Significance
Protein location and function are dependent on diverse cell states. We develop a conditional function predictor (CoFP) for proteome-wide prediction of condition-specific locations and functions of proteins. In addition to highly accurate retrieval of condition-dependent locations and functions in individual conditions, CoFP successfully discovers dynamic function changes of yeast proteins, including Tsr1, Caf120, Dip5, Skg6, Lte1, and Nnf2, under DNA-damaging stresses. Beyond specific predictions, CoFP reveals a global landscape of changes in protein location and function, highlighting a surprising number of proteins that translocate from the mitochondria to the nucleus or from endoplasmic reticulum to Golgi apparatus under stress. CoFP has the potential to discover previously unidentified condition-specific locations and functions under diverse conditions of cellular growth.
Keywords: dynamic function prediction, protein translocation, DTT and MMS, systems biology, bioinformatics
Abstract
Protein location and function can change dynamically depending on many factors, including environmental stress, disease state, age, developmental stage, and cell type. Here, we describe an integrative computational framework, called the conditional function predictor (CoFP; http://nbm.ajou.ac.kr/cofp/), for predicting changes in subcellular location and function on a proteome-wide scale. The essence of the CoFP approach is to cross-reference general knowledge about a protein and its known network of physical interactions, which typically pool measurements from diverse environments, against gene expression profiles that have been measured under specific conditions of interest. Using CoFP, we predict condition-specific subcellular locations, biological processes, and molecular functions of the yeast proteome under 18 specified conditions. In addition to highly accurate retrieval of previously known gold standard protein locations and functions, CoFP predicts previously unidentified condition-dependent locations and functions for nearly all yeast proteins. Many of these predictions can be confirmed using high-resolution cellular imaging. We show that, under DNA-damaging conditions, Tsr1, Caf120, Dip5, Skg6, Lte1, and Nnf2 change subcellular location and RNA polymerase I subunit A43, Ino2, and Ids2 show changes in DNA binding. Beyond specific predictions, this work reveals a global landscape of changing protein location and function, highlighting a surprising number of proteins that translocate from the mitochondria to the nucleus or from endoplasmic reticulum to Golgi apparatus under stress.
A cellular response can induce striking changes in the subcellular location and function of proteins. As a recent example, the activating transcription factor-2 (ATF2) plays an oncogenic role in the nucleus, whereas genotoxic stress-induced localization within the mitochondria gives ATF2 the ability to play tumor suppressor, resulting in promotion of cell death (1). Changes in protein location are typically identified using a variety of experimental methods [e.g., protein tagging (2), immunolabeling (3), or cellular subfractionation of target organelles followed by mass spectrometry (4)]. Although highly successful, such measurements can be laborious and time-consuming, even for a single protein (all methods except mass spectrometry) and condition (all methods).
For these reasons and others, computational prediction of protein location and function has been a very active area of bioinformatic research. Early methods attempted to infer protein function based mainly on individual protein features, such as sequence similarity or structural homology (3, 5–17). These methods range from simple sequence–sequence comparisons to profile- or pattern-based supervised learning methods. Other methods predicted protein function using gene expression data (18, 19) based on the observation that proteins with similar patterns of expressions share similar functions (20). Another class of methods is based on text mining (21, 22).
Although such methods are still widely used for annotating general protein locations or functions, the recent availability of data about large-scale molecular networks, such as protein–protein interactions, has changed the functional prediction paradigm (7, 23). In reality, proteins seldom function alone (24). Therefore, a number of network-based methods have been developed that predict location or function based on a protein’s physically interacting or functionally related partners (25). Network-based methods follow either of two distinct approaches, which we call direct vs. module-based annotation schemes. Direct annotation methods propagate protein location or function annotations over a biological network based on the assumption that nearby proteins in the network have similar functions. Module-based methods first identify groups of functionally related genes or gene products using unsupervised clustering methods and then assign a representative function to each module based on the known locations or functions of its members (25).
Notably, all of these previous methods have difficulty predicting condition-specific or dynamic behavior. The main difficulty in predicting such dynamics is the lack of known protein locations and functions under the target condition(s), which are required for generating a prediction model in the training stage. One possible solution is to find dynamic network modules in gene expression networks constructed under specific conditions (26). However, it is difficult to assign representative locations or functions to the dynamic module, and one cannot assign a location or function to other proteins not belonging to the module.
Here, we describe a general approach for predicting the proteome-wide, condition-dependent locations and functions of proteins under diverse conditions. By integrating expression data measured under a specific condition with a protein interaction network pooling data from many studies, our method indicates the probable location and function of each protein under that condition. We apply the classification method to 17 different stresses, revealing a landscape of dynamic changes in location and function across the yeast proteome.
Results
Genome-Wide Prediction of Condition-Dependent Location and Function Under Diverse Stresses.
To predict condition-dependent location and function under diverse stresses, we developed a protein network-based prediction framework that uses diverse features of both individual proteins and interacting neighbors called a conditional function predictor (CoFP). We first accessed the known locations and functions of all yeast proteins as determined from previous high-throughput experiments and gene ontology (GO) terms (Fig. 1A), focusing on 18 distinct locations (locations), 33 high-level biological processes (processes), and 22 general molecular functions (functions) (SI Appendix, Table S1). We next downloaded and combined the contents of the BioGRID (27), DIP (28), and SGD (29) databases in addition to in vivo physical protein–protein interactions (30). In total, 77,364 protein interactions between 5,778 yeast proteins were prepared (Fig. 1B). For individual protein features, we used sequences, chemical properties, motifs, and specific GO terms of each yeast protein (Fig. 1C). Predictions of protein location or function were based on an established approach that integrates information about interacting neighbors in addition to single proteins as indicators of protein function (31). Previously, we observed that the co-occurrence of sequence, structure, or function between a protein and its interacting partners is a strong predictor of joint subcellular location (32). We applied forward selection to choose feasible feature sets of high predictive power from the pool of generated individual and network protein features, with a network neighborhood restricted to nearest neighbors within distance 2, using a divide-and-conquer k-nearest-neighbor method (Fig. 1D). A dynamic context for condition-dependent location and function was achieved through the concept of conditional network neighborhoods, in which expression profiles gathered for conditions of interest are projected onto protein–protein interaction networks (Methods).
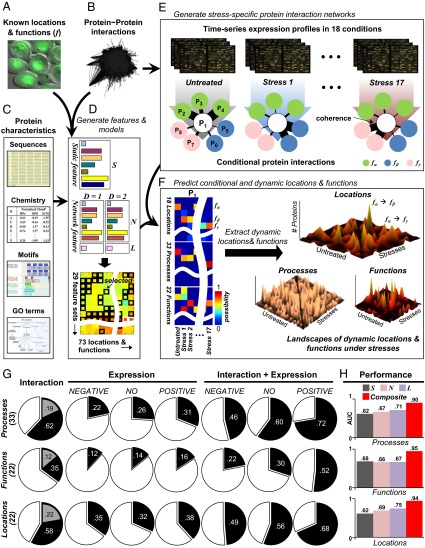
Fig. 1.
