Significance
The unpredictable recurrences of prion epidemics, their incurable lethality, and the capacity of animal prions to infect humans provide significant motivation to ascertain the parameters governing disease transmission. The unprecedented spread and uncertain zoonotic potential of chronic wasting disease, a contagious epidemic among deer, elk, and other cervids, is of particular concern. Here we demonstrate that naturally occurring primary structural differences in cervid PrPs differentially impact the efficiency of intra- and interspecies prion transmission. Our results not only deliver information about the role of primary structural variation on prion susceptibility, but also provide functional support to a mechanism in which plasticity of a tertiary structural epitope governs prion protein conversion and intra- and interspecies susceptibility to prions.
Keywords: protein structure, prion replication, protective polymorphisms, structural plasticity
Abstract
Understanding the molecular parameters governing prion propagation is crucial for controlling these lethal, proteinaceous, and infectious neurodegenerative diseases. To explore the effects of prion protein (PrP) sequence and structural variations on intra- and interspecies transmission, we integrated studies in deer, a species naturally susceptible to chronic wasting disease (CWD), a burgeoning, contagious epidemic of uncertain origin and zoonotic potential, with structural and transgenic (Tg) mouse modeling and cell-free prion amplification. CWD properties were faithfully maintained in deer following passage through Tg mice expressing cognate PrP, and the influences of naturally occurring PrP polymorphisms on CWD susceptibility were accurately reproduced in Tg mice or cell-free systems. Although Tg mice also recapitulated susceptibility of deer to sheep prions, polymorphisms that provided protection against CWD had distinct and varied influences. Whereas substitutions at residues 95 and 96 in the unstructured region affected CWD propagation, their protective effects were overridden during replication of sheep prions in Tg mice and, in the case of residue 96, deer. The inhibitory effects on sheep prions of glutamate at residue 226 in elk PrP, compared with glutamine in deer PrP, and the protective effects of the phenylalanine for serine substitution at the adjacent residue 225, coincided with structural rearrangements in the globular domain affecting interaction between α-helix 3 and the loop between β2 and α-helix 2. These structure–function analyses are consistent with previous structural investigations and confirm a role for plasticity of this tertiary structural epitope in the control of PrP conversion and strain propagation.
That prion propagation occurs through corruption of host-encoded cellular prion protein (PrPC) by abnormally conformed PrPSc, where PrPSc refers to the scrapie isoform of PrP, is not only generally accepted, but also increasingly acknowledged as a canonical mechanism of protein-based information transfer. Variant Creutzfeldt–Jakob disease (CJD), resulting from exposure to bovine spongiform encephalopathy, exemplifies prion zoonosis (1), and the emergence of further epidemics, particularly chronic wasting disease (CWD) of deer, elk, and other cervids, raises additional concerns. The unprecedented contagious spread of CWD, the only known prion disease of wild animals, appears inexorable. The disease is currently recognized in 22 US states, the Canadian provinces of Saskatchewan and Alberta, and the Republic of Korea, and the host range among cervid species is also expanding. Although CWD has been experimentally transmitted to noncervid species, the long incubation times and low attack rates in these situations reflect the generally lower efficiencies of interspecies transmission. The efficiency with which prions traverse species is dictated, in part, by the extent to which the primary structures of PrPSc comprising the prion and the PrPC expressed in the infected host are related (2). The transmission characteristics of prions, including time to onset of disease and neuropathological profiles, are measures by which prion strains are defined. Once again PrP structure is implicated, but in this case information is enciphered in the tertiary configuration of PrPSc (3, 4). Underscoring the importance of PrP structure in disease susceptibility, single amino acid variations within species influence susceptibility and disease presentation.
The PrP-coding sequence of white-tailed deer encodes either glutamine (Q) or histidine (H) at codon 95 and glycine (G) or serine (S) at codon 96, and these polymorphisms have been associated with susceptibility to CWD. Mule deer PrP is also polymorphic at codon 225, encoding either S or phenylalanine (F). The immediately adjacent position 226 encodes the singular primary structural difference between Rocky Mountain elk and deer PrP. Elk PrP contains glutamate (E) and deer PrP Q at this position. To analyze the effects of these amino acid variations on CWD propagation and susceptibility to prions from different species, we generated transgenic (Tg) mouse models and incorporated studies in the natural host with cell-free prion amplification and molecular modeling approaches.
Results
Structural Effects of Substitutions at Residues 225 and 226.
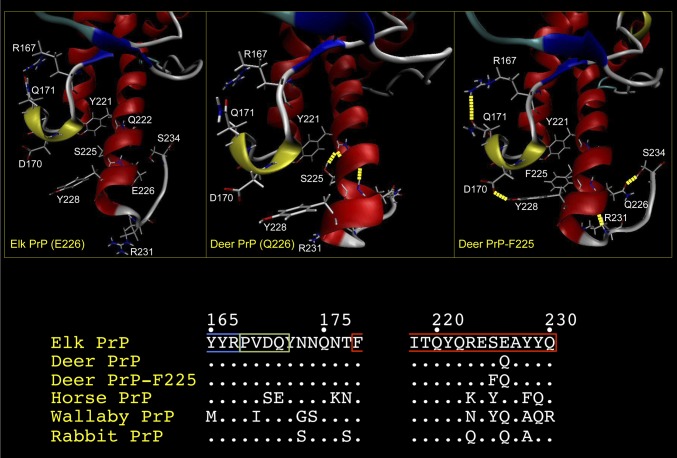
Our previous studies signified a critical role for residue 226 in strain stability (5, 6). Its adjacent proximity to the mule deer PrP residue 225 polymorphism, and the suggested protective role of F225 on CWD in free-ranging deer (7), motivated us to further explore the effects of these substitutions. We performed molecular dynamics (MD) simulations on the effects of these substitutions on the structure of elk PrP derived from NMR spectroscopic analyses (8). All three structures exhibited similar global architectures (Fig. 1). We paid particular attention to the loop region connecting the second strand of the β-sheet (β2) and α-helix 2 (the β2–α2 loop) because it is extremely well defined in elk PrP (8) and has been implicated in modulating interspecies prion transmission (9). In all three structures, residues 168–172 formed the previously reported, distinct 310-helical turn (8). In deer PrP-F225, we observed long-range interactions between the β2–α2 loop and α-helix 3, resulting from hydrogen bonding between tyrosine (Y) at residue 228 and aspartate (D) at residue 170 in the 310-helical turn. In contrast, the spatial separation of D170 from Y228 precluded hydrogen bond formation in deer and elk PrP structures. The distances between these two residues are 3.8 and 3.3 Å in elk and deer PrP, respectively, and conformational change resulting from occupancy of F at residue 225 reduces this distance to 1.9 Å, sufficiently close for the hydrogen bond formation. This rearrangement is consistent with a concomitant increase in the stability of the discontinuous epitope formed by the β2–α2 loop and α-helix 3. In addition, hydrogen bonding of Q226 with two individual carbonyls in the main chain of the C-terminal loop occurred in deer PrP-F225, but not in elk or deer PrP.
