(7448):254–257. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hand J, Scott-Waldron C, Balsamo G. Outbreak of Francisella novicida infections among occupants at a long-term residential facility - Louisiana, April-July, 2011. Louisiana Morbidity Rep. 2012;23(1):1–6. [Google Scholar]
- 13.Birdsell DN, et al. Francisella tularensis subsp. novicida isolated from a human in Arizona. BMC Res Notes. 2009;2:1–6. doi: 10.1186/1756-0500-2-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leelaporn A, Yongyod S, Limsrivanichakorn S, Yungyuen T, Kiratisin P. Francisella novicida Bacteremia, Thailand. Emerg Infect Dis. 2008;14(12):1935–1937. doi: 10.3201/eid1412.080435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones CL, et al. Subversion of host recognition and defense systems by Francisella spp. Microbiol Mol Biol Rev. 2012;76(2):383–404. doi: 10.1128/MMBR.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han S, Bishop BM, van Hoek ML. Antimicrobial activity of human beta-defensins and induction by Francisella. Biochem Biophys Res Commun. 2008;371(4):670–674. doi: 10.1016/j.bbrc.2008.04.092. [DOI] [PubMed] [Google Scholar]
- 17.Mohapatra NP, et al. Identification of an orphan response regulator required for the virulence of Francisella spp. and transcription of pathogenicity island genes. Infect Immun. 2007;75(7):3305–3314. doi: 10.1128/IAI.00351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urban C, Tiruvury H, Mariano N, Colon-Urban R, Rahal JJ. Polymyxin-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 2011;55(1):388–389. doi: 10.1128/AAC.01088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjöstedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003;71(10):5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole LE, et al. Phagosomal retention of Francisella tularensis results in TIRAP/Mal-independent TLR2 signaling. J Leukoc Biol. 2010;87(2):275–281. doi: 10.1189/jlb.0909619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandes-Alnemri T, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11(5):385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones JW, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci USA. 2010;107(21):9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rathinam VA, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11(5):395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13(10):1586–1600. doi: 10.1111/j.1462-5822.2011.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones CL, Sampson TR, Nakaya HI, Pulendran B, Weiss DS. Repression of bacterial lipoprotein production by Francisella novicida facilitates evasion of innate immune recognition. Cell Microbiol. 2012;14(10):1531–1543. doi: 10.1111/j.1462-5822.2012.01816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202(8):1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Word Health Organization (2007) WHO Guidelines on Tularaemia. WHO Library Cataloguing-in Publication Data (World Health Organization, Geneva), 978 92 4 154737 6. [Google Scholar]
- 28.Jones CL, Weiss DS. TLR2 signaling contributes to rapid inflammasome activation during F. novicida infection. PLoS ONE. 2011;6(6):e20609. doi: 10.1371/journal.pone.0020609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louwen R, Staals RH, Endtz HP, van Baarlen P, van der Oost J. The role of CRISPR-Cas systems in virulence of pathogenic bacteria. Microbiol Mol Biol Rev. 2014;78(1):74–88. doi: 10.1128/MMBR.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunderson FF, Cianciotto NP. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. MBio. 2013;4(2):e00074–e13. doi: 10.1128/mBio.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer KL, Gilmore MS. Multidrug-resistant enterococci lack CRISPR-cas. MBio. 2010;1(4):e00227–e10. doi: 10.1128/mBio.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang TN, et al. Uropathogenic Escherichia coli are less likely than paired fecal E. coli to have CRISPR loci. Infect Genet Evol. 2013;19:212–218. doi: 10.1016/j.meegid.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 33.Burley KM, Sedgley CM. CRISPR-Cas, a prokaryotic adaptive immune system, in endodontic, oral, and multidrug-resistant hospital-acquired Enterococcus faecalis. J Endod. 2012;38(11):1511–1515. doi: 10.1016/j.joen.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12(2):177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Louwen R, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barré syndrome. Eur J Clin Microbiol Infect Dis. 2013;32(2):207–226. doi: 10.1007/s10096-012-1733-4. [DOI] [PubMed] [Google Scholar]
- 36.Allos BM. Campylobacter jejuni infections: Update on emerging issues and trends. Clin Infect Dis. 2001;32(8):1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- 37.Cady KC, O’Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193(14):3433–3445. doi: 10.1128/JB.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zegans ME, et al. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191(1):210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J Bacteriol. 2007;189(10):3738–3750. doi: 10.1128/JB.00187-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boysen A, Ellehauge E, Julien B, Søgaard-Andersen L. The DevT protein stimulates synthesis of FruA, a signal transduction protein required for fruiting body morphogenesis in Myxococcus xanthus. J Bacteriol. 2002;184(6):1540–1546. doi: 10.1128/JB.184.6.1540-1546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thöny-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175(22):7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss DS, et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci USA. 2007;104(14):6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brotcke A, et al. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect Immun. 2006;74(12):6642–6655. doi: 10.1128/IAI.01250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anthony LS, Gu MZ, Cowley SC, Leung WW, Nano FE. Transformation and allelic replacement in Francisella spp. J Gen Microbiol. 1991;137(12):2697–2703. doi: 10.1099/00221287-137-12-2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.