Proteome-wide prediction of conditional locations and functions under stresses. (A) Generally known protein functions, including 18 subcellular locations, 33 biological processes, and 22 molecular functions. (B) Yeast protein–protein interactions accumulated from several databases. (C) Static information of proteins, including sequence, chemical properties, motifs, and GO terms (single-protein features). (D) Model generation after generating static single-protein (denoted S) and network features (denoted N and L) up to network distance D = 2. The best combination of features is selected for each functional category using a divide-and-conquer k-nearest-neighbor method classifier. (E) Stress-specific interaction networks in individual conditions are generated by assigning different functional coherence scores to each interaction of a protein depending on the interactor’s similarity in time series gene expression profiles. (F) After generating the selected features from D using the condition-dependent networks from E, the prepared 73 classifiers compute a conditional functional map for the protein, indicating the quantitative possibility that the protein is in each function under each condition. Dynamic functions under stressful conditions are identified by calculation of significant differences in the possibility degree in a stress condition. (G) The fraction of protein pairs having the same process, function, or location (rows) shown for protein pairs involved in physical protein interaction (column 1), high coexpression (columns 2–4; NEGATIVE, negative correlation; NO, no correlation; POSITIVE, positive correlation), and both physical interaction and high coexpression (columns 5–7). Gray sectors indicate random expectation resulting from 100 permutation tests. (H) The average performance (area under the ROC curve (AUC) value) of S, N, or L feature sets for process, function, and location. Composite indicates the performance of the selected feature sets for individual functional categories.
As a proof of principle, we applied this approach to predict conditional locations, processes, and functions of 5,778 yeast proteins under diverse stress conditions. To provide expression profiles in different conditions, we used time series microarray experiments from the Stanford Microarray Database (www.tbdb.org) categorized into 17 stresses in addition to an untreated stress-free condition (SI Appendix, Table S2). By assigning coherence scores based on coexpression degree (Methods), we generated stress-dependent protein interaction networks for individual conditions (Fig. 1E). The stress-dependent coherence scores under 18 conditions yielded stress-dependent protein network features, resulting in a conditional function map with degrees of possibility assigned to individual locations or functions under distinct conditions (Fig. 1F). By comparing the predicted functions between each stress condition with the stress-free condition, we extracted dynamic protein states regarding location, process, and function.
Protein Interaction and Expression Are both Useful in Protein Location and Function Prediction.
CoFP is based on the assumption that interacting proteins that are also coexpressed have similar locations and functions. To test this assumption, we examined the functional agreement between protein pairs divided into various groups: (i) all interacting protein pairs; (ii) all protein pairs with POSITIVE (Pearson correlation coefficient γ ≥ 0.3), NEGATIVE (γ ≤ −0.3), and NO correlation (−0.3 < γ < 0.3) between expression patterns; and (iii) interacting pairs with POSITIVE, NEGATIVE, and NO correlation between the patterns (Fig. 1G). Many interacting proteins share the same process (>62%), function (>35%), or location (>58%) in contrast to random expectation (P values of Z tests are almost zero using 100 permutation tests). Among the three types, process showed the highest correlation, although it is the largest category, whereas function showed the lowest correlation. However, the function-sharing fractions of all protein pairs with POSITIVE expression correlation were much less than those of all interacting pairs. However, if we consider only the interacting pairs of the POSITIVE expression pairs, then the function-sharing fraction increases dramatically (Interaction + Expression in Fig. 1G). We also observed these phenomena between all function–category pairs (SI Appendix, Figs. S1–S3).
We next analyzed the predictive power of network features generated by using (i) physically interacting partners only; (ii) positively coexpressed partners only; and (iii) those features together. The best performance was achieved when both network feature sets were incorporated; using just network features from the gene expression network showed the worst performance (SI Appendix, Fig. S4). Moreover, network features generated using more than network distance 2 were less informative. Thus, we integrated 9 kinds of individual protein static feature sets Ss and 20 kinds of network feature sets Ns (using neighbors’ S features) and Ls (using neighbors’ locations or functions) up to network distance D = 2. However, the whole 29 feature sets were not required for function prediction (SI Appendix, Fig. S5). Using the feature sets, therefore, we optimized prediction models by finding feasible feature sets for each location or function using the divide-and-conquer k-nearest-neighbor method framework (in total, 73 models: 33 models for process, 18 models for location, and 22 models for function) (Fig. 1H and SI Appendix, Figs. S6 and S7). For process and location, the network features Ls, based on known locations or functions of the neighborhood, were better than both Ss and Ns, mainly because of the previous high degrees of sharing of the same function that occurs between interacting pairs in process and location compared with function (shown in Fig. 1G). For function, however, Ss showed the best performance compared with other kinds of feature sets. Moreover, GO-based features were more informative than other features; thus, GO-based static and network features were widely selected (SI Appendix, Fig. S6), and this high performance was not achieved with randomized GO annotations (SI Appendix, Fig. S8). Motif-based features and network features using neighbors’ generally known functions were also useful for diverse kinds of function and widely selected. Furthermore, the importance of static or network features was somewhat dependent on functional purity within (SI Appendix, Fig. S9) or between (SI Appendix, Figs. S1–S3) proteins (details in Discussion). We observed that selecting different features per location or function by using individual protein and network features resulted in a dramatic increase in performance (SI Appendix, Fig. S10 shows feature set selection, and SI Appendix, Figs. S11–S13 shows the receiver operating characteristic curves). The performance of the selected feature sets outperformed other kinds of simple feature selection methods, including regression and entropy-based methods (SI Appendix, Fig. S14).
Expression-Combined Network Models Revise Previously Known Locations and Functions.
We first generated a weighted protein interaction network by using a functional coherence scoring scheme on the time series expression profiles from the untreated normal condition (SI Appendix, Table S2). Using this network, we predicted 9,301 processes, 4,419 functions, and 4,336 locations with high possibility among the proteins with known location or function (SI Appendix, Fig. S15). In addition to the strong agreement with the previously known locations or functions (SI Appendix, Fig. S15 G–I), some proteins had different but strong signals. For example, cysteine-three-histidine-1 (Cth1) was originally mapped to DNA binding-related functions at the beginning of this study. However, the prediction showed a higher possibility for RNA binding than DNA binding (SI Appendix, Fig. S16 A–C). Interestingly, the function of Cth1 has been recently recurated as RNA binding rather than DNA binding in SGD (www.yeastgenome.org) based on a recently discovered role in mRNA degradation (33). Regarding process, we also observed similar cases (SI Appendix, Fig. S16 D and E). After reassigning function and process using more recent GO terms from the SGD, 99 (51 previously unidentified and 48 previously mislabeled) cases for function and 608 (202 previously unidentified and 406 previously mislabeled) cases for process were additionally found to be correct (SI Appendix, Table S3).