Fig. 1.
Structural comparisons of elk PrP, deer PrP, and deer PrP-F225. Average conformations showing both the secondary structure and the backbone structure of elk PrP (E226), deer PrP (Q226), and deer PrP-F225 obtained from 5-ns MD trajectories. Images of structures were generated using VMD. The figure focuses on the region of the protein encompassing the β2–α2 loop and α-helix 3. α-Helices are colored red; 310 helices, apple green; β-sheets, blue; turns, cyan; and coils, white. Hydrogen bonds are indicated as dashed yellow lines. Amino acid residues involved in structural differences between the three PrPs, or that vary between them, are shown in white. Only the side chains for these residues are shown. (Lower) The primary structures in the region of the β2–α2 loop and α-helix 3 of elk, deer, and deer-F225 as well as PrP from species analyzed in previous structural studies are shown.
Transgenic Modeling of the Effects of Residues 225 and 226 on Prion Pathogenesis.
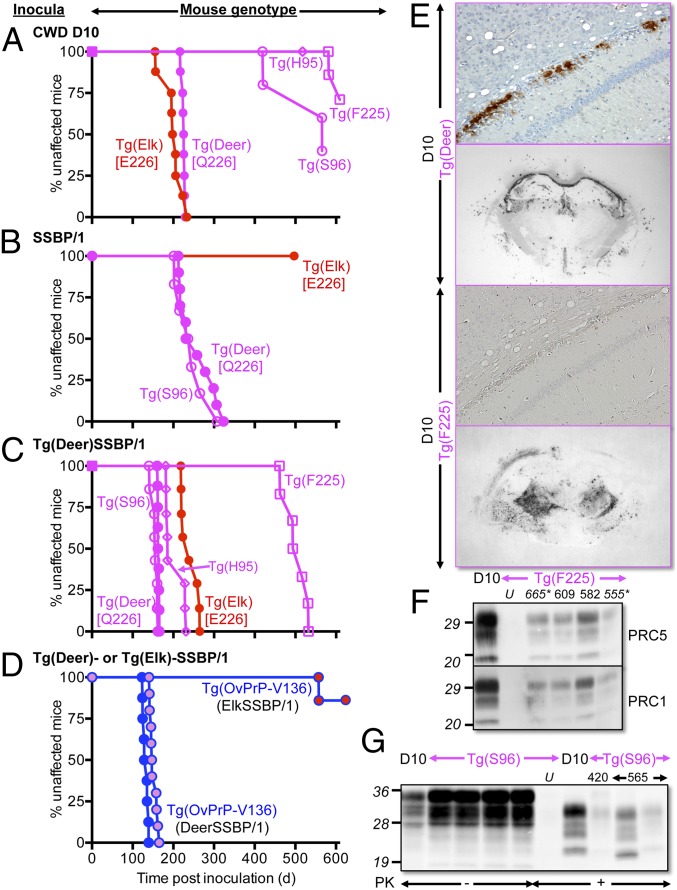
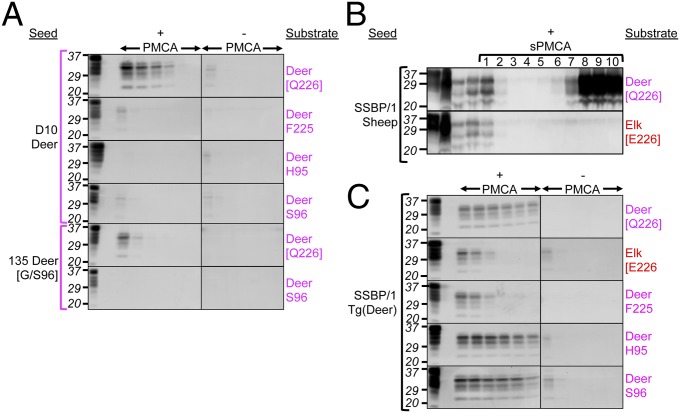
To address the effect of residue 226 on the propagation of prions from a different species, we inoculated mice with SSBP/1, a frequently used experimental source of sheep scrapie (reviewed in ref. 10), which has been used in several related transgenic studies (11–13). Whereas we previously showed that SSBP/1 prions caused disease in Tg(DeerPrP)1536+/− mice with incubation times similar to CWD (11), in sharp contrast, SSBP/1-inoculated Tg(ElkPrP)5037+/− mice failed to develop neurological signs up to ∼500 d post inoculation (Fig. 2B and Table S1). Because serial protein misfolding cyclic amplification (sPMCA), an in vitro technique to amplify prions, has been used to overcome prion species barriers in vitro (14), we used it to address the relative efficiencies of deer and elk PrPC to support propagation of sheep prions. SSBP/1 was diluted 10-fold into 10% brain homogenates prepared from perfused Tg(DeerPrP)1536+/− or Tg(ElkPrP)5037+/− mice. Following a round of PMCA consisting of alternating periods of sonication and incubation for 36 cycles, the product was diluted 10-fold into another tube containing Tg(DeerPrP)1536+/− or Tg(ElkPrP)5037+/− brain homogenate for a further round of PMCA. This process was repeated for 10 rounds. Consistent with a species barrier (11), PrPSc was not produced in early rounds of sPMCA using deer PrPC as substrate, but was detected by round 6 with a maximum plateau occurring between rounds 8 and 10 (Fig. 3B), whereas SSBP/1 adapted in Tg(DeerPrP)1536+/− mice amplified deer PrPC within one round of PMCA, even at high seed dilutions (Fig. 3C). By comparison, elk PrPC failed to amplify SSBP/1 during any round of sPMCA (Fig. 3B). Resistance to SSBP/1 was overcome when Tg(ElkPrP)5037+/− mice were inoculated with SSBP/1 passaged in Tg(DeerPrP)1536+/− mice (Fig. 2C and Table S1), an effect that was recapitulated during PMCA (Fig. 3C). Effects of residue 226 were also observed during passage of SSBP/1 from Tg(DeerPrP)1536+/− or Tg(ElkPrP)5037+/− mice to Tg(OvPrP-V136)4166+/− mice expressing sheep PrP with the valine (V) polymorphism at residue 136 (12). Whereas SSBP/1 from Tg(DeerPrP) mice produced complete attack rates, and mean incubation times that were only 14% longer than SSBP/1 from sheep (Fig. 2D and Table S1), the majority of Tg(OvPrP-V136)4166+/− mice inoculated with SSBP/1 from Tg(ElkPrP) mice remained free of disease for 630 d (Fig. 2D and Table S1).