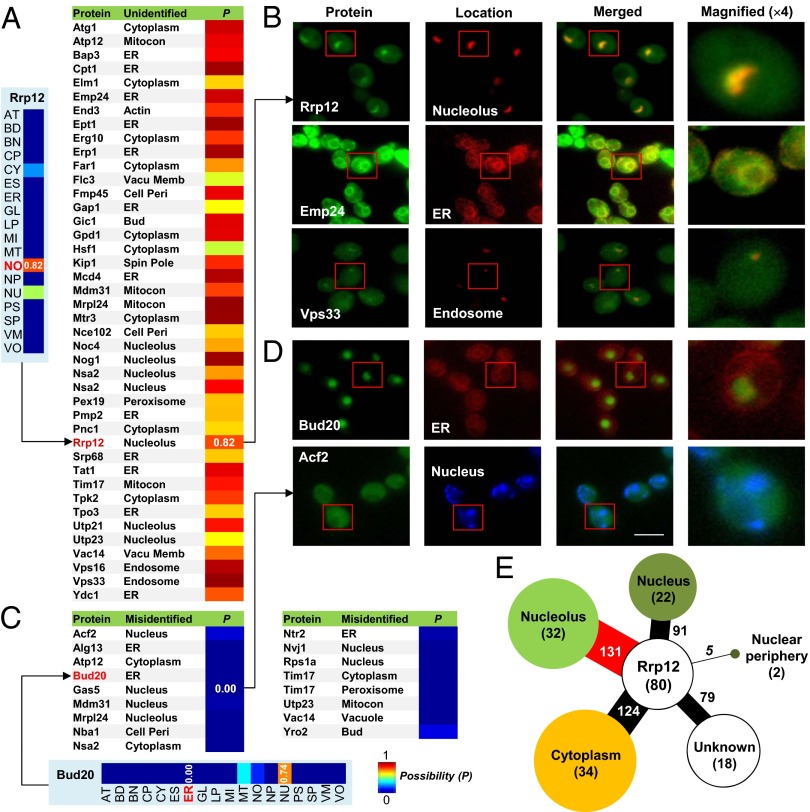
Regarding location, we observed qualitatively similar results. For example, ribosomal RNA processing-12 (Rrp12) was originally localized to the nucleus and cytoplasm by previous high-throughput experiments (2), whereas our predictions produced a higher signal at the nucleolus than other locations (Fig. 2A). We, therefore, retested the localization of Rrp12 to find that it, indeed, does accumulate strongly at the nucleolus (Fig. 2B). As additional examples, we discovered a previously missed endoplasmic reticulum (ER) location for endomembrane protein-24 (Emp24) and an endosome location for vacuolar protein sorting-33 (Vps33) (Fig. 2B). In the case of bud site selection-20 (Bud20), it was originally localized to the nucleus and ER, but our prediction (Fig. 2C) and a new experiment (Fig. 2D) show that Bud20 is almost completely absent at the ER. In another example, the method correctly points out a previously misidentified nuclear location for assembly complementing factor-2 (Acf2) (Fig. 2D). In some cases, therefore, it seems that CoFP can complement or revise the image readouts of single high-throughput experiments. This power is observed mainly because CoFP synthesizes evidence from multiple key interacting players under a specific condition. For example, although many Rrp12 interactors localize to the cytoplasm, the summed functional coherence score in normal conditions is higher in the nucleolus than other locations (Fig. 2E), and location purity is highest in the nucleolus (SI Appendix, Fig. S1A). In total, we revised 59 locations for 50 proteins; these predictions differed from a previous localization attempt but were correctly confirmed here by new GFP experiments (Fig. 2 and SI Appendix, Fig. S17).
Fig. 2.
Experimental validation of predicted locations in a stress-free condition. (A) Forty-two previously unidentified locations but correctly predicted cases as shown in new validation experiments. (Left) The heat map is the location prediction for Rrp12. By prediction, Rrp12 had the strongest signal (0.82 possibility) at nucleolus (NO). (B) Example validation experiments for 42 cases, including Rrp12, Emp24, and Vps33. Protein is marked in green, and location is marked in red. Yellow indicates high overlap between the corresponding proteins and the location markers. Red squares indicate the area that is magnified 4×. RFP-tagged Nop56, Sec66, and Snf7 were used as location markers for the nucleolus, ER, and endosome, respectively. (C) Seventeen previously misidentified locations but correctly predicted cases as shown in new validation experiments. (Lower) The heat map is the prediction for Bud20. Although originally localized to the ER, Bud20 had an almost zero possibility in this location, a prediction that we validated. (D) Some examples of the new experiments for 17 cases, including Bud20 and Acf2. Protein is marked in green, ER location is marked in red, and nucleus is marked in blue. (Scale bar: 5 μm.) (E) The coherence-mapped interaction network in location prediction of Rrp12. The node and edge sizes are proportional to the summed coherence score. The numbers in parentheses of the nodes indicate the numbers of neighbors in corresponding localizations. Full names for the abbreviations of locations and functions are shown in SI Appendix, Table S1.
Expression-Combined Network Models Can Discover Unknown Locations and Functions.
The locations of 1,931 yeast proteins could not be clearly mapped by previous genome-wide experiments (2), primarily because of low GFP signals. Similarly, with process and function, there were 1,683 and 2,207 proteins, respectively, not annotated by GO as of January of 2008. Of these uncharacterized proteins, 990 processes, 670 functions, and 1,277 locations were predicted with high possibility (SI Appendix, Fig. S18 and Tables S4–S6). To investigate the performance of these predictions, we remapped their locations and functions using updated GO terms (SI Appendix, Tables S7–S9). In summary, the performance for unannotated proteins was 0.51 precision, 0.99 specificity, and 0.95 accuracy for process; 0.65 precision, 0.99 specificity, and 0.95 accuracy for function; and 0.64 precision, 0.98 specificity, and 0.93 accuracy for location (SI Appendix, Fig. S19).
Discovering Stress-Dependent Locations and Functions of Yeast Proteins.
Next, we analyzed the time series expression profiles performed under each of 17 different stresses (SI Appendix, Fig. S20 and Table S2) to calculate functional coherence scores between interacting protein pairs under each condition (Methods), similar to those calculated for the untreated stress-free condition. This process resulted in a dynamic interaction network, in which each interaction is assigned a different score in each stress condition. We then predicted conditional locations and functions for 5,778 yeast proteins subject to 17 different stress conditions. In most cases, the predicted locations or functions under stress were consistent with their locations or functions under untreated conditions (details in SI Appendix, Fig. S21).