Fig. 2.
Transgenic modeling of cervid PrP polymorphisms. (A–D) Survival curves of various inoculated Tg mice. (D) Tg(OvPrP-V136) mice expressing sheep PrP with V at residue 136. Red filled circles, SSBP/1 passaged in Tg(ElkPrP); magenta filled circles, SSBP/1 passaged in Tg(DeerPrP); blue filled circles, SSBP/1. Graphs in A–D have the same timescales. (E) PrPSc deposition in the region of the hippocampus and thalamus of D10-infected Tg(Deer) and Tg(F225) mice assessed by immunohistochemistry and histoblotting. (F and G) Western blots showing PrPSc in brains of diseased D10-infected Tg(F225) and Tg(S96) mice. Numbers above lanes in F and G refer to days post inoculation when mice were killed. Asterisks indicate asymptomatic mice. U, uninfected mice. In F and G, “D10” indicates D10 CWD brain. Western blots were probed with mAbs PRC5 and PRC1 as indicated.
Fig. 3.
Effects of cervid PrP substitutions on PMCA of CWD and SSBP/1. (A and C) PMCA using PrPC substrates from Tg mice. (A) Brain extracts from D10 deer or G/S96 deer 135 were used as seeds. (C) SSBP/1 adapted in Tg(DeerPrP) mice was used as seed. “+PMCA” and “–PMCA” indicate whether or not samples were subject to sonication. The first two lanes of the +PMCA panels are brain homogenates from uninfected Tg mice, untreated or treated with PK, respectively. All other samples were PK-treated. (B) Serial PMCA using PrPC from Tg(DeerPrP) or Tg(ElkPrP) mice seeded with sheep SSBP/1 prions. Numbers refer to rounds of serial PMCA. (Upper and Lower) The left four lanes contain undigested substrate; undigested SSBP/1; PK-treated SSBP/1; and unsonicated material from round 1. All other samples were PK-treated. Blots were probed with mAb 6H4.
To address the effects of residue 225, we produced Tg(DeerPrP-F225)5107+/−. Inoculation with D10 CWD prions produced disease in only two mice after 582 and 609 d (Fig. 2A). Although Western blotting confirmed the presence of PrPSc in the brains of these mice, as well as two mice with ambiguous clinical signs (Fig. 2F), levels were decreased compared with diseased Tg(DeerPrP)1536+/− mice. Histoblots showed discernibly different global patterns of PrPSc deposition in the brains of diseased D10-inoculated Tg(DeerPrP-F225)5107+/− and Tg(DeerPrP)1536+/− mice (Fig. 2E). Consistent with our previous studies (5), immunohistochemically (IHC) stained sections of D10-infected Tg(DeerPrP)1536+/− mice were characterized by continuous, symmetrical plaque deposits of PrPSc throughout the hippocampal alveus. This pattern did not occur in D10-inoculated Tg(DeerPrP-F225)5107+/− mice; instead, PrPSc staining was diffuse and more widespread with histoblots showing deposition in the region of the thalamus, not the corpus callosum. When seeded with D10 CWD, DeerPrPC-F225 had impaired PMCA efficiency compared with wild-type PrPC (Fig. 3A). This barrier to CWD transmission did not appear to result from mismatched primary structures at residue 225 between PrPSc and PrPC because CWD prions from brain extracts of diseased Tg(DeerPrP-F225)5107+/− mice or a diseased mule deer heterozygous at 225, referred to as 04–7951, failed to cause disease in Tg(DeerPrP-F225)5107+/− mice. These sources did, however, transmit to Tg(DeerPrP)1536+/− mice (Table S2).
Transgenic Modeling of Additional Cervid PrP Polymorphisms.
All D10 CWD-inoculated Tg(DeerPrP-H95)7505+/− mice remained free of clinical signs for ∼520 d (Fig. 2A and Table S2). Attack rates were incomplete when Tg(DeerPrP-S96)7511+/− mice were challenged, and incubation times of mice that did succumb were >300 d longer than the mean D10 CWD incubation time in Tg(DeerPrP)1536+/− mice (Fig. 2A and Table S2). Variable amounts of PrPSc accumulated in the brains of the three diseased Tg(DeerPrP-S96)7511+/− mice killed between 420 and 565 d (Fig. 2G). Interestingly, brain extracts of one mouse that succumbed to disease after 565 d had the highest levels of protease-resistant PrP, which migrated on Western blots more rapidly than the other samples, including the D10 inoculum. PMCA using deer PrPC-H95 or deer PrPC-S96 as substrates also failed to produce PrPSc even at low CWD dilutions (Fig. 3A). All PMCA reactions were normalized to contain equal amounts of mutant and wild-type PrP substrates.