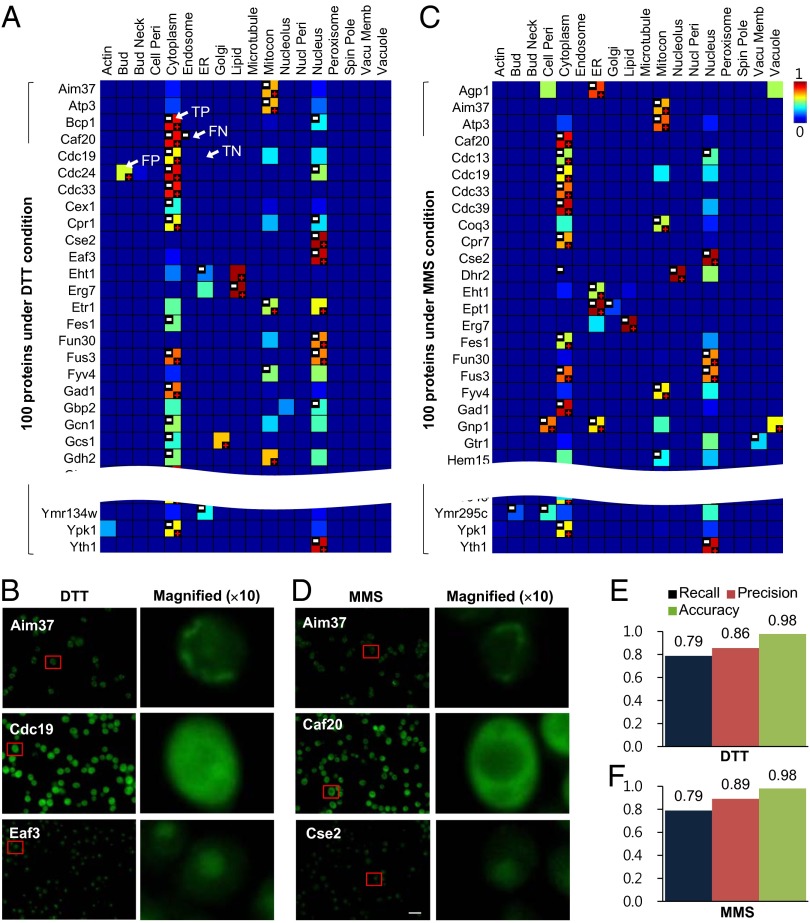
To test the performance of the conditional location predictions, we experimentally tested the predicted locations of 100 randomly selected proteins under dithiothreitol (DTT) and another 100 proteins under methyl methanesulfonate (MMS). DTT is a strong reducing agent that promotes reductive stress, and the intracellular redox state can influence many protein functions and activities (34). MMS is an alkylating agent inducing DNA damage, and mutations in many DNA damage response genes have been implicated in neurological diseases and cancer (35). For example, the predicted locations of Aim37 (Mic27; mitochondrial contact site and cristae organizing system-27), cell division cycle-19 (Cdc19), and Esa1p-associated factor-3 (Eaf3) under DTT treatment were mitochondrion, cytoplasm, and nucleus, respectively (Fig. 3A), all of which were validated by follow-up imaging (Fig. 3B). Aim37, cap associated factor-20 (Caf20), and chromosome segregation-2 (Cse2) were predicted to be in the mitochondrion, cytoplasm, and nucleus, respectively, under the MMS condition (Fig. 3C), which were also correctly validated (Fig. 3D). In total, the precision and recall were 0.86 and 0.79, respectively, under DTT treatment and 0.89 and 0.79, respectively, under MMS treatment for the randomly chosen proteins (Fig. 3 E and F). We observed that this performance was reasonably stable, even when errors were introduced into the underlying protein–protein interaction network (SI Appendix, Fig. S22). Even when 30% of all known interactions were replaced by erroneous interactions, the average decrease was only 0.02 and 0.0006 in sensitivity and specificity, respectively, and these decreases were not statistically significant. Moreover, CoFP, which uses features from both individual proteins and network neighbors, showed gradual decrease in performance with less network coverage, indicating potential applicability to other organisms with fewer known protein interactions, including human, worm, fly, and plant (SI Appendix, Fig. S23).
Fig. 3.
Performance of predicted conditional locations under DTT and MMS conditions. (A) The tested 100 proteins under DTT condition. The heat map indicates the possibility degrees of the proteins for individual locations. The predicted locations are indicated by red +, and validated locations are white −. FN, false negative; FP, false positive; TN, true negative; TP, true positive. (B) Some examples of validated proteins under DTT condition. Green, corresponding proteins; red squares, 10× magnified. (C) The tested 100 proteins under MMS condition. (D) Some examples of validated proteins under MMS condition. (Scale bar: 10 μm.) (E and F) The summarized performance of the experimentally tested 100 proteins under the (E) DTT and (F) MMS conditions shown in A and C, respectively.
Mapping a Global Landscape of Dynamic Locations and Functions.
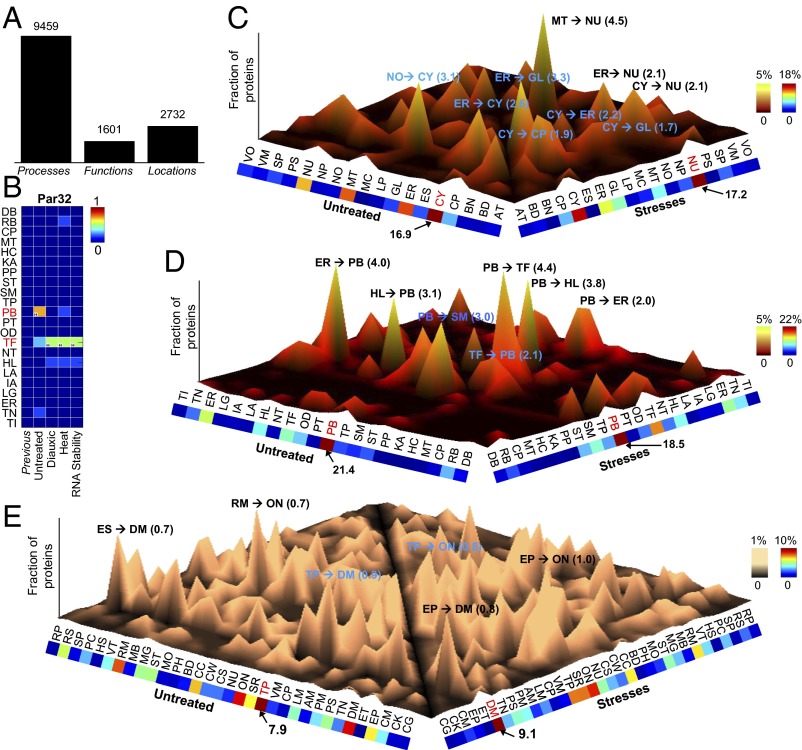
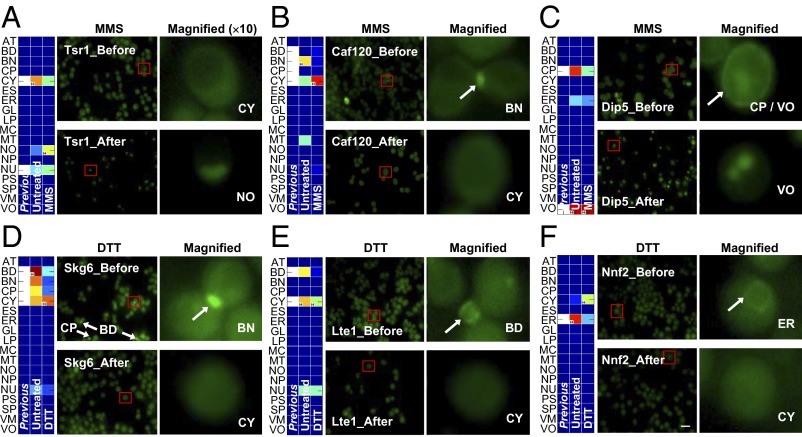
Next, we sought to identify and study all proteins with locations or functions that differed between stress and normal conditions. In total, we identified 9,459 significant differences for process, 1,601 significant differences for function, and 2,732 significant differences for location for all 17 stresses (Methods and Fig. 4A). For example, one prediction showed that phosphorylated after rapamycin-32 (Par32) had a protein-binding molecular function in the untreated condition (Fig. 4B) but transferase activity when subject to diauxic shift, heat, and RNA stability stresses. Twenty S rRNA accumulation-1 (Tsr1), a protein required for processing of 20S precursor of ribosomal RNA (pre-rRNA) in the cytoplasm, was predicted to be most likely located within the cytoplasm and the nucleus under the stress-free condition, whereas under MMS, our method indicated a highly increased signal at the nucleolus but a decreased signal at the cytoplasm (Fig. 5A). As shown in the fluorescence images (Fig. 5A), the conditional locations of Tsr1 changed in accordance with our predictions: Tsr1, a mainly cytoplasmic protein under stress-free conditions, was predominantly localized to the nucleolus under the MMS condition. Moreover, CCR4 associated factor-120 (Caf120), a part of the CCR4-NOT transcriptional regulatory complex, and dicarboxylic amino acid permease-5 (Dip5), a dicarboxylic amino acid permease, also changed their locations under the MMS condition, which is consistent with the predictions (disappearance from the bud neck and cell periphery, respectively) (Fig. 5 B and C). Under the DTT condition, suppressor of lethality of kex2 gas1 double null mutant-6 (Skg6), a potential Cdc28 substrate (36), was predicted to disappear from the bud, bud neck, and cell periphery (Fig. 5D). Correlated with this prediction, we observed that most Skg6 disappeared from locations, including bud, bud neck, and cell periphery, under DTT treatment. Low temperature essential-1 (Lte1), a putative GDP/GTP exchange factor, and necessary for nuclear function-2 (Nnf2), with unknown function, also changed their subcellular locations under the DTT condition in accordance with the predictions (disappearance from bud and ER, respectively) (Fig. 5 E and F). These validated locations were less well-predicted using expression profiles with additional (simulated) noise, implying that good expression data are critical in identification of dynamic functions and locations (SI Appendix, Fig. S24).
Fig. 4.
The landscape of dynamic locations and functions of yeast proteins under stresses. (A) The numbers of predicted dynamic functional events regarding biological processes, molecular functions, and locations integrated over 17 stresses. (B) Example showing the predicted functions of Par32 across diverse conditions, including untreated stress-free normal, diauxic shift, heat shock, and RNA stability conditions. The prediction shows that Par32 is likely to have a protein-binding (PB) function under stress-free conditions and transferase activity (TF) in all three stresses. H in the heat map indicates the function with the highest signal for each condition. The blue cells in the Previous column indicate that this protein had no previously known functions in GO. (C–E) The landscape of changing protein state in terms of (C) location, (D) function, and (E) process across all dynamic yeast proteins. Proteins were considered dynamic if they were significantly different (P value < 0.01) between untreated and stress conditions. Each peak (z axis) corresponds to the percentage of these proteins disappearing from one function (x axis) and appearing in another function (y axis). Colors along the x and y margins represent the total percentage of proteins with functions disappearing or appearing, respectively. Full names for the abbreviations of locations and functions are shown in SI Appendix, Table S1.
Fig. 5.
Experimental validation of dynamic locations under DTT and MMS conditions. The validated dynamic locations of (A) Tsr1, (B) Caf120, and (C) Dip5 under MMS condition and (D) Skg6, (E) Lte1, and (F) Nnf2 under DTT condition. All white cells in the “Previous” columns indicate previously known locations. The red squares indicate the areas that are magnified 10×. Full names for the abbreviations of locations and functions are shown in SI Appendix, Table S1. BD, bud; BN, bud neck; CP, cell periphery; CY, cytoplasm; NO, nucleolus; VO, vacuole. (Scale bar: 10 μm.)
The global landscapes of dynamic locations and functions are shown in Fig. 4 C–E. The most frequent changes in location were from mitochondrion to nucleus (4.54%) or ER to Golgi (3.27%) (Fig. 4C). With function, many proteins were related to protein binding, like Par32 (Fig. 4D). Moreover, many proteins (7.92%) related to transport lost this role with respect to process (Fig. 4E). Instead, many proteins (9.06%) acquired DNA metabolic process when subjected to stress.
Changes in Protein–DNA Binding of RNA Polymerase I Subunit A43, Inositol Requiring-2, and Ime2-Dependent Signaling-2 Under Stress.
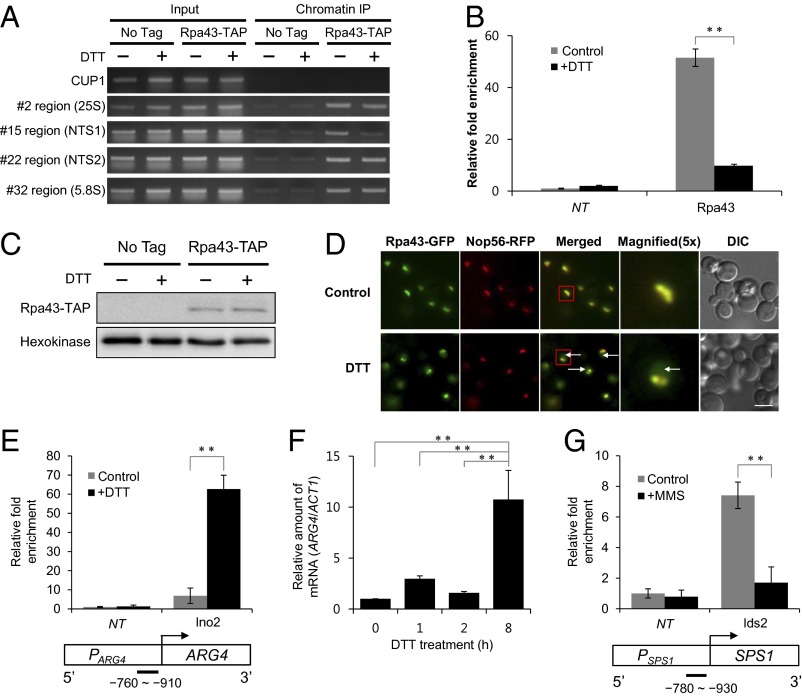
Finally, we sought to validate proteins having molecular functions that were predicted to change under DTT and MMS stress conditions. We selected the DNA-binding (GO:0003677) category for experimental follow-up because of its biological relevance to transcription. GO terms define the DNA binding as any molecular function by which a gene product interacts selectively and noncovalently with DNA, and it includes various subontologies, such as the sequence-specific DNA binding (GO:0043565) and the positive/negative regulation of DNA binding (GO:0043392/0043388; www.geneontology.org). CoFP predicted that RNA polymerase I subunit A43 (Rpa43) decreases in the DNA-binding functionality under DTT. Because RNA polymerase I is an rDNA-binding protein complex for rRNA transcription, we tested the reduction in rDNA binding of Rpa43 under DTT treatment. After 2 h incubation with the treatment of 2.5 mM DTT, the association of Rpa43 with the NTS1 region of rDNA was considerably decreased (Fig. 6A). A quantitative real-time PCR assay confirmed the decrease in the DNA-binding affinity of Rpa43 under the DTT condition (Fig. 6B). The expression level of Rpa43 was similar before and after the DTT treatment (Fig. 6C), indicating that the observed decrease in DNA-binding affinity did not result from a lower expression of Rpa43. Interestingly, we also found that some of Rpa43 was translocated to the nucleoplasm from the nucleolus under DTT (Fig. 6D).
Fig. 6.