Effects of the S96 and H95 polymorphisms on SSBP/1 propagation were less severe. To minimize effects of primary structural differences between sheep and cervid PrP, we challenged Tg mice with SSBP/1 passaged in Tg(DeerPrP) mice. Disease onset in Tg(DeerPrP-S96)7511+/− mice was slightly (5%), but significantly (P ≤ 0.05), more rapid than in Tg(DeerPrP)1536+/− mice, whereas inoculation of Tg(DeerPrP-H95)7505+/− mice resulted in an ∼25% prolonged mean incubation time (P ≤ 0.05). Direct inoculation of Tg(DeerPrP-S96)7511+/− mice with SSBP/1 from sheep produced survival curves and mean incubation times indistinguishable from SSBP/1-inoculated Tg(DeerPrP)1536+/− mice (Fig. 2B and Table S2). Consistent with these in vivo effects, although the S96 and H95 substitutions had negligible effects on conversion to PrPSc by PMCA, the F225 and E226 substitutions resulted in reduced PMCA efficiency compared with wild-type deer PrPC (Fig. 3C).
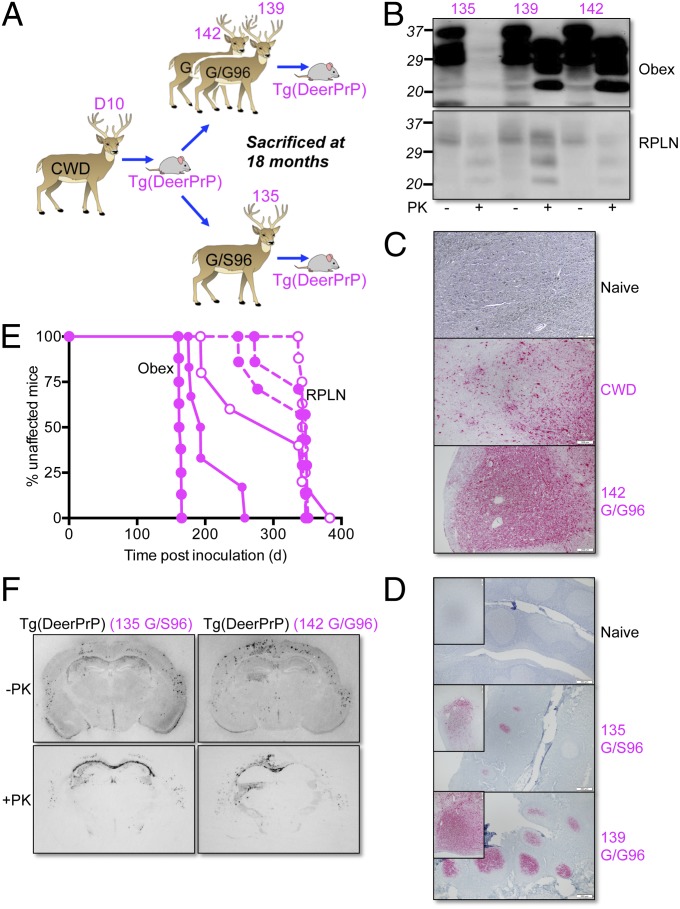
Transgenic Mice Recapitulate Prion Pathogenesis in the Natural Host.
Three white-tailed deer were inoculated with brain material from Tg(DeerPrP)1536+/− mice that succumbed to infection with D10 CWD (15) (Fig. 4A). Deer 135 was heterozygous G/S at residue 96, and deer 139 and 142 were homozygous G/G. Tonsil biopsies performed at 3-mo intervals showed deer 139 and 142 to be PrPSc-positive at 6 mo and beyond, whereas deer 135 remained negative to the 12-mo time point. By termination at 18 mo, there was abundant PrPSc deposition in tonsil follicles and obex of deer 139 and 142, but appreciably less PrPSc in both tissues from deer 135 (Fig. 4 B and D). Deposition patterns of PrPSc in the obex of wild-type deer were equivalent to deer inoculated with a natural case of CWD (Fig. 4C). When passaged to Tg(DeerPrP)1536+/− mice, prions from the obex of the 96G/S 135 deer produced ∼100-d-longer mean incubation times and wider variances of incubation times than samples from inoculated wild-type deer (P ≤ 0.05 in both cases) (Fig. 4E and Table S3). Histoblotting showed comparable PrPSc deposition patterns of prions originating from the obex of the wild-type 142 deer and G/S96 135 deer (Fig. 4F), suggesting that the longer incubation periods of prions from G/S96 135 deer were not the result of altered strain properties. Retropharyngeal lymph node (RPLN) samples from all deer had similar PrPSc levels, regardless of genotype, albeit substantially lower than PrPSc in the brains of wild-type deer (Fig. 4B). Accordingly, incubation times of RPLN materials in Tg(DeerPrP)1536+/− mice were consistently longer (Fig. 4E and Table S3). Inoculated Tg(DeerPrP-S96)7511+/− and Tg(DeerPrP-H95)7505+/− remained free of clinical signs for >530 d when inoculated with obex material from the G/S96 135 deer (Table S2).
Fig. 4.
Transmission of Tg mouse-adapted CWD prions to deer. (A) Passage history of CWD from D10 CWD deer to Tg(DeerPrP)1536+/− mice and thence to hand-raised white-tailed deer. (B) Western blots showing levels of total PrP and PrPSc in obex and RPLN of autopsied deer at 18 mo. Fifty micrograms of total protein from obex of 139 and 142 was digested; for 135, the amount was 500 μg. Western blots were probed with mAb 6H4. (C) PrPSc deposition patterns in obex assessed by immunohistochemistry. Naive, uninfected deer; CWD, deer infected with naturally occurring CWD; 142 G/G96, deer infected with Tg(DeerPrP)-passaged D10 CWD. (D) PrPSc deposition patterns in tonsil of deer 135 and 139 assessed by IHC. (Insets) Higher magnifications of individual follicles. (Scale bars in C and D, 200 μM.) (E) Survival curves of Tg(DeerPrP)1536+/− mice infected with obex (solid lines) or RPLN (dashed lines). Solid symbols, WT infected deer; open symbols, G/S96 deer. (F) PrPSc deposition patterns assessed by histoblotting in the hippocampus/thalamus of Tg(DeerPrP)1536+/− mice infected with brain homogenates from 135 and 142 deer. Blots were treated with PK as indicated.
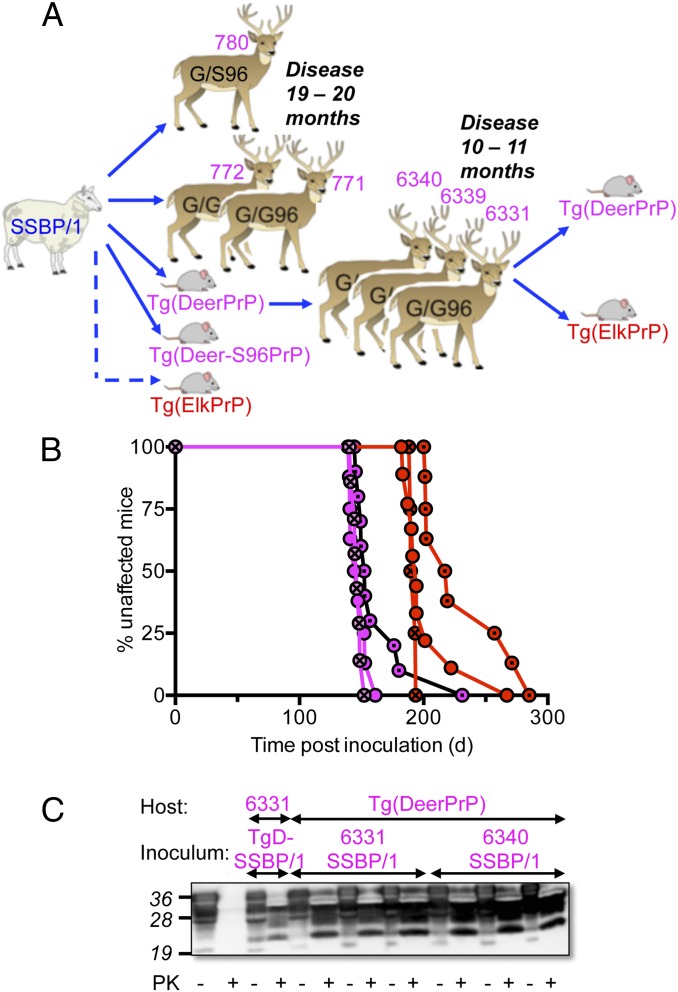
Three white-tailed deer (771, 772, and 780) were inoculated with SSBP/1 (Fig. 5A). Deer 780 was heterozygous G/S at residue 96, and deer 771 and 772 were homozygous G/G. Consistent with results in Tg mice (Fig. 2B), SSBP/1 propagation was unaffected by substitution of S for G at residue 96, with all deer, regardless of genotype, developing disease between 19 and 20 mo. Additional white-tailed deer (6331, 6339, and 6340) were inoculated with brain homogenates of diseased Tg(DeerPrP)1536+/− mice infected with SSBP/1 (11) (Fig. 5A). Times to onset of disease were 10–11 mo (Fig. 5A), ∼50% more rapid than deer inoculated with SSBP/1. Similar to SSBP/1 from diseased Tg(DeerPrP) mice, SSBP/1 passaged in white-tailed deer produced disease in Tg(ElkPrP)5037+/− mice with incubation times 30–40% longer than in Tg(DeerPrP)1537+/− mice (Fig. 5B and Table S1). Examples of PrPSc accumulation in diseased deer and Tg mice are shown in Fig. 5C.
Fig. 5.
Transmisssion of SSBP/1 to deer. (A) Passage history of SSBP/1 prions from sheep to deer and Tg mice. (B) Brain extracts from deer 6331 (circles), 6339 (circles with dots), and 6340 (circles with crosses) infected with SSBP/1 prions from Tg(DeerPrP) mice were transmitted to Tg(DeerPrP) (magenta) or Tg(ElkPrP) (red) mice. (C) Western blots of representative transmissions of SSBP/1 to deer and Tg mice.
Discussion
PrP Polymorphisms Operate by Distinct Mechanisms Depending on Occupancy Within Particular Structural Domains.
The strain-specific effects of the H95Q and G96S amino acid substitutions reported here parallel previous Tg studies in which we showed that the elk PrP leucine (L) polymorphism at residue 132, although inhibiting CWD, had negligible effects on the propagation of SSBP/1 (11). It therefore appears that amino acid substitutions in the unstructured region of PrP affect PrPC-to-PrPSc conversion during CWD propagation, but their adverse effects are overridden during replication of SSBP/1. Previous studies described a similar phenomenon when comparing the transmission of sporadic and variant CJD in Tg mice expressing human PrP V129 (16). Our findings show that residues 225 and 226 play a critical role in PrPC-to-PrPSc conversion and strain propagation, but their effects are distinct from those produced by the H95Q, G95S, and M132L polymorphisms. Structural analyses show that residues 225 and 226 are located in the distal region of α-helix 3 and that under certain conditions this domain participates with the β2–α2 loop to form a solvent-accessible contiguous epitope (17). Consistent with a role for this epitope in PrP conversion, these polymorphisms severely impact replication of both SSBP/1 and, to variable degrees, CWD. In the case of Tg mice expressing deerPrP-F225, SSBP/1 incubation times were prolonged threefold, whereas inoculation with CWD produced incomplete attack rates or prolonged and variable incubation times in small numbers of mice. In those Tg(DeerPrP-F225) mice that did succumb to CWD, PrPSc distribution patterns were altered compared with Tg(DeerPrP) mice (Fig. 2D). In the case of substitution of E for Q at residue 226, inhibited propagation of SSBP/1 was absolute. Our previous studies showed that residue 226 also affected CWD replication, but to a lesser degree, with disease onset prolonged by 20–46% in CWD-inoculated Tg(DeerPrP) compared with Tg(ElkPrP) mice, with PrPSc distribution and neuropathology varying in each case (18). Additional findings support the notion that residue 226 plays a critical role in prion pathogenesis. Although the resistance of elk PrPC to SSBP/1 was overcome following adaptation in deer or Tg(DeerPrP) mice, incubation times of deer-adapted SSBP/1 were longer in Tg mice expressing E226 compared with Q226. Moreover, passage in Tg mice expressing E226 or Q226 profoundly affected the ability of SSBP/1 to reinfect Tg mice expressing sheep PrPC. The results reported here parallel aspects of other previously published studies indicating that species-specific amino acid differences at residue 226 controlled the manifestation of CWD quasi-species or closely related strains (5). Our previous and current findings therefore collectively point to an important role for residues 225 and 226 in the manifestation of prion strain properties.