Experimental validation of dynamic functions under DTT and MMS conditions. (A) PCR analysis to check the association of Rpa43 with the rDNA regions (25S, NTS1, NTS2, and 5.8S regions of the rDNA). The Rpa43-associated DNA was prepared by ChIP and amplified using primer sets located in the indicated regions. PCR with the primer set within the CUP1 region is used as an internal control. No tag indicates samples from nontagged cells. (B) The association of Rpa43 with the NTS1 region of rDNA under DTT treatment using quantitative real-time PCR. Amplification efficiencies were validated and normalized against CUP1, and fold increases were calculated using the comparative cycle threshold (Ct) method. Values are the mean of three independent experiments, and error bars indicate SDs. (C) Western blot analysis showing the protein levels of Rpa43 under DTT treatment. (D) Fluorescence images showing the localization changes of Rpa43 under DTT treatment. Arrows indicate Rpa43 spreading out from the nucleolus. RFP-tagged Nop56 was used as a nucleolar marker. Red squares indicate the area that is magnified 5×. DIC, differential interference contrast images. (Scale bar: 5 μm.) (E) The association of Ino2 with the promoter region of ARG4 was measured using a ChIP assay under DTT treatment. Relative fold enrichment refers to the relative ratio of PCR products amplified from immunoprecipitated DNA to products from input DNA. PCR amplicons used in ChIP assays are indicated below the promoter region of the target gene. (F) The expression of ARG4 was measured under DTT treatment. Cells were treated with 2.5 mM DTT for the indicated times at 25 °C, and the amount of ARG4 was analyzed by quantitative real-time RT-PCR. (G) The association of Ids2 with the promoter region of SPS1 was measured using a ChIP assay under MMS treatment. NT, no tag. **P value < 0.01 using a Student t test.
In contrast to Rpa43, inositol requiring-2 (Ino2) was predicted to increase in the DNA-binding function under DTT. To validate this prediction, we measured the binding of Ino2 to the promoter region of arginine requiring-4 (ARG4), which encodes an argininosuccinate lyase, catalyzing the final step in the arginine biosynthesis pathway. As predicted, the binding of Ino2 to the promoter region of ARG4 was increased under the DTT condition (Fig. 6E). Consistent with this observation, the expression of ARG4 increased more than 10 times with DTT treatment (Fig. 6F). In addition, under the MMS condition, we found that Ime2-dependent signaling-2 (Ids2), a protein involved in the modulation of Ime2 activity during meiosis, exhibited decreased association with the promoter region of sporulation specific-1 (SPS1), a putative protein serine/threonine kinase required for correct localization of enzymes involved in spore wall synthesis (Fig. 6G). Consistent with these findings, MMS treatment has been found to reduce the expression of SPS1 (37).
Discussion
To predict the condition-specific localization and function of proteins, CoFP adds context to static measurements of protein–protein interactions by integrating these networks with condition-specific gene expression profiles. We have shown that CoFP can discover dynamic changes in protein location and function under diverse stresses on a proteome-wide scale (Figs. 4–6) as well as condition-dependent location and function (Figs. 2 and 3). The core concept of CoFP is that physically interacting proteins with high coherence scores in mRNA expression share similar functionalities (Fig. 1G). One then need only to look at the interacting partners that are highly coherent in a condition to make inferences regarding dynamic localization and other functionality. Similar ideas have recently been applied to predict proteomic changes in glioma (38), in which conditional network neighbors were helpful for predicting the conditional localization of cancer proteins. In principle, CoFP can map conditional functionality for any condition for which gene expression profiles are produced, such as stem cell differentiation, response to drugs, or external stress on the system. A web server for the prediction of condition-dependent and dynamic locations and functions is available at http://nbm.ajou.ac.kr/cofp/.
CoFP uses possibility rather than probability theory. Unlike probability theory, in possibility theory, the sum of possibilities for all possible outcomes may be >1 or <1. Here, this property allows us to express the fact that there can be several simultaneous functions or locations for a single protein in a single condition (the sum of all possibilities is >1). However, sometimes, there is no most likely function for a protein, and we wish to capture this general uncertainty (in which case, the sum of all possibilities is <1).
We observed that the importance of static or network features in CoFP was somewhat dependent on functional purity within or between proteins. In case of peroxisome location, for example, network features are more informative (SI Appendix, Fig. S7A) owing to higher functional purity of interacting proteins in peroxisome (SI Appendix, Fig. S1). For ligase molecular function, however, single-protein features were more informative than network features (SI Appendix, Fig. S7C), mainly owing to its higher functional purity of individual proteins in ligase (SI Appendix, Fig. S9A) compared with its relative lower functional purity of between interacting neighbors in ligase (SI Appendix, Fig. S3).
In the feature set selection for individual processes and functions, we used GO annotations as parts of both training and evaluation. To reduce a circularity problem, we used newly assigned GO terms to check the performance of proteins without the GO terms used previously. Moreover, we here experimentally validated some of conditional and dynamic functions, including locations under DTT and MMS conditions in addition to a stress-free yeast extract/peptone/dextrose (YPD) condition, for performance assessment of conditional and dynamic functions, including locations. For example, we experimentally validated previously unidentified locations or previous experimental errors of yeast proteins under the YPD condition (Fig. 2 and SI Appendix, Fig. S17) and checked the performance of conditional locations using 100 proteins under MMS and DTT conditions (Fig. 3). We also experimentally tested the dynamic locations and functions under the MMS and DTT conditions (Figs. 5 and 6). Other predicted functions under diverse stresses should be tested in a near feature.
Among the dynamic changes in localization (Fig. 4), we correctly confirmed that Tsr1, Caf120, and Dip5 changed locations under the MMS condition (Fig. 5). Tsr1 moved from the cytosol to the nucleolus after MMS treatment. This protein is required for processing 20S pre-rRNA in the cytoplasm and associates with pre-40S ribosomal particles (39). Dynamic localization of Tsr1 was also previously observed in cells depleted of ribosomal protein of the small subunit-15 (Rps15), a protein component of the 40S ribosomal subunit. Rps15 depletion leads to retention of 20S pre-rRNA-containing late pre-40S particles and its associating protein Tsr1 in the nucleolus (39). Given that MMS is a DNA-damaging agent and that it induces stalled replication forks (40), our data suggest that the MMS-induced DNA damage pathway is linked to ribosome biogenesis. Consistent with this notion, treatment of cells with MMS reduces the expression of proteins involved in the synthesis and assembly of ribosomes, including the DEAD box helicase Dbp3, the H/ACA box protein Nhp2, the late preribosomal RNA processing and particle assembly factors Nip7 and Nsr1 (nucleolin), and the C/D box proteins Nop1 (fibrillarin), Nop56, and Nop58, in addition to several ribosomal proteins (41). Taken together, it seems clear that DNA damage induces a multifaceted inhibition of ribosome biogenesis. Moreover, Caf120, a component of the evolutionarily conserved CCR4-NOT transcriptional regulatory complex involved in controlling mRNA metabolism (23), and Dip5, a dicarboxylic amino acid permease mediating high-affinity and high-capacity transport of l-glutamate and l-aspartate (26), also changed their locations under the MMS condition. Given that MMS causes DNA damage and consequent cell cycle arrest at multiple checkpoints, it is plausible that mRNA metabolism and amino acid transport are dysregulated under the MMS condition. Presumably, the localization changes of Caf120 and Dip5 reflect the functional changes involved in the dysregulation of mRNA metabolism and amino acid transport. We also correctly confirmed that Skg6, Lte1, and Nnf2 changed their locations under the DTT condition. However, previous knowledge related to these proteins is limited, and whether the molecular functions of Skg6, Lte1, and Nnf2 and the biological processes involving them are regulated in a redox-dependent manner is not clear at present.