Our studies of α-helix 3 resident polymorphic variants provide functional support for previous high-resolution structural analyses of PrP. Whereas initial studies suggested rigidity of the β2–α2 loop to be a key determinant of prion pathogenesis (8), subsequent work implied a more complex mechanism in which the distal region of α-helix 3 participated with the β2–α2 loop to form a solvent-accessible contiguous epitope (17). These and subsequent studies (19) ascribed greater importance to the plasticity of this discontinuous epitope. For example, substitution of D found in horse by S at residue 170 of mouse (elk PrP numbering) (Fig. 1) increased not only the structural order of the β2–α2 loop, but also the long-range interaction with Y228 in α-helix 3. Underscoring the importance of long-range β2–α2 loop/α-helix 3 interactions, similar structural connections occur between this residue, which is alanine (A) in Tammar wallaby PrP, and residue 169 in the 310 helix, which is isoleucine (I) in this species (19) (Fig. 1). Stabilizing long-range interactions between the β2–α2 loop and α-helix 3 also occur in rabbit PrP, a species generally regarded as resistant to prion infection (reviewed in ref. 20). X-ray crystallographic analyses showed the rabbit β2–α2 loop to be clearly ordered and indicated that hydrophobic interactions between the side chains of V169 and, in this case Y221 (elk PrP numbering) of α-helix 3, contributed to the stability of the β2–α2 loop/α helix 3 epitope (21) (Fig. 1).
Our results substantiate the view that long-range interactions between the β2–α2 loop and α-helix 3 provide protection against prion infection and suggest a likely mechanism to account for the protective effects of the F225 polymorphism. Our MD analyses show that the combined S225F and E226Q substitutions in deer PrP-F225 collectively alter the orientations of D170 and Y228 and that this structural change allows hydrogen bonding between the side chains of these residues and reduced plasticity of the β2–α2 loop/α-helix 3 epitope compared with deer or elk PrP structures. We propose that the increased stability of this tertiary structural epitope precludes PrPC-to-PrPSc conversion of deerPrP-F225.
Authentic Recapitulation of Intra- and Interspecies Prion Transmission in Tg Mice.
To our knowledge, these integrated studies are the first to conclusively demonstrate that the parameters controlling intraspecies propagation of CWD and interspecies adaptation of sheep prions in deer are faithfully recapitulated in Tg mouse models. This applies not only to CWD propagated by the wild-type deer PrP sequence, but also to the effects of polymorphic variation, because the inhibitory effects of the S96 polymorphism on CWD prion pathogenesis in deer are recapitulated in Tg mice. Our results demonstrating inhibitory effects of CWD in deer with the G/S96 genotype are in accordance with previous studies (22, 23). Our results also build on our previous findings that sheep SSBP/1 caused disease in Tg(DeerPrP)1536+/− mice with incubation times similar to CWD prions (11). We show here that deer are likewise susceptible to SSBP/1 and, in accordance with our previous transmission studies in Tg(DeerPrP) mice (11), that SSBP/1 adapted in Tg(DeerPrP) mice is transmissible to deer with ∼50% shorter incubation times than the same prions from sheep. We show the effects of the G96S substitution on SSBP/1 replication to be inconsequential in either Tg(DeerPrP) mice or deer, which is consistent with transmission studies to deer of a US scrapie isolate (24).
This is the first report, to our knowledge, in which the effects of the white-tailed deer PrP H95 and mule deer PrP F225 polymorphisms have been modeled in Tg mice. When assessing the results of incubation time experiments in Tg mice, it is important to consider the effects of variations in transgene expression between lines. Frequently an inverse relationship exists between transgene expression and time to onset of disease (2), although this is not always the case (25). The Tg mice used here express deer, elk, and deer-S96 PrP at approximately fivefold greater than wild type, whereas levels are lower in Tg mice expressing deer-H95 and deer-F225 (Table S4), making it difficult to completely correlate variations in times to onset of disease among lines with the effects of polymorphic variation. Nonetheless, our studies on the susceptibility of Tg(DeerPrP-S96)7511+/− and Tg(DeerPrP-H95)7505+/− mice to CWD are consistent with the observed effects of experimentally infected deer expressing substitutions at positions 95 and 96 (23), and our results also confirm a protective role for F225 in CWD, previously suggested based on the rarity of the substitution in free-ranging deer with disease (7). Finally, the in vivo effects of polymorphisms on CWD and scrapie propagation were recapitulated in PMCA when levels of PrPC from the brains of various Tg mice were normalized to equivalent levels.
Previous studies consistently showed Tg60 mice expressing deer PrP-S96 to be entirely resistant to CWD, and investigators were unable to identify PrPSc in the brains of asymptomatic Tg60 mice (26, 27). Our results show that after long incubation times CWD-infected Tg(DeerPrP-S96)7511+/− mice were ultimately susceptible to disease, albeit with incomplete attack rates. Moreover, deer PrPSc-S96 was present in the brains of diseased as well as asymptomatic inoculated Tg(DeerPrP-S96)7511+/− mice, although at levels lower than deer PrPSc in the brains of diseased Tg(DeerPrP)1536+/− mice. The discrepancy is most likely related to the low transgene expression in Tg60 mice, reported to be 70% the levels found in deer. CWD occurs naturally in deer homozygous for the PrP-S96 allele (28), which is clearly inconsistent with a completely protective effect of this substitution, suggesting that Tg(DeerPrP-S96)7511+/− mice represent an accurate Tg model in which to assess the effects of the S96 substitution.
Finally, our findings showing that Tg(DeerPrP) mice, but not Tg(ElkPrP) mice, are sensitive to infection with SSBP/1 belie previously published results showing that SSBP/1 of the same provenance caused disease in two lines of Tg mice expressing elk PrP (13). However, our results appear to be consistent with the reported susceptibilities of elk and deer to sheep prions. In previous studies, of six elk inoculated with scrapie, three presented with neurological signs and neuropathology, but only after long and variable times to disease onset ranging from 25 to 46 mo (29). In contrast, our results with SSBP/1 demonstrate relatively facile transmission of scrapie to deer, with all inoculated animals developing within 19–20 mo, which is in accordance with susceptibility of deer to a US scrapie isolate with a similar time-to-disease onset (24). Polymorphisms of ovine PrP add a further level of complexity because they control the propagation scrapie strains. Occupancy of residue 136 by A or V is of particular importance. Our previous results indicated that SSBP/1 is composed of a dominant strain that is preferentially propagated by sheep PrP encoding V at 136 (12). In contrast, the scrapie prions used in the deer transmission studies of Greenlee and colleagues (24) were isolated from a sheep encoding A136, raising the possibility that deer may be susceptible to multiple scrapie strains.
Materials and Methods
Inocula.
CNS or lymphoreticular system (LRS) samples were obtained either from naturally CWD affected or experimentally inoculated, indoor-housed deer. SSBP/1 was derived from infected Cheviot sheep at the Neuropathogenesis Unit (Edinburgh). Deer CNS and LRS and mouse brains were homogenized to 10% (wt/vol) in sterile PBS lacking Ca2+ and Mg2+ ions.
Transgenic Mice.
All work with animals was performed in compliance with University of Kentucky and Colorado State University Institutional Animal Care and Use Committees. The generation and characterization of Tg(DeerPrP)1536+/−, Tg(ElkPrP)5037+/−, and Tg(OvPrP-V136)4166+/− mice has been described previously (5, 12, 15). Newly derived Tg lines were generated using MoPrP.Xho (30). Site-directed mutagenesis (Quick Change, Stratagene) was used to make single amino acid changes in the wild-type mule deer PRNP ORF (GenBank accession no. AF009180). Tg mice were produced and characterized as previously described (5, 12, 15) (Table S4).
Determination of Incubation Times.
Incubation times were determined as previously described (5, 12, 15).
Infected Cervids.
CWD-naive, hand-raised white-tailed deer fawns from Georgia in the United States were inoculated and maintained in dedicated restricted-access indoor facilities. Deer, 139, 142, and 135 were inoculated with brain material from D10 CWD-infected Tg(DeerPrP)1536+/− mice. Deer 139 was inoculated i.v., whereas deer 142 and 135 were challenged intracranially. Deer were monitored for disease by serial tonsil and recto-anal mucosa-associated lymphoid tissue biopsy performed at 0, 3, 6, and 12 mo post inoculation and at termination (18 mo). Equal portions of tissue were collected and stored [−70 °C or fixed in 10% (vol/vol) formalin] at each serial collection time point and at study termination, at which point palantine tonsil, brain, and RPLN were also collected for the detection of PrPSc by IHC and Western blotting. Deer 771, 772, and 780 were inoculated i.v. with SSBP/1, and deer 6331, 6339, and 6340 were inoculated i.v. with brain homogenates of diseased Tg(DeerPrP)1536+/− mice infected with SSBP/1.
Western Blotting.
Western blotting was performed and analyzed as previously described (12).
Protein Misfolding Cyclic Amplification.
PMCA reactions were performed and analyzed as previously described (31). We used equal amounts of substrate PrPC based on the relative levels of expression in each Tg line and amplified with increasing fourfold dilutions of seeds for a set number of cycles. For serial PMCA, 6 μL of amplified material was diluted into 54 μL of fresh normal brain homogenate. Western blots were developed using anti-PrP mAbs 6H4 (Prionics AG) or PRC5 (32).
Histoblotting.
Histoblots were produced and analyzed according to previously described protocols (31). Images were captured with a NikonDMX 1200F digital camera and analyzed using MetaMorph imaging software (Molecular Devices).
Immunohistochemistry.
IHC was performed as previously described (31) using mAb 6H4 as primary antibody, and IgG1 biotinylated goat anti-mouse as secondary antibody (SouthernBiotech). Digitized images were obtained by light microscopy using a Nikon Eclipse E600 microscope equipped with a Nikon DMX 1200F digital camera.
Molecular Dynamics Simulation.
Explicit solvent Particle Mesh Ewald (33) MD simulations were carried out in AMBER 12 (34) using the ff99SB force field with a time step of 2 fs. For each protein, Na+ ions were added to neutralize the system, and the protein was then immersed in a rectangular box of TIP3P water (18.8 Å along each axis). An initial energy equilibration was performed for 2 ps at constant volume with positional restraints on the protein to allow solvent molecules to relax. The system was then heated from 0 to 300 K for 10,000 steps (20 ps) with reduced positional restraints on the protein and all bonds involving hydrogen fixed via SHAKE (35). The protein-solvent system was then minimized at constant volume for 2,500 cycles of steepest descent followed by conjugate gradient minimization for 1,000 cycles. Langevin dynamics (36) at a constant pressure of 1 atm and a nonbonded cutoff of 12.0 Å was then carried out on the solvated protein with a collision frequency of 1 ps−1 with temperature maintained constant at 300 K for a total time of 5 ns. Both the trajectories from each MD simulation and the individual PrPs were visualized in VMD (37).
Supplementary Material
Acknowledgments
We thank Tanya Seward for technical assistance; Jeanette Hayes-Klug and Kelly Anderson for overseeing white-tailed deer; and our colleagues at Prionics for mAb 6H4. This work was supported by National Institutes of Health Grants R01 NS040334 and P01 AI077774.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404739111/-/DCSupplemental.