For molecular functions, we correctly validated the decrease or increase in the DNA-binding function of Rpa43, Ino2, and Ids2 under the MMS or DTT condition. Rpa43 showed decreased association with the NTS1 region of rDNA under the DTT condition. Rpa43 was also translocated to the nucleoplasm from the nucleolus under this stress. The decrease in the DNA-binding function of Rpa43 under the DTT condition did not result from a lower expression of Rpa43. These observations suggest that DTT treatment may reduce rRNA transcription and consequently, compromise ribosome biogenesis. Consistent with this notion, several ribosomal proteins exhibit a decrease in expression under the DTT condition (42). Ino2 is a component of the heteromeric Ino2/Ino4 basic helix–loop–helix transcription activator that binds inositol/choline-responsive elements (43, 44). We observed that the binding of Ino2 to the promoter region of ARG4 was increased under the DTT condition and that the expression level of ARG4 increased more than 10 times. A similar expression pattern of ARG4 was reported previously (42). It seems that the binding of Ino2 to the promoter region of ARG4 is directly related to the activation of ARG4 expression.
In summary, the network-based location and function prediction framework can correctly discover previously unidentified condition-dependent and dynamic locations and functions of yeast proteins under diverse stresses. Additional investigation to confirm the predictions derived from our analysis will provide valuable information for functional annotation of unknown proteins and lead to a deeper understanding of cellular dynamics under stress.
Methods
Known Locations, Functions, and Interactions.
For subcellular locations (location) of yeast proteins, we downloaded the localization data in the work by Huh et al. (2), which used GFP-tagging experiments to annotate 3,919 proteins with up to 22 distinct locations. Of 22 locations, we excluded punctate composite and combined the Golgi apparatus-related locations, including Golgi apparatus, late Golgi, early Golgi, and ER to Golgi, into one location (SI Appendix, Table S1A). For test of predicted locations using GO cellular component terms, we used the mapping relationship shown in SI Appendix, Table S1A. For other kinds of general protein function, we downloaded the GO annotations from the AmiGO database (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi) and assigned biological process (process) and molecular function (function) to yeast proteins based on the GO Slim categories (33 biological processes and 22 molecular functions) (SI Appendix, Table S1 B and C). Note that we only used GO annotations supported by experimental evidence codes. For protein interaction data, we combined the contents of the BioGRID (27), DIP (28), and SGD (29) databases and recent in vivo interactions (30).
Analysis of Gene Expression Profiles.
For yeast gene expression profiles, we downloaded the microarray experiments from the expression Stanford Microarray Database (www.tbdb.org) and categorized them by 17 major stresses, including leucine starvation, uracil starvation, nitrogen starvation, DTT treatment, γ-radiation, H2O2 oxidative stress, heat shock, cold shock, menadione treatment, MMS treatment, phosphate starvation, calcium osmotic, salt treatment, hypoosmotic, hyperosmotic, and RNA stability, in addition to a stress-free normal condition (SI Appendix, Table S2). From the downloaded expression, we used the experiment sets with high coverage of the proteins in the protein interaction data. When replicated samples are available, we calculated the median values among the expression levels of the replicates. In the cases of two-channel microarray platforms, we extracted the total expression values for a specific channel of interest to obtain the RNA abundances. Within a platform, we applied quantile normalization with median values across samples.
Functional Coherence Score Scheme.
We calculated the functional coherence score between interacting protein pairs to generate a condition-dependent interaction network under a specific condition where time series microarray expression profiles are produced. A functional partnership score between proteins a and b under a specific condition is
where is the correlation degree of a Pearson correlation coefficient between gene expression abundances (of a) and (of b) and is the right area under the probability distribution of correlation degrees of all interacting protein pairs from an input value. If a and b are not a direct interaction, then we multiply all in a path between a and b. With multiple paths, we choose the one with the maximum value.
Prediction of Condition-Dependent and Dynamic Location and Function.
In total, 29 kinds of single-protein or network feature sets were generated for each protein (31). Instead of using whole features, we selected a feasible feature set for each functional category owing to diversity of feature performance across different kinds of functionality. However, there might exist an overfitting problem to training data. To reduce the problem, as also discussed in our previous studies (31), we applied a leave-two-out cross-validation approach with an AUC measure. The feature selection scheme was well-applicable to diverse cases of multiple species, including human, worm, fly, and yeast as well as plant (31, 32). In total, 73 models are generated for individual functional categories. After generating good future sets for individual functional categories, we further generate a possibility degree of a protein for each functional category under a condition. Similar to the work by Lee et al. (38), the possibility degree Pf of a function f with a confidence degree c is defined as
where and are the total areas under the distribution of condition degrees of positives and negatives, respectively, regarding a function f and and are the areas under the distribution of confidence degrees of positives and negatives until c, respectively. A higher value of (∼0–1) means a higher degree of possibility that a protein has the function f. A significant difference on the possibility degrees between a stress condition and a stress-free normal condition is regarded as a dynamic functionality (P value < 0.01 of Z tests using the results of 30 sample permutation tests from all expression data used here).
DTT and MMS Treatment and Microscopic Analysis of Yeast Proteins.
Yeast cells were grown to midlogarithmic phase in synthetic complete (SC) medium at 30 °C, and one-half of the cells were collected to serve as the untreated control (regarded as a normal condition). Remaining cells were exposed to 2.5 mM DTT (D1037; BIOSESANG) or 0.01% MMS (M4016; Sigma) and then incubated for 2 h. For microscopic analysis, we used 96-well glass-bottom microplates (Whatman) pretreated with concanavalin A (L7647; Sigma) to ensure cell adhesion. Microscopy was performed using a Zeiss Axiovert 200M Inverted Microscope as previously described (45). Fluorescence images of individual samples were taken using an FITC filter set (excitation band pass filter, 450–490 nm; beam splitter, 510 nm; emission band pass filter, 515–565 nm), a Rhodamine filter set (excitation band pass filter, 546 nm; beam splitter, 580 nm; emission long pass filter, 590 nm), and a DAPI filter set (excitation band pass filter, 365 nm; beam splitter, 395 nm; emission long pass filter, 397 nm). At least 50 cells were analyzed per each experiment. We analyzed subcellular localization of GFP-fused proteins by visual inspection of images and then reconfirmed it by colocalization assay as described previously (2).
ChIP Assay and Quantitative Real-Time PCR Analysis.
ChIP assays were performed as previously described (46). For ChIP experiments using tandem affinity purification (TAP)-tagged proteins/strains, prewashed IgG Sepharose Beads (17-0969-01; GE Healthcare) were used. ChIP samples were analyzed by quantitative real-time PCR using SYBR Green and the Applied Biosystems 7300 Real-Time PCR System. Relative fold enrichment was determined by calculating the ratio of the target region to CUP1, an internal control, as follows: [target region (IP)/CUP1 (IP)]/[target region (input)/CUP1 (input)]. The sequences of PCR primers used in ChIP experiments are shown in SI Appendix, Table S10. Each set of experiments was performed at least three times.
Western Blot Analysis.
Yeast cells grown to midlogarithmic phase in SC medium were harvested, washed three times with PBS, and disrupted by bead beating in 5 volumes lysis buffer (20 mM Tris⋅Cl, pH 7.5, 1 mM EDTA, 1 mM PMSF, 1 mM benzamidine, 1 μg/mL leupetin, 1 μg/mL pepstatin). Cell debris was removed by centrifuging at 5,000 × g for 5 min, and the remaining cell extract was centrifuged at 14,000 × g for 30 min. The supernatant was transferred to a new tube and mixed with SDS/PAGE sample buffer. SDS/PAGE and Western blot analysis were performed by standard methods using HRP-conjugated antibodies.