References
- 1.Will RG, et al. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347(9006):921–925. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 2.Prusiner SB, et al. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 1990;63(4):673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 3.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68(12):7859–7868. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telling GC, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274(5295):2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 5.Angers RC, et al. Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science. 2010;328(5982):1154–1158. doi: 10.1126/science.1187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bian J, Kang HE, Telling GC. Quinacrine promotes replication and conformational mutation of chronic wasting disease prions. Proc Natl Acad Sci USA. 2014;111(16):6028–6033. doi: 10.1073/pnas.1322377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jewell JE, Conner MM, Wolfe LL, Miller MW, Williams ES. Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J Gen Virol. 2005;86(Pt 8):2127–2134. doi: 10.1099/vir.0.81077-0. [DOI] [PubMed] [Google Scholar]
- 8.Gossert AD, Bonjour S, Lysek DA, Fiorito F, Wüthrich K. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci USA. 2005;102(3):646–650. doi: 10.1073/pnas.0409008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurt TD, et al. Prion transmission prevented by modifying the β2-α2 loop structure of host PrPC. J Neurosci. 2014;34(3):1022–1027. doi: 10.1523/JNEUROSCI.4636-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groschup MH, Gretzschel A, Kuczius T. Prion Strains. Berlin: Walter de Gruyter; 2006. [Google Scholar]
- 11.Green KM, et al. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. J Gen Virol. 2008;89(Pt 2):598–608. doi: 10.1099/vir.0.83168-0. [DOI] [PubMed] [Google Scholar]
- 12.Saijo E, et al. Epigenetic dominance of prion conformers. PLoS Pathog. 2013;9(10):e1003692. doi: 10.1371/journal.ppat.1003692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamgüney G, et al. Transmission of scrapie and sheep-passaged bovine spongiform encephalopathy prions to transgenic mice expressing elk prion protein. J Gen Virol. 2009;90(Pt 4):1035–1047. doi: 10.1099/vir.0.007500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121(2):195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Browning SR, et al. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J Virol. 2004;78(23):13345–13350. doi: 10.1128/JVI.78.23.13345-13350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wadsworth JD, et al. Human prion protein with valine 129 prevents expression of variant CJD phenotype. Science. 2004;306(5702):1793–1796. doi: 10.1126/science.1103932. [DOI] [PubMed] [Google Scholar]
- 17.Pérez DR, Damberger FF, Wüthrich K. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J Mol Biol. 2010;400(2):121–128. doi: 10.1016/j.jmb.2010.04.066. [DOI] [PubMed] [Google Scholar]
- 18.Angers RC, et al. Chronic wasting disease prions in elk antler velvet. Emerg Infect Dis. 2009;15(5):696–703. doi: 10.3201/eid1505.081458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christen B, Hornemann S, Damberger FF, Wüthrich K. Prion protein NMR structure from tammar wallaby (Macropus eugenii) shows that the beta2-alpha2 loop is modulated by long-range sequence effects. J Mol Biol. 2009;389(5):833–845. doi: 10.1016/j.jmb.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Zhang Y. Molecular dynamics studies on the NMR and X-ray structures of rabbit prion proteins. J Theor Biol. 2014;342:70–82. doi: 10.1016/j.jtbi.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Khan MQ, et al. Prion disease susceptibility is affected by beta-structure folding propensity and local side-chain interactions in PrP. Proc Natl Acad Sci USA. 2010;107(46):19808–19813. doi: 10.1073/pnas.1005267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathiason CK, et al. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science. 2006;314(5796):133–136. doi: 10.1126/science.1132661. [DOI] [PubMed] [Google Scholar]
- 23.Johnson CJ, et al. Prion protein polymorphisms affect chronic wasting disease progression. PLoS ONE. 2011;6(3):e17450. doi: 10.1371/journal.pone.0017450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenlee JJ, Smith JD, Kunkle RA. White-tailed deer are susceptible to the agent of sheep scrapie by intracerebral inoculation. Vet Res. 2011;42:107. doi: 10.1186/1297-9716-42-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korth C, et al. Abbreviated incubation times for human prions in mice expressing a chimeric mouse-human prion protein transgene. Proc Natl Acad Sci USA. 2003;100(8):4784–4789. doi: 10.1073/pnas.2627989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meade-White K, et al. Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. J Virol. 2007;81(9):4533–4539. doi: 10.1128/JVI.02762-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Race B, Meade-White K, Miller MW, Fox KA, Chesebro B. In vivo comparison of chronic wasting disease infectivity from deer with variation at prion protein residue 96. J Virol. 2011;85(17):9235–9238. doi: 10.1128/JVI.00790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keane DP, et al. Chronic wasting disease in a Wisconsin white-tailed deer farm. J Vet Diagn Invest. 2008;20(5):698–703. doi: 10.1177/104063870802000534. [DOI] [PubMed] [Google Scholar]
- 29.Hamir AN, et al. Transmission of sheep scrapie to elk (Cervus elaphus nelsoni) by intracerebral inoculation: Final outcome of the experiment. J Vet Diagn Invest. 2004;16(4):316–321. doi: 10.1177/104063870401600410. [DOI] [PubMed] [Google Scholar]
- 30.Borchelt DR, et al. A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genet Anal. 1996;13(6):159–163. doi: 10.1016/s1050-3862(96)00167-2. [DOI] [PubMed] [Google Scholar]
- 31.Green KM, et al. Accelerated high fidelity prion amplification within and across prion species barriers. PLoS Pathog. 2008;4(8):e1000139. doi: 10.1371/journal.ppat.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang HE, et al. Characterization of conformation-dependent prion protein epitopes. J Biol Chem. 2012;287(44):37219–37232. doi: 10.1074/jbc.M112.395921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Essmann U, et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 34.Case DA, et al. AMBER 12. San Francisco: University of California; 2012. [Google Scholar]
- 35.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 36.Brunger A, Brooks CL, Karplus M. Stochastic boundary conditions for molecular dynamics simulations of St2 water. Chem Phys Lett. 1984;105(5):495–500. [Google Scholar]
- 37.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14(1):33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.