Quantification of ARG4 mRNA.
Total RNA was isolated from yeast cells using the RNeasy MiniKit (Qiagen). cDNA for RT-PCR was generated using the ProtoScript First Strand cDNA Synthesis Kit (New England Biolabs). The amounts of ARG4 and ACT1 mRNA were analyzed by quantitative real-time RT-PCR using the Applied Biosystems 7300 Real-Time PCR System. Amplification efficiencies were validated and normalized against ACT1, and fold increases were calculated using the 2−∆∆CT method (47). The primers used for the amplification of ARG4 were 5′-CTGAAAGACTTGGTCTAAGC-3′ and 5′-CAATTGCTTCAATACAGCAG-3′, and those used for ACT1 were 5′-TGACTGACTACTTGATGAAG-3′ and 5′-TGCATTTCTTGTTCGAAGTC-3′. All reactions were carried out in triplicate.
Supplementary Material
Acknowledgments
This research was supported by National Research Foundation of Korea Grants 2013R1A2A2A04013317 (to K.L.), 2010-0028631 (to K.L.), and 2012R1A2A2A01047175 (to W.-K.H.) and National Institutes of Health Grants GM085764 (to T.I.) and ES014811 (to T.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318881111/-/DCSupplemental.
References
- 1.Lau E, et al. PKCε promotes oncogenic functions of ATF2 in the nucleus while blocking its apoptotic function at mitochondria. Cell. 2012;148(3):543–555. doi: 10.1016/j.cell.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 3.Scott MS, Calafell SJ, Thomas DY, Hallett MT. Refining protein subcellular localization. PLOS Comput Biol. 2005;1(6):e66. doi: 10.1371/journal.pcbi.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilchrist A, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127(6):1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Gardy JL, et al. PSORT-B: Improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 2003;31(13):3613–3617. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton P, et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35(Web Server Issue):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- 8.Scott MS, Thomas DY, Hallett MT. Predicting subcellular localization via protein motif co-occurrence. Genome Res. 2004;14(10A):1957–1966. doi: 10.1101/gr.2650004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K, Kim DW, Na D, Lee KH, Lee D. PLPD: Reliable protein localization prediction from imbalanced and overlapped datasets. Nucleic Acids Res. 2006;34(17):4655–4666. doi: 10.1093/nar/gkl638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402(6757):83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 11.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Curr Opin Struct Biol. 2005;15(3):275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Forslund K, Sonnhammer EL. Predicting protein function from domain content. Bioinformatics. 2008;24(15):1681–1687. doi: 10.1093/bioinformatics/btn312. [DOI] [PubMed] [Google Scholar]
- 13.Lee D, Redfern O, Orengo C. Predicting protein function from sequence and structure. Nat Rev Mol Cell Biol. 2007;8(12):995–1005. doi: 10.1038/nrm2281. [DOI] [PubMed] [Google Scholar]
- 14.Minshull J, Ness JE, Gustafsson C, Govindarajan S. Predicting enzyme function from protein sequence. Curr Opin Chem Biol. 2005;9(2):202–209. doi: 10.1016/j.cbpa.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vinayagam A, et al. Applying Support Vector Machines for Gene Ontology based gene function prediction. BMC Bioinformatics. 2004;5:116. doi: 10.1186/1471-2105-5-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zehetner G. OntoBlast function: From sequence similarities directly to potential functional annotations by ontology terms. Nucleic Acids Res. 2003;31(13):3799–3803. doi: 10.1093/nar/gkg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker MG, Volkmuth W, Sprinzak E, Hodgson D, Klingler T. Prediction of gene function by genome-scale expression analysis: Prostate cancer-associated genes. Genome Res. 1999;9(12):1198–1203. doi: 10.1101/gr.9.12.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatri P, Drăghici S. Ontological analysis of gene expression data: Current tools, limitations, and open problems. Bioinformatics. 2005;21(18):3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fyshe A, Liu Y, Szafron D, Greiner R, Lu P. Improving subcellular localization prediction using text classification and the gene ontology. Bioinformatics. 2008;24(21):2512–2517. doi: 10.1093/bioinformatics/btn463. [DOI] [PubMed] [Google Scholar]
- 22.Wong A, Shatkay H. Protein function prediction using text-based features extracted from the biomedical literature: The CAFA challenge. BMC Bioinformatics. 2013;14(Suppl 3):S14. doi: 10.1186/1471-2105-14-S3-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee K, et al. Protein networks markedly improve prediction of subcellular localization in multiple eukaryotic species. Nucleic Acids Res. 2008;36(20):e136. doi: 10.1093/nar/gkn619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barabási AL, Oltvai ZN. Network biology: Understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 25.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor IW, et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27(2):199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 27.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41(Database Issue):D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xenarios I, et al. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30(1):303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christie KR, et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32(Database Issue):D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarassov K, et al. An in vivo map of the yeast protein interactome. Science. 2008;320(5882):1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 31.Lee K, et al. Protein networks markedly improve prediction of subcellular localization in multiple eukaryotic species. Nucleic Acids Res. 2008;36(20):e136. doi: 10.1093/nar/gkn619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K, Thorneycroft D, Achuthan P, Hermjakob H, Ideker T. Mapping plant interactomes using literature curated and predicted protein-protein interaction data sets. Plant Cell. 2010;22(4):997–1005. doi: 10.1105/tpc.109.072736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, et al. A protein domain co-occurrence network approach for predicting protein function and inferring species phylogeny. PLoS ONE. 2011;6(3):e17906. doi: 10.1371/journal.pone.0017906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rietsch A, Beckwith J. The genetics of disulfide bond metabolism. Annu Rev Genet. 1998;32:163–184. doi: 10.1146/annurev.genet.32.1.163. [DOI] [PubMed] [Google Scholar]
- 35.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubersax JA, et al. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425(6960):859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- 37.Takagi Y, et al. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol Cell. 2005;18(2):237–243. doi: 10.1016/j.molcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Lee K, et al. Proteome-wide discovery of mislocated proteins in cancer. Genome Res. 2013;23(8):1283–1294. doi: 10.1101/gr.155499.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Léger-Silvestre I, et al. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 2004;23(12):2336–2347. doi: 10.1038/sj.emboj.7600252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lundin C, et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33(12):3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96(4):1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambroziak J, Henry SA. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269(21):15344–15349. [PubMed] [Google Scholar]
- 44.Schüller HJ, Richter K, Hoffmann B, Ebbert R, Schweizer E. DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix transcription factor: Structural requirements as defined by saturation mutagenesis. FEBS Lett. 1995;370(1-2):149–152. doi: 10.1016/0014-5793(95)00818-t. [DOI] [PubMed] [Google Scholar]
- 45.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24(9):767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 46.Ha CW, Huh WK. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011;39(4):1336–1350. doi: 10.1093/nar/gkq895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlatzer D, et al. Novel urinary protein biomarkers predicting the development of microalbuminuria and renal function decline in type 1 diabetes. Diabetes Care. 2012;35(3):549–555. doi: 10.2337/dc11-